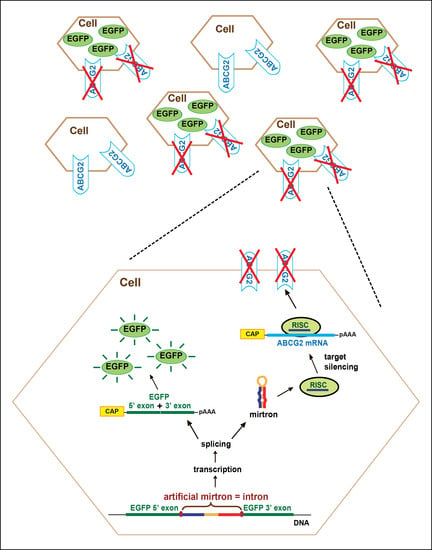
Posttranscriptional Regulation of the Human ABCG2 Multidrug Transporter Protein by Artificial Mirtrons
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatics, Statistical Analysis
2.2. Plasmid Constructs
2.3. Cell Cultures and Manipulation
2.4. RNA Analysis
2.5. Luciferase Assay
2.6. Western Blot (Immunoblot)
3. Results
3.1. Artificial Mirtron Design
3.2. Investigating Splicing Ability of the Selected Artificial Mirtrons
3.3. Functional Testing of Artificial Mirtrons by Luciferase Reporter Assay
3.4. Targeting ABCG2 Expression by Artificial Mirtrons
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doyle, L.A.; Yang, W.; Abruzzo, L.V.; Krogmann, T.; Gao, Y.; Rishi, A.K.; Ross, D.D. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl. Acad. Sci. USA 1998, 95, 15665–15670. [Google Scholar] [CrossRef] [Green Version]
- Allikmets, R.; Schriml, L.M.; Hutchinson, A.; Romano-Spica, V.; Dean, M. A human placenta-specific ATP-binding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Res. 1998, 58, 5337–5339. [Google Scholar]
- Miyake, K.; Mickley, L.; Litman, T.; Zhan, Z.; Robey, R.; Cristensen, B.; Brangi, M.; Greenberger, L.; Dean, M.; Fojo, T.; et al. Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistant cells: Demonstration of homology to ABC transport genes. Cancer Res. 1999, 59, 8–13. [Google Scholar]
- Krishnamurthy, P.; Schuetz, J.D. Role of ABCG2/BCRP in biology and medicine. Annu. Rev. Pharmacol. Toxicol. 2006, 46, 381–410. [Google Scholar] [CrossRef]
- Sarkadi, B.; Homolya, L.; Szakacs, G.; Varadi, A. Human multidrug resistance ABCB and ABCG transporters: Participation in a chemoimmunity defense system. Physiol. Rev. 2006, 86, 1179–1236. [Google Scholar] [CrossRef]
- Apati, A.; Orban, T.I.; Varga, N.; Nemeth, A.; Schamberger, A.; Krizsik, V.; Erdelyi-Belle, B.; Homolya, L.; Varady, G.; Padanyi, R.; et al. High level functional expression of the ABCG2 multidrug transporter in undifferentiated human embryonic stem cells. Biochim. Biophys. Acta 2008, 1778, 2700–2709. [Google Scholar] [CrossRef] [Green Version]
- Apati, A.; Szebenyi, K.; Erdei, Z.; Varady, G.; Orban, T.I.; Sarkadi, B. The importance of drug transporters in human pluripotent stem cells and in early tissue differentiation. Expert Opin. Drug Metab. Toxicol. 2016, 12, 77–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.; Schuetz, J.D.; Bunting, K.D.; Colapietro, A.M.; Sampath, J.; Morris, J.J.; Lagutina, I.; Grosveld, G.C.; Osawa, M.; Nakauchi, H.; et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat. Med. 2001, 7, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Hegedus, C.; Szakacs, G.; Homolya, L.; Orban, T.I.; Telbisz, A.; Jani, M.; Sarkadi, B. Ins and outs of the ABCG2 multidrug transporter: An update on in vitro functional assays. Adv. Drug Deliv. Rev. 2009, 61, 47–56. [Google Scholar] [CrossRef]
- Gameiro, M.; Silva, R.; Rocha-Pereira, C.; Carmo, H.; Carvalho, F.; Bastos, M.L.; Remiao, F. Cellular models and in vitro assays for the screening of MODULATORS of P-gp, MRP1 and BCRP. Molecules 2017, 22, 600. [Google Scholar] [CrossRef] [Green Version]
