Antagonising Chromatin Remodelling Activities in the Regulation of Mammalian Ribosomal Transcription
Abstract
:1. Introduction
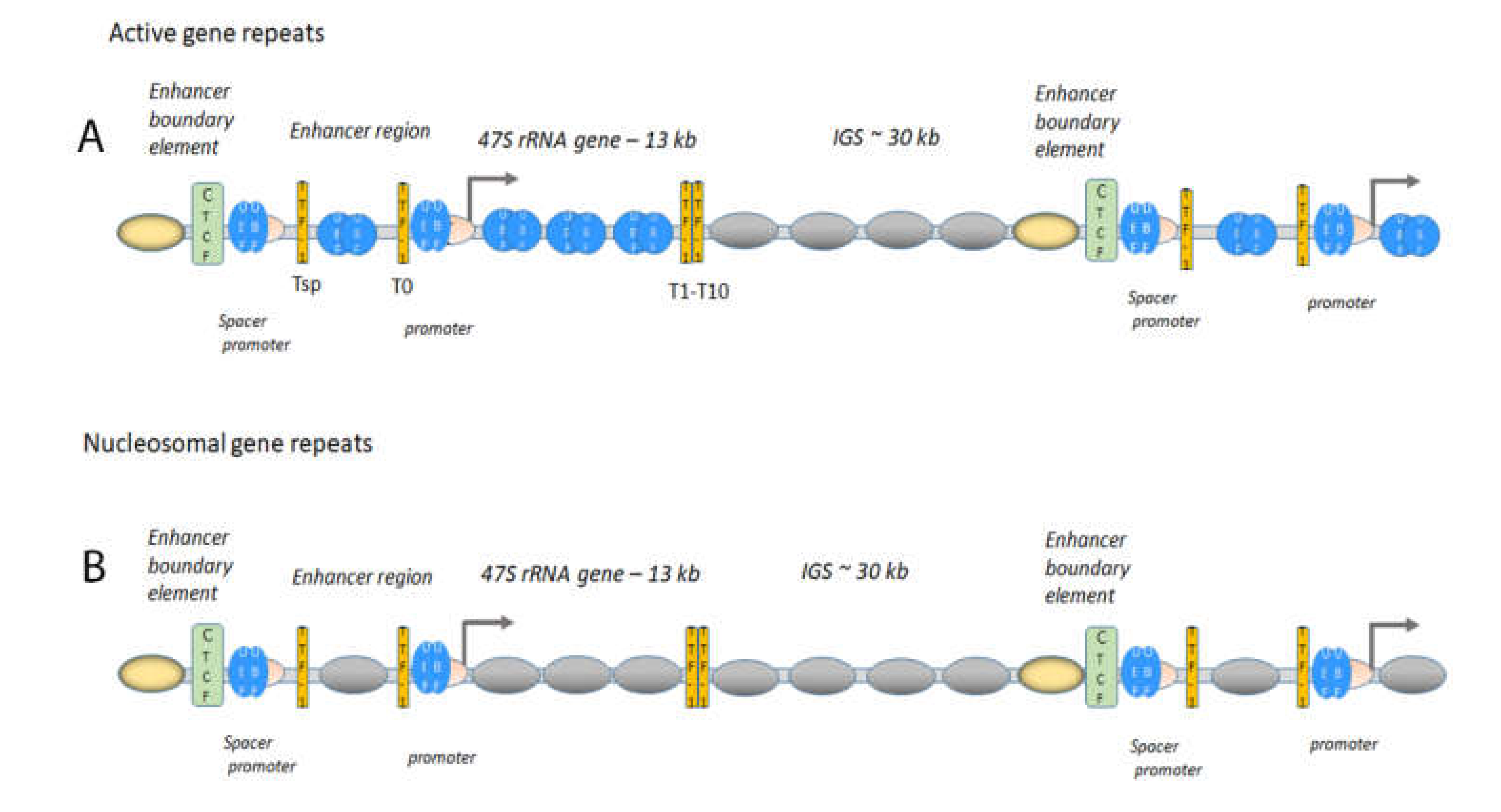
1.1. The Active 47S rRNA Gene Transcription Is Organised in an Open Chromatin State
1.2. The Boundary Element, a Form of Insulator, Is Important in the Maintenance of Chromatin States and Factor Loading
1.3. Gene Expression of Active Gene Repeats Requires Transcription Factors
1.4. Active Gene Repeats Form Loops
1.5. Acetylation of UBF Enhances 47S rRNA Gene Transcription
2. The Inactive 47S rRNA Gene Repeats Are DNA Methylated and Organised in Constitutive Heterochromatin
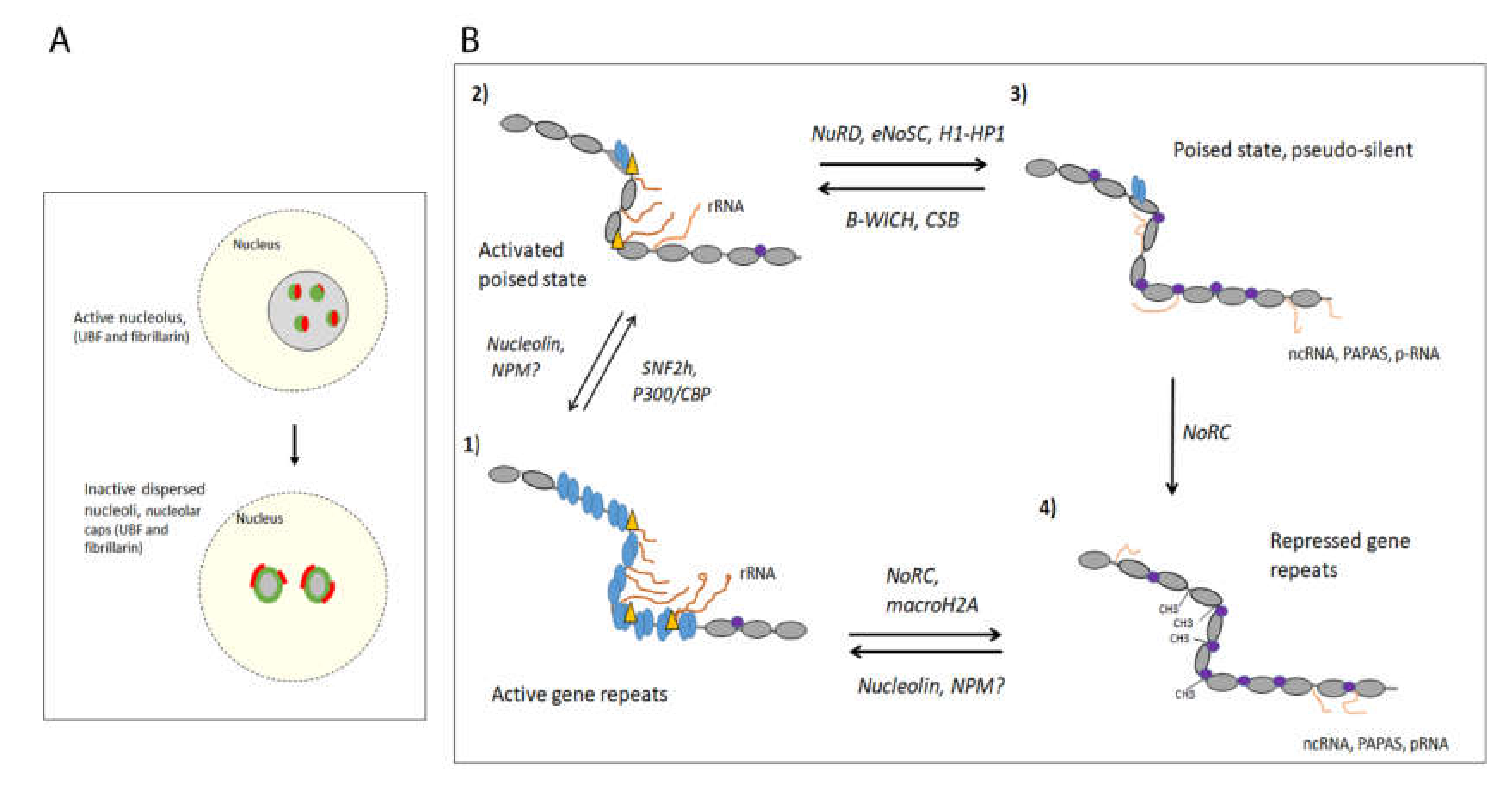
3. The Switch between Active and DNA-Methylated Inactive 47S rRNA Gene Repeats
3.1. The Permissive Poise Chromatin State Is Organised in Bivalent Chromatin on Unmethylated DNA
3.2. The Activation of the Permissive Poised State Involves Establishing an Active Nucleosome State
3.3. Transcriptions Factors, c-MYC, Induce Chromatin Changes and Activate Gene Expression
3.4. HATs and HDACs Are Involved in Chromatin Changes
3.5. eNOSC Induces Silencing in Response to Low NADH-Levels
4. Acute Stress Responses and the Regulation of rRNA Transcription
4.1. lncRNA Originating from the Human IGS Are INVOLVED in Stress Responses with Reduced 47S rRNA Gene Transcription
4.2. Nuclear Integrity Is Important for 47S rRNA Gene Transcription
5. The Trigger to Switch between Different States
TTF-1 Appears to Be a Determining Factor in the Switch
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kusnadi, E.P.; Hannan, K.M.; Hicks, R.J.; Hannan, R.D.; Pearson, R.B.; Kang, J. Regulation of rDNA transcription in response to growth factors, nutrient and energy. Gene 2015, 556, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.; Schneekloth, J.S., Jr.; Panov, K.I.; Hannan, K.M.; Hannan, R.D. Targeting the RNA Polymerase I Transcription for Cancer Therapy Comes of Age. Cells 2020, 9, 266. [Google Scholar] [CrossRef] [Green Version]
- Gaviraghi, M.; Vivori, C.; Tonon, G. How Cancer Exploits Ribosomal RNA Biogenesis: A Journey beyond the Boundaries of rRNA Transcription. Cells 2019, 17, 1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanij, E.; Hannan, K.M.; Xuan, J.; Yan, S.; Ahern, J.E.; Trigos, A.S.; Brajanovski, N.; Son, J.; Chan, K.T.; Kondrashova, O.; et al. CX-5461 activates the DNA damage response and demonstrates therapeutic efficacy in high-grade serous ovarian cancer. Nat. Commun. 2020, 11, 2641. [Google Scholar] [CrossRef]
- Low, J.Y.; Sirajuddin, P.; Moubarek, M.; Agarwal, S.; Rege, A.; Guner, G.; Liu, H.; Yang, Z.; De Marzo, A.M.; Bieberich, C.; et al. Effective targeting of RNA polymerase I in treatment-resistant prostate cancer. Prostate 2019, 79, 1837–1851. [Google Scholar] [CrossRef]
- Catez, F.; Dalla Venezia, N.; Marcel, V.; Zorbas, C.; Lafontaine, D.L.J.; Diaz, J.J. Ribosome biogenesis: An emerging druggable pathway for cancer therapeutics. Biochem. Pharmacol. 2019, 159, 74–81. [Google Scholar] [CrossRef] [Green Version]
- Sharifi, S.; Bierhoff, H. Regulation of RNA Polymerase I Transcription in Development, Disease, and Aging. Annu. Rev. Biochem. 2018, 87, 51–73. [Google Scholar] [CrossRef]
- Piazzi, M.; Bavelloni, A.; Gallo, A.; Faenza, I.; Blalock, W.L. Signal Transduction in Ribosome hamdaneBiogenesis: A Recipe to Avoid Disaster. Int. J. Mol. Sci. 2019, 20, 2718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Li, X. Transcriptional regulation in eukaryotic ribosomal protein genes. Genomics 2007, 90, 421–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guimaraes, J.C.; Zavolan, M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 2016, 17, 236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zheng, Y.; Hu, H.; Li, X. Integrative analyses shed new light on human ribosomal protein gene regulation. Sci. Rep. 2016, 6, 28619. [Google Scholar] [CrossRef] [Green Version]
- Simsek, D.; Tiu, G.C.; Flynn, R.A.; Byeon, G.W.; Leppek, K.; Xu, A.F.; Chang, H.Y.; Barna, M. The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell 2017, 169, 1051–1065. [Google Scholar] [CrossRef] [Green Version]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez-Verdun, D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2011, 2, 189–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moss, T.; Langlois, F.; Gagnon-Kugler, T.; Stefanovsky, V. A housekeeper with power of attorney: The rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 2007, 64, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Grob, A.; Colleran, C.; McStay, B. Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division. Genes Dev. 2014, 28, 220–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farley, K.I.; Surovtseva, Y.; Merkel, J.; Baserga, S.J. Determinants of mammalian nucleolar architecture. Chromosoma 2015, 124, 323–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McStay, B. Nucleolar organizer regions: Genomic ‘dark matter’ requiring illumination. Genes Dev. 2016, 30, 1598–1610. [Google Scholar] [CrossRef] [Green Version]
- Cerqueira, A.V.; Lemos, B. Ribosomal DNA and the Nucleolus as Keystones of Nuclear Architecture, Organization, and Function. Trends Genet. 2019, 35, 710–723. [Google Scholar] [CrossRef]
- Rodriguez-Corona, U.; Sobol, M.; Rodriguez-Zapata, L.C.; Hozak, P.; Castano, E. Fibrillarin from Archaea to human. Biol Cell. 2015, 107, 159–174. [Google Scholar] [CrossRef]
- Durut, N.; Sáez-Vásquez, J. Nucleolin: Dual roles in rDNA chromatin transcription. Gene 2015, 556, 7–12. [Google Scholar] [CrossRef]
- Scott, D.D.; Oeffinger, M. Nucleolin and nucleophosmin: Nucleolar proteins with multiple functions in DNA repair. Biochem. Cell Biol. 2016, 94, 419–432. [Google Scholar] [CrossRef]
- López, D.J.; Rodríguez, J.A.; Bañuelos, S. Nucleophosmin, a multifunctional nucleolar organizer with a role in DNA repair. Biochim. Biophys Acta Proteins Proteom. 2020, 1868, 140532. [Google Scholar] [CrossRef]
- Mangan, H.; Gailín, M.Ó.; McStay, B. Integrating the genomic architecture of human nucleolar organizer regions with the biophysical properties of nucleoli. FEBS J. 2017, 284, 3977–3985. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.W.; Xu, G.; Wang, Y.; Shan, L.; Luan, P.F.; Wang, Y.; Wu, M.; Yang, L.Z.; Xing, Y.H.; Yang, L.; et al. Nascent Pre-rRNA Sorting via Phase Separation Drives the Assembly of Dense Fibrillar Components in the Human Nucleolus. Mol. Cell 2019, 76, 767–783. [Google Scholar] [CrossRef]
- Sen Gupta, A.; Joshi, G.; Pawar, S.; Sengupta, K. Nucleolin modulates compartmentalization and dynamics of histone 2B-ECFP in the nucleolus. Nucleus 2018, 9, 350–367. [Google Scholar] [CrossRef] [PubMed]
- Frottin, F.; Schueder, F.; Tiwary, S.; Gupta, R.; Körner, R.; Schlichthaerle, T.; Cox, J.; Jungmann, R.; Hartl, F.U.; Hipp, M.S. The nucleolus functions as a phase-separated protein quality control compartment. Science 2019, 365, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Salvetti, A.; Greco, A. Viruses and the nucleolus: The fatal attraction. Biochim. Biophys Acta. 2014, 1842, 840–847. [Google Scholar] [CrossRef]
- Schöfer, C.; Weipoltshammer, K. Nucleolus and chromatin. Histochem. Cell Biol. 2018, 150, 209–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bersaglieri, C.; Santoro, R. Genome Organization in and around the Nucleolus. Cells 2019, 8, 579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moss, T.; Mars, J.C.; Tremblay, M.G.; Sabourin-Felix, M. The chromatin landscape of the ribosomal RNA genes in mouse and human. Chromosome Res. 2019, 27, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Sanij, E.; Poortinga, G.; Sharkey, K.; Hung, S.; Holloway, T.P.; Quin, J.; Robb, E.; Wong, L.H.; Thomas, W.G.; Stefanovsky, V.; et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 2008, 183, 1259–1274. [Google Scholar] [CrossRef] [Green Version]
- Zentner, G.E.; Saiakhova, A.; Manaenkov, P.; Adams, M.D.; Scacheri, P.C. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 2011, 39, 4949–4960. [Google Scholar] [CrossRef] [Green Version]
- Hamdane, N.; Stefanovsky, V.Y.; Tremblay, M.G.; Németh, A.; Paquet, E.; Lessard, F.; Sanij, E.; Hannan, R.; Moss, T. Conditional inactivation of Upstream Binding Factor reveals its epigenetic functions and the existence of a somatic nucleolar precursor body. PLoS Genet. 2014, 10, e1004505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herdman, C.; Mars, J.C.; Stefanovsky, V.Y.; Tremblay, M.G.; Sabourin-Felix, M.; Lindsay, H.; Robinson, M.D.; Moss, T. A unique enhancer boundary complex on the mouse ribosomal RNA genes persists after loss of Rrn3 or UBF and the inactivation of RNA polymerase I transcription. PLoS Genet. 2017, 13, e1006899. [Google Scholar] [CrossRef] [PubMed]