- Kovacsics, D.; Brozik, A.; Tihanyi, B.; Matula, Z.; Borsy, A.; Meszaros, N.; Szabo, E.; Nemeth, E.; Fothi, A.; Zambo, B.; et al. Precision-engineered reporter cell lines reveal ABCG2 regulation in live lung cancer cells. Biochem. Pharmacol. 2020, 175, 113865. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landthaler, M.; Yalcin, A.; Tuschl, T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004, 14, 2162–2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [Green Version]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Ketting, R.F.; Fischer, S.E.; Bernstein, E.; Sijen, T.; Hannon, G.J.; Plasterk, R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001, 15, 2654–2659. [Google Scholar] [CrossRef] [Green Version]
- Park, J.E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef]
- Tian, Y.; Simanshu, D.K.; Ma, J.B.; Park, J.E.; Heo, I.; Kim, V.N.; Patel, D.J. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol. Cell 2014, 53, 606–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macrae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacRae, I.J.; Zhou, K.; Doudna, J.A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007, 14, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Katsura, A.; Yasuda, T.; Ueno, T.; Mano, H.; Sugimoto, K.; Miyazono, K. Small-RNA asymmetry is directly driven by mammalian Argonautes. Nat. Struct. Mol. Biol. 2015, 22, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef]
- Pasquinelli, A.E. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012, 13, 271–282. [Google Scholar] [CrossRef]
- Miyoshi, K.; Miyoshi, T.; Siomi, H. Many ways to generate microRNA-like small RNAs: Non-canonical pathways for microRNA production. Mol. Genet. Genom. 2010, 284, 95–103. [Google Scholar] [CrossRef]
- Yang, J.S.; Lai, E.C. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell 2011, 43, 892–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtis, H.J.; Sibley, C.R.; Wood, M.J. Mirtrons, an emerging class of atypical miRNA. Wiley Interdiscip. Rev. RNA 2012, 3, 617–632. [Google Scholar] [CrossRef] [PubMed]
- Daugaard, I.; Hansen, T.B. Biogenesis and function of ago-associated RNAs. Trends Genet. 2017, 33, 208–219. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef] [Green Version]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef] [Green Version]
- Sibley, C.R.; Seow, Y.; Saayman, S.; Dijkstra, K.K.; El Andaloussi, S.; Weinberg, M.S.; Wood, M.J. The biogenesis and characterization of mammalian microRNAs of mirtron origin. Nucleic Acids Res. 2012, 40, 438–448. [Google Scholar] [CrossRef]
- Havens, M.A.; Reich, A.A.; Duelli, D.M.; Hastings, M.L. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012, 40, 4626–4640. [Google Scholar] [CrossRef] [Green Version]
- Schamberger, A.; Sarkadi, B.; Orban, T.I. Human mirtrons can express functional microRNAs simultaneously from both arms in a flanking exon-independent manner. RNA Biol. 2012, 9, 1177–1185. [Google Scholar] [CrossRef] [Green Version]
- Seow, Y.; Sibley, C.R.; Wood, M.J. Artificial mirtron-mediated gene knockdown: Functional DMPK silencing in mammalian cells. RNA 2012, 18, 1328–1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sibley, C.R.; Seow, Y.; Curtis, H.; Weinberg, M.S.; Wood, M.J. Silencing of Parkinson’s disease-associated genes with artificial mirtron mimics of miR-1224. Nucleic Acids Res. 2012, 40, 9863–9875. [Google Scholar] [CrossRef]