- Hamdane, N.; Tremblay, M.G.; Dillinger, S.; Stefanovsky, V.Y.; Németh, A.; Moss, T. Disruption of the UBF gene induces aberrant somatic nucleolar bodies and disrupts embryo nucleolar precursor bodies. Gene 2017, 612, 5–11. [Google Scholar] [CrossRef]
- Mars, J.C.; Sabourin-Felix, M.; Tremblay, M.G.; Moss, T. A Deconvolution Protocol for ChIP-Seq Reveals Analogous Enhancer Structures on the Mouse and Human Ribosomal RNA Genes. G3 (Bethesda) 2018, 8, 303–314. [Google Scholar] [CrossRef] [Green Version]
- Stefanovsky, V.Y.; Bazett-Jones, D.P.; Pelletier, G.; Moss, T. The DNA supercoiling architecture induced by the transcription factor xUBF requires three of its five HMG-boxes. Nucleic Acids Res. 1996, 24, 3208–3215. [Google Scholar] [CrossRef] [Green Version]
- Tuan, J.C.; Zhai, W.; Comai, L. Recruitment of TATA-binding protein-TAFI complex SL1 to the human ribosomal DNA promoter is mediated by the carboxy-terminal activation domain of upstream binding factor (UBF) and is regulated by UBF phosphorylation. Mol. Cell. Biol. 1999, 19, 2872–2879. [Google Scholar] [CrossRef] [Green Version]
- Friedrich, J.K.; Panov, K.I.; Cabart, P.; Russell, J.; Zomerdijk, J.C. TBP-TAF complex SL1 directs RNA polymerase I pre-initiation complex formation and stabilizes upstream binding factor at the rDNA promoter. J. Biol. Chem. 2005, 280, 29551–29558. [Google Scholar] [CrossRef] [Green Version]
- Panov, K.I.; Friedrich, J.K.; Russell, J.; Zomerdijk, J.C. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 2006, 25, 3310–3322. [Google Scholar] [CrossRef] [Green Version]
- Sanij, E.; Hannan, R.D. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics 2009, 4, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, A.; Grummt, I. A novel promoter in the mouse rDNA spacer is active in vivo and in vitro. EMBO J. 1987, 6, 3487–3492. [Google Scholar] [CrossRef] [PubMed]
- Paalman, M.H.; Henderson, S.L.; Sollner-Webb, B. Stimulation of the mouse rRNA gene promoter by a distal spacer promoter. Mol. Cell. Biol. 1995, 15, 4648–4656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, C.; Schmitz, K.M.; Li, J.; Grummt, I.; Santoro, R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell 2006, 22, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Neubert, M.; Grummt, I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008, 9, 774–780. [Google Scholar] [CrossRef]
- Van de Nobelen, S.; Rosa-Garrido, M.; Leers, J.; Heath, H.; Soochit, W.; Joosen, L.; Jonkers, I.; Demmers, J.; Van der Reijden, M.; Torrano, V.; et al. CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenet. Chromatin. 2010, 3, 19. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.; Jia, J.; Wu, C.; Yao, M.; Li, M.; Jin, J.; Jiang, C.; Cai, Y.; Pei, D.; Pan, G.; et al. Ribosomal RNA gene transcription mediated by the master genome regulator protein CCCTC-binding factor (CTCF) is negatively regulated by the condensin complex. J. Biol. Chem. 2013, 288, 26067–26077. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Shen, X.; Fan, L.; Yu, Z. Analysis of histone modifications at human ribosomal DNA in liver cancer cell. Sci. Rep. 2015, 5, 18100. [Google Scholar] [CrossRef]
- Sadova, A.A.; Kupriyanova, N.S.; Pavlova, G.V. Mapping and Quantification of Non-Coding RNA Originating from the rDNA in Human Glioma Cells. Cancers 2020, 12, 2090. [Google Scholar] [CrossRef]
- Goodfellow, S.J.; Zomerdijk, J.C. Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell Biochem. 2013, 61, 211–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, S.S.; Lesmana, A.; Peck, A.; Lee, R.; Tchoubrieva, E.; Hannan, K.M.; Lin, J.; Sheppard, K.E.; Jastrzebski, K.; Quinn, L.M.; et al. Cell cycle and growth stimuli regulate different steps of RNA polymerase I transcription. Gene 2017, 612, 36–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diesch, J.; Bywater, M.J.; Sanij, E.; Cameron, D.P.; Schierding, W.; Brajanovski, N.; Son, J.; Sornkom, J.; Hein, N.; Evers, M.; et al. Changes in long-range rDNA-genomic interactions associate with altered RNA polymerase II gene programs during malignant transformation. Commun. Biol. 2019, 2, 39. [Google Scholar] [CrossRef] [PubMed]
- Stefanovsky, V.; Langlois, F.; Gagnon-Kugler, T.; Rothblum, L.I.; Moss, T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodelling. Mol Cell. 2006, 21, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Son, J.; Hannan, K.M.; Poortinga, G.; Hein, N.; Cameron, D.P.; Ganley, A.R.D.; Sheppard, K.E.; Pearson, R.B.; Hannan, R.D.; Sanij, E. rDNA Chromatin Activity Status as a Biomarker of Sensitivity to the RNA Polymerase I Transcription Inhibitor CX-5461. Front. Cell Dev. Biol. 2020, 8, 568. [Google Scholar] [CrossRef]
- Németh, A.; Strohner, R.; Grummt, I.; Längst, G. The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res. 2004, 32, 4091–4099. [Google Scholar] [CrossRef] [Green Version]
- López-estraño, C.; Schvartzman, J.B.; Krimer, D.B.; Hernández, P. Co-localization of polar replication fork barriers and rRNA transcription terminators in mouse rDNA. J. Mol. Biol. 1998, 277, 249–256. [Google Scholar] [CrossRef]
- Akamatsu, Y.; Kobayashi, T. The Human RNA Polymerase I Transcription Terminator Complex Acts as a Replication Fork Barrier That Coordinates the Progress of Replication with rRNA Transcription Activity. Mol. Cell. Biol. 2015, 35, 1871–1881. [Google Scholar] [CrossRef] [Green Version]
- Németh, A.; Guibert, S.; Tiwari, V.K.; Ohlsson, R.; Längst, G. Epigenetic regulation of TTF-I-mediated promoterterminator interactions of rRNA genes. EMBO J. 2008, 27, 1255–1265. [Google Scholar] [CrossRef]
- Németh, A.; Längst, G. Genome organization in and around the nucleolus. Trends Genet. 2011, 27, 149–156. [Google Scholar] [CrossRef]
- Denissov, S.; Lessard, F.; Mayer, C.; Stefanovsky, V.; Van Driel, M.; Grummt, I.; Moss, T.; Stunnenberg, H.G. A model for the topology of active ribosomal RNA genes. EMBO Rep. 2011, 12, 231–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maiser, A.; Dillinger, S.; Längst, G.; Schermelleh, L.; Leonhardt, H.; Németh, A. Super-resolution in situ analysis of active ribosomal DNA chromatin organization in the nucleolus. Sci. Rep. 2020, 10, 7462. [Google Scholar] [CrossRef]
- Ghirlando, R.; Felsenfeld, G. CTCF: Making the right connections. Genes Dev. 2016, 30, 881–891. [Google Scholar] [CrossRef] [Green Version]
- Pelletier, G.; Stefanovsky, V.Y.; Faubladier, M.; Hirschler-Laszkiewicz, I.; Savard, J.; Rothblum, L.I.; Côté, J.; Moss, T. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol. Cell 2000, 6, 1059–1066. [Google Scholar] [CrossRef]
- Hirschler-Laszkiewicz, I.; Cavanaugh, A.; Hu, Q.; Catania, J.; Avantaggiati, M.L.; Rothblum, L.I. The role of acetylation in rDNA transcription. Nucleic Acids Res. 2001, 29, 4114–4124. [Google Scholar] [CrossRef] [Green Version]
- Meraner, J.; Lechner, M.; Loidl, A.; Goralik-Schramel, M.; Voit, R.; Grummt, I.; Loidl, P. Acetylation of UBF changes during the cell cycle and regulates the interaction of UBF with RNA polymerase I. Nucleic Acids Res. 2006, 34, 1798–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Hwang, Y.J.; Boo, J.H.; Han, D.; Kwon, O.K.; Todorova, K.; Kowall, N.W.; Kim, Y.; Ryu, H. Dysregulation of upstream binding factor-1 acetylation at K352 is linked to impaired ribosomal DNA transcription in Huntington’s disease. Cell Death Differ. 2011, 18, 1726–1735. [Google Scholar] [CrossRef] [Green Version]
- Peng, Q.; Wu, J.; Zhang, Y.; Liu, Y.; Kong, R.; Hu, L.; Du, X.; Ke, Y. 1A6/DRIM, a novel t-UTP, activates RNA polymerase I transcription and promotes cell proliferation. PLoS ONE 2010, 5, e14244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, R.; Zhang, L.; Hu, L.; Peng, Q.; Han, W.; Du, X.; Ke, Y. hALP, a novel transcriptional U three protein (t-UTP), activates RNA polymerase I transcription by binding and acetylating the upstream binding factor (UBF). J. Biol. Chem. 2011, 286, 7139–7148. [Google Scholar] [CrossRef] [Green Version]
- Kresoja-Rakic, J.; Santoro, R. Nucleolus and rRNA Gene Chromatin in Early Embryo Development. Trends Genet. 2019, 35, 868–879. [Google Scholar] [CrossRef] [Green Version]
- Romanova, L.; Korobova, F.; Noniashvilli, E.; Dyban, A.; Zatsepina, O. High resolution mapping of ribosomal DNA in early mouse embryos by fluorescence in situ hybridization. Biol. Reprod. 2006, 74, 807–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grummt, I.; Längst, G. Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim. Biophys. Acta 2013, 1829, 393–404. [Google Scholar] [CrossRef]