- Kock, K.H.; Kong, K.W.; Hoon, S.; Seow, Y. Functional VEGFA knockdown with artificial 3′-tailed mirtrons defined by 5′ splice site and branch point. Nucleic Acids Res. 2015, 43, 6568–6578. [Google Scholar] [CrossRef] [Green Version]
- Curtis, H.J.; Seow, Y.; Wood, M.J.A.; Varela, M.A. Knockdown and replacement therapy mediated by artificial mirtrons in spinocerebellar ataxia 7. Nucleic Acids Res. 2017, 45, 7870–7885. [Google Scholar] [CrossRef] [Green Version]
- Burset, M.; Seledtsov, I.A.; Solovyev, V.V. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000, 28, 4364–4375. [Google Scholar] [CrossRef]
- Burset, M.; Seledtsov, I.A.; Solovyev, V.V. SpliceDB: Database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res. 2001, 29, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Desmet, F.O.; Hamroun, D.; Lalande, M.; Collod-Beroud, G.; Claustres, M.; Beroud, C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009, 37, e67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Schamberger, A.; Orban, T.I. Experimental validation of predicted mammalian microRNAs of mirtron origin. Methods Mol. Biol. 2014, 1182, 245–263. [Google Scholar] [CrossRef]
- Lacy-Hulbert, A.; Thomas, R.; Li, X.P.; Lilley, C.E.; Coffin, R.S.; Roes, J. Interruption of coding sequences by heterologous introns can enhance the functional expression of recombinant genes. Gene Ther. 2001, 8, 649–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orban, T.I.; Seres, L.; Ozvegy-Laczka, C.; Elkind, N.B.; Sarkadi, B.; Homolya, L. Combined localization and real-time functional studies using a GFP-tagged ABCG2 multidrug transporter. Biochem. Biophys. Res. Commun. 2008, 367, 667–673. [Google Scholar] [CrossRef]
- Orban, T.I.; Apati, A.; Nemeth, A.; Varga, N.; Krizsik, V.; Schamberger, A.; Szebenyi, K.; Erdei, Z.; Varady, G.; Karaszi, E.; et al. Applying a “double-feature” promoter to identify cardiomyocytes differentiated from human embryonic stem cells following transposon-based gene delivery. Stem Cells 2009, 27, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Sarkadi, B.; Orban, T.I.; Szakacs, G.; Varady, G.; Schamberger, A.; Erdei, Z.; Szebenyi, K.; Homolya, L.; Apati, A. Evaluation of ABCG2 expression in human embryonic stem cells: Crossing the same river twice? Stem Cells 2010, 28, 174–176. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schamberger, A.; Várady, G.; Fóthi, Á.; Orbán, T.I. Posttranscriptional Regulation of the Human ABCG2 Multidrug Transporter Protein by Artificial Mirtrons. Genes 2021, 12, 1068. https://doi.org/10.3390/genes12071068
Schamberger A, Várady G, Fóthi Á, Orbán TI. Posttranscriptional Regulation of the Human ABCG2 Multidrug Transporter Protein by Artificial Mirtrons. Genes. 2021; 12(7):1068. https://doi.org/10.3390/genes12071068
Chicago/Turabian StyleSchamberger, Anita, György Várady, Ábel Fóthi, and Tamás I. Orbán. 2021. "Posttranscriptional Regulation of the Human ABCG2 Multidrug Transporter Protein by Artificial Mirtrons" Genes 12, no. 7: 1068. https://doi.org/10.3390/genes12071068
APA StyleSchamberger, A., Várady, G., Fóthi, Á., & Orbán, T. I. (2021). Posttranscriptional Regulation of the Human ABCG2 Multidrug Transporter Protein by Artificial Mirtrons. Genes, 12(7), 1068. https://doi.org/10.3390/genes12071068