- Savić, N.; Bär, D.; Leone, S.; Frommel, S.C.; Weber, F.A.; Vollenweider, E.; Ferrari, E.; Ziegler, U.; Kaech, A.; Shakhova, O.; et al. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 2014, 15, 720–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santoro, R.; Grummt, I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell 2001, 8, 719–725. [Google Scholar] [CrossRef]
- Schmitz, K.M.; Mayer, C.; Postepska, A.; Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010, 24, 2264–2269. [Google Scholar] [CrossRef] [Green Version]
- Strohner, R.; Németh, A.; Nightingale, K.P.; Grummt, I.; Becker, P.B.; Längst, G. Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol. Cell. Biol. 2004, 24, 1791–1798. [Google Scholar] [CrossRef] [Green Version]
- Strohner, R.; Nemeth, A.; Jansa, P.; Hofmann-Rohrer, U.; Santoro, R.; Längst, G.; Grummt, I. NoRC–a novel member of mammalian ISWI-containing chromatin remodelling machines. EMBO J. 2001, 20, 4892–4900. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Grummt, I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr. Biol. 2005, 15, 1434–1438. [Google Scholar] [CrossRef] [Green Version]
- Tallant, C.; Valentini, E.; Fedorov, O.; Overvoorde, L.; Ferguson, F.M.; Filippakopoulos, P.; Svergun, D.I.; Knapp, S.; Ciulli, A. Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC. Structure 2015, 23, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Längst, G.; Grummt, I. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 2006, 25, 5735–5741. [Google Scholar] [CrossRef] [Green Version]
- Manelyte, L.; Strohner, R.; Gross, T.; Längst, G. Chromatin targeting signals, nucleosome positioning mechanism and non-coding RNA-mediated regulation of the chromatin. PLoS Genet. 2014, 10, e1004157. [Google Scholar] [CrossRef] [Green Version]
- Bierhoff, H.; Dammert, M.A.; Brocks, D.; Dambacher, S.; Schotta, G.; Grummt, I. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol. Cell 2014, 54, 675–682. [Google Scholar] [CrossRef] [Green Version]
- Cong, R.; Das, S.; Douet, J.; Wong, J.; Buschbeck, M.; Mongelard, F.; Bouvet, P. macroH2A1 histone variant represses rDNA transcription. Nucleic Acids Res. 2014, 42, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalashnikova, A.A.; Winkler, D.D.; McBryant, S.J.; Henderson, R.K.; Herman, J.A.; DeLuca, J.G.; Luger, K.; Prenni, J.E.; Hansen, J.C. Linker histone H1.0 interacts with an extensive network of proteins found in the nucleolus. Nucleic Acids Res. 2013, 41, 4026–4035. [Google Scholar] [CrossRef]
- Szerlong, H.J.; Herman, J.A.; Krause, C.M.; DeLuca, J.G.; Skoultchi, A.; Winger, Q.A.; Prenni, J.E.; Hansen, J.C. Proteomic characterization of the nucleolar linker histone H1 interaction network. J. Mol. Biol. 2015, 427, 2056–2071. [Google Scholar] [CrossRef] [Green Version]
- Mayor, R.; Izquierdo-Bouldstridge, A.; Millan-Arino, L.; Bustillos, A.; Sampaio, C.; Luque, N.; Jordan, A. Genome distribution of replication-independent histone H1 variants shows H1.0 associated with nucleolar domains and H1X with RNA polymerase II-enriched regions. J. Biol. Chem. 2015, 290, 7474–7491. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Teo, B.H.D.; Cai, Y.; Wee, S.Y.K.; Lu, J. The linker histone H1.2 is a novel component of the nucleolar organizer regions. J. Biol. Chem. 2018, 293, 2358–2369. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; John, S.; Pesavento, J.J.; Schultz-Norton, J.R.; Schiltz, R.L.; Baek, S.; Nardulli, A.M.; Hager, G.L.; Kelleher, N.L.; Mizzen, C.A. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J. Cell Biol. 2010, 189, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.; Giri, S.; Wang, Y.; Chakraborty, A.; Ghosh, A.K.; Anantharaman, A.; Aggarwal, V.; Sathyan, K.M.; Ha, T.; Prasanth, K.V.; et al. BEND3 represses rDNA transcription by stabilizing a NoRC component via USP21 deubiquitinase. Proc. Natl. Acad. Sci. USA 2015, 112, 8338–8343. [Google Scholar] [CrossRef] [Green Version]
- Rajarajacholan, U.K.; Thalappilly, S.; Riabowol, K. ING1 regulates rRNA levels by altering nucleolar chromatin structure and mTOR localization. Nucleic Acids Res. 2017, 45, 1776–1792. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.A.; Dobson, J.R.; Lian, J.B.; Stein, J.L.; Van Wijnen, A.J.; Zaidi, S.K.; Stein, G.S. A RUNX2-HDAC1 co-repressor complex regulates rRNA gene expression by modulating UBF acetylation. J. Cell Sci. 2012, 125, 2732–2739. [Google Scholar] [CrossRef] [Green Version]
- Prakash, V.; Carson, B.B.; Feenstra, J.M.; Dass, R.A.; Sekyrova, P.; Hoshino, A.; Petersen, J.; Guo, Y.; Parks, M.M.; Kurylo, C.M.; et al. Ribosome biogenesis during cell cycle arrest fuels EMT in development and disease. Nat. Commun. 2019, 10, 2110. [Google Scholar] [CrossRef]
- Cong, R.; Das, S.; Ugrinova, I.; Kumar, S.; Mongelard, F.; Wong, J.; Bouvet, P. Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res. 2012, 40, 9441–9454. [Google Scholar] [CrossRef] [Green Version]
- Angelov, D.; Bondarenko, V.A.; Almagro, S.; Menoni, H.; Mongélard, F.; Hans, F.; Mietton, F.; Studitsky, V.M.; Hamiche, A.; Dimitrov, S.; et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006, 25, 1669–1679. [Google Scholar] [CrossRef]
- Kobayashi, J.; Fujimoto, H.; Sato, J.; Hayashi, I.; Burma, S.; Matsuura, S.; Chen, D.J.; Komatsu, K. Nucleolin participates in DNA double-strand break-induced damage response through MDC1-dependent pathway. PLoS ONE 2012, 7, e49245. [Google Scholar] [CrossRef] [Green Version]
- Birch, J.L.; Tan, B.C.; Panov, K.I.; Panova, T.B.; Andersen, J.S.; Owen-Hughes, T.A.; Russell, J.; Lee, S.C.; Zomerdijk, J.C. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 2009, 28, 854–865. [Google Scholar] [CrossRef] [Green Version]
- Tessarz, P.; Santos-Rosa, H.; Robson, S.C. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 2014, 505, 564–568. [Google Scholar] [CrossRef] [Green Version]
- Hisaoka, M.; Ueshima, S.; Murano, K.; Nagata, K.; Okuwaki, M. Regulation of nucleolar chromatin by B23/nucleophosmin jointly depends upon its RNA binding activity and transcription factor UBF. Mol. Cell. Biol. 2010, 30, 4952–4964. [Google Scholar] [CrossRef] [Green Version]
- Murano, K.; Okuwaki, M.; Hisaoka, M.; Nagata, K. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol. Cell. Biol. 2008, 28, 3114–3126. [Google Scholar] [CrossRef] [Green Version]
- Kowalski, A. Nuclear and nucleolar activity of linker histone variant H1.0. Cell. Mol. Biol. Lett. 2016, 21, 15. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.; Ling, T.; Zhou, Y.; Feng, W.; Zhu, Q.; Stunnenberg, H.G.; Grummt, I.; Tao, W. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc. Natl. Acad. Sci. USA 2012, 109, 8161–8166. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Feng, W.; Imhof, A.; Grummt, I.; Zhou, Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol. Cell 2007, 27, 585–595. [Google Scholar] [CrossRef]
- Ling, T.; Xie, W.; Luo, M.; Shen, M.; Zhu, Q.; Zong, L.; Zhou, T.; Gu, J.; Lu, Z.; Zhang, F.; et al. CHD4/NuRD maintains demethylation state of rDNA promoters through inhibiting the expression of the rDNA methyltransferase recruiter TIP5. Biochem. Biophys. Res. Commun. 2013, 437, 101–107. [Google Scholar] [CrossRef]
- Shen, M.; Zhou, T.; Xie, W.; Ling, T.; Zhu, Q.; Zong, L.; Lyu, G.; Gao, Q.; Zhang, F.; Tao, W. The chromatin remodeling factor CSB recruits histone acetyltransferase PCAF to rRNA gene promoters in active state for transcription initiation. PLoS ONE 2013, 8, e62668. [Google Scholar] [CrossRef] [Green Version]
- Okur, M.N.; Lee, J.H.; Osmani, W.; Kimura, R.; Demarest, T.G.; Croteau, D.L.; Bohr, V.A. Cockayne syndrome group A and B proteins function in rRNA transcription through nucleolin regulation. Nucleic Acids Res. 2020, 48, 2473–2485. [Google Scholar] [CrossRef]
- Percipalle, P.; Fomproix, N.; Cavellán, E.; Voit, R.; Reimer, G.; Krüger, T.; Thyberg, J.; Scheer, U.; Grummt, I.; Farrants, A.K. The chromatin remodelling complex WSTF-SNF2h interacts with nuclear myosin 1 and has a role in RNA polymerase I transcription. EMBO Rep. 2006, 7, 525–530. [Google Scholar] [CrossRef]
- Cavellán, E.; Asp, P.; Percipalle, P.; Farrants, A.K. The WSTF-SNF2h chromatin remodeling complex interacts with several nuclear proteins in transcription. J. Biol. Chem. 2006, 281, 16264–16271. [Google Scholar] [CrossRef] [Green Version]
- Vintermist, A.; Böhm, S.; Sadeghifar, F.; Louvet, E.; Mansén, A.; Percipalle, P.; Ostlund Farrants, A.K. The chromatin remodelling complex B-WICH changes the chromatin structure and recruits histone acetyl-transferases to active rRNA genes. PLoS ONE 2011, 6, e19184. [Google Scholar] [CrossRef] [Green Version]
- Rolicka, A.; Guo, Y.; Gañez Zapater, A.; Tariq, K.; Quin, J.; Vintermist, A.; Sadeghifar, F.; Arsenian-Henriksson, M.; Östlund Farrants, A.K. The chromatin-remodeling complexes B-WICH and NuRD regulate ribosomal transcription in response to glucose. FASEB J. 2020, 34, 10818–10834. [Google Scholar] [CrossRef] [PubMed]
- Arabi, A.; Wu, S.; Ridderstråle, K.; Bierhoff, H.; Shiue, C.; Fatyol, K.; Fahlén, S.; Hydbring, P.; Söderberg, O.; Grummt, I.; et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005, 7, 303–310. [Google Scholar] [CrossRef]
- Grandori, C.; Gomez-Roman, N.; Felton-Edkins, Z.A.; Ngouenet, C.; Galloway, D.A.; Eisenman, R.N.; White, R.J. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005, 7, 311–318. [Google Scholar] [CrossRef]
- Li, Z.; Hann, S.R. Nucleophosmin is essential for c-Myc nucleolar localization and c-Myc-mediated rDNA transcription. Oncogene 2013, 32, 1988–1994. [Google Scholar] [CrossRef] [Green Version]
- McMahon, S.B.; Wood, M.A.; Cole, M.D. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 2000, 20, 556–562. [Google Scholar] [CrossRef] [Green Version]
- Von Walden, F.; Casagrande, V.; Östlund Farrants, A.K.; Nader, G.A. Mechanical loading induces the expression of a Pol I regulon at the onset of skeletal muscle hypertrophy. Am. J. Physiol. Cell Physiol. 2012, 302, C1523–C1530. [Google Scholar] [CrossRef] [Green Version]
- Sadeghifar, F.; Böhm, S.; Vintermist, A.; Östlund Farrants, A.K. The B-WICH chromatin-remodelling complex regulates RNA polymerase III transcription by promoting Max-dependent c-Myc binding. Nucleic Acids Res. 2015, 43, 4477–4490. [Google Scholar] [CrossRef]
- Poortinga, G.; Wall, M.; Sanij, E.; Siwicki, K.; Ellul, J.; Brown, D.; Holloway, T.P.; Hannan, R.D.; McArthur, G.A. c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res. 2011, 39, 3267–3281. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, Z.; Wang, Z.; Yu, F.; Li, J.; Wong, J. SUMOylation down-regulates rDNA transcription by repressing expression of upstream-binding factor and proto-oncogene c-Myc. J. Biol. Chem. 2019, 294, 19155–19166. [Google Scholar] [CrossRef]
- Dunn, S.; Lombardi, O.; Cowling, V.H. c-Myc co-ordinates mRNA cap methylation and ribosomal RNA production. Biochem. J. 2017, 474, 377–384. [Google Scholar] [CrossRef] [Green Version]
- Poortinga, G.; Quinn, L.M.; Hannan, R.D. Targeting RNA polymerase I to treat MYC-driven cancer. Oncogene 2015, 34, 403–412. [Google Scholar] [CrossRef]
- Sanij, E.; Diesch, J.; Lesmana, A.; Poortinga, G.; Hein, N.; Lidgerwood, G.; Cameron, D.P.; Ellul, J.; Goodall, G.J.; Wong, L.H.; et al. A novel role for the Pol I transcription factor UBTF in maintaining genome stability through the regulation of highly transcribed Pol II genes. Genome Res. 2015, 25, 201–212. [Google Scholar] [CrossRef] [Green Version]
- Trinh, D.A.; Shirakawa, R.; Kimura, T.; Sakata, N.; Goto, K.; Horiuchi, H. Inhibitor of Growth 4 (ING4) is a positive regulator of rRNA synthesis. Sci. Rep. 2019, 9, 17235. [Google Scholar] [CrossRef] [Green Version]
- Izumikawa, K.; Ishikawa, H.; Yoshikawa, H.; Fujiyama, S.; Watanabe, A.; Aburatani, H.; Tachikawa, H.; Hayano, T. LYAR potentiates rRNA synthesis by recruiting BRD2/4 and the MYST-type acetyltransferase KAT7 to rDNA. Nucleic Acids Res. 2019, 47, 10357–10372. [Google Scholar] [CrossRef] [Green Version]
- Muth, V.; Nadaud, S.; Grummt, I.; Voit, R. Acetylation of TAFI68, a subunit of TIF-IB/ SL1, activates RNA polymerase I transcription. EMBO J. 2001, 20, 1353–1362. [Google Scholar] [CrossRef] [Green Version]
- Voit, R.; Seiler, J.; Grummt, I. Cooperative action of Cdk1/cyclin B and SIRT1 is required for mitotic repression of rRNA synthesis. PLoS Genet. 2015, 11, e1005246. [Google Scholar] [CrossRef] [Green Version]
- Blank, M.F.; Grummt, I. The seven faces of SIRT7. Transcription 2017, 8, 67–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ford, E.; Voit, R.; Liszt, G.; Magin, C.; Grummt, I.; Guarente, L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006, 20, 1075–1080. [Google Scholar] [CrossRef] [Green Version]
- Grob, A.; Roussel, P.; Wright, J.E.; McStay, B.; Hernandez-Verdun, D.; Sirri, V. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J. Cell Sci. 2009, 122, 489–498. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Seiler, J.; Santiago-Reichelt, M.; Felbel, K.; Grummt, I.; Voit, R. Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol. Cell 2013, 52, 303–313. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Hotz-Wagenblatt, A.; Voit, R.; Grummt, I. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017, 31, 1370–1381. [Google Scholar] [CrossRef]
- Chen, S.; Blank, M.F.; Iyer, A.; Huang, B.; Wang, L.; Grummt, I.; Voit, R. SIRT7-dependent deacetlyation of the U3-55k protein controls pre-rRNA processing. Nat. Commun. 2016, 7, 10734. [Google Scholar] [CrossRef] [Green Version]
- Sirri, V.; Grob, A.; Berthelet, J.; Jourdan, N.; Roussel, P. Sirtuin 7 promotes 45S pre-rRNA cleavage at site 2 and determines the processing pathway. J. Cell Sci. 2019, 132, jcs228601. [Google Scholar] [CrossRef] [Green Version]
- Iyer-Bierhoff, A.; Krogh, N.; Tessarz, P.; Ruppert, T.; Nielsen, H.; Grummt, I. SIRT7-dependent deacetylation of fibrillarin controls histone H2A methylation and rRNA synthesis during the cell cycle. Cell Rep. 2018, 25, 2946–2954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer-Bierhoff, A.; Grummt, I. Stop-and-Go: Dynamics of Nucleolar Transcription During the Cell Cycle. Epigene. Insights 2019, 12, 2516865719849090. [Google Scholar] [CrossRef] [Green Version]
- Frescas, D.; Guardavaccaro, D.; Bassermann, F.; Koyama-Nasu, R.; Pagano, M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 2007, 450, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Yano, H.; Ogasawara, S.; Yoshioka, S.; Imamura, H.; Okamoto, K.; Tsuneoka, M. Mild glucose starvation induces KDM2A-mediated H3K36me2 demethylation through AMPK to reduce rRNA transcription and cell proliferation. Mol. Cell. Biol. 2015, 35, 4170–4184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galbiati, A.; Penzo, M.; Bacalini, M.G.; Onofrillo, C.; Guerrieri, A.N.; Garagnani, P.; Franceschi, C.; Treré, D.; Montanaro, L. Epigenetic up-regulation of ribosome biogenesis and more aggressive phenotype triggered by the lack of the histone demethylase JHDM1B in mammary epithelial cells. Oncotarget 2017, 8, 37091–37103. [Google Scholar] [CrossRef] [Green Version]
- De Nicola, I.; Guerrieri, A.N.; Penzo, M.; Ceccarelli, C.; De Leo, A.; Trerè, D.; Montanaro, L. Combined expression levels of KDM2A and KDM2B correlate with nucleolar size and prognosis in primary breast carcinomas. Histol. Histopathol. 2020, 35, 1181–1187. [Google Scholar] [CrossRef]
- Okamoto, K.; Tanaka, Y.; Ogasawara, S.; Obuse, C.; Nakayama, J.I.; Yano, H.; Tsuneoka, M. KDM2A-dependent reduction of rRNA transcription on glucose starvation requires HP1 in cells, including triple-negative breast cancer cells. Oncotarget 2019, 10, 4743–4760. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Yonezawa, M.; Ye, J.; Jenuwein, T.; Grummt, I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat. Struct Mol. Biol. 2010, 17, 445–450. [Google Scholar] [CrossRef]
- Ulicna, L.; Kalendova, A.; Kalasova, I.; Vacik, T.; Hozák, P. PIP2 epigenetically represses rRNA genes transcription interacting with PHF8. Biochim. Biophys Acta Mol. Cell Biol. Lipids 2018, 1863, 266–275. [Google Scholar] [CrossRef]
- Ke, R.; Xu, Q.; Li, C.; Luo, L.; Huang, D. Mechanisms of AMPK in the maintenance of ATP balance during energy metabolism. Cell Biol. Int. 2018, 42, 384–392. [Google Scholar] [CrossRef] [Green Version]
- Hannan, K.M.; Brandenburger, Y.; Jenkins, A.; Sharkey, K.; Cavanaugh, A.; Rothblum, L.; Moss, T.; Poortinga, G.; McArthur, G.A. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell. Biol. 2003, 23, 8862–8877. [Google Scholar] [CrossRef] [Green Version]
- Mayer, C.; Zhao, J.; Yuan, X.; Grummt, I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004, 18, 423–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannan, K.M.; Sanij, E.; Hein, N.; Hannan, R.D.; Pearson, R.B. Signaling to the ribosome in cancer--It is more than just mTORC1. IUBMB Life 2011, 63, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Von Walden, F. Ribosome biogenesis in skeletal muscle: Coordination of transcription and translation. J. Appl. Physiol. 2019, 127, 591–598. [Google Scholar] [CrossRef]
- Hoppe, S.; Bierhoff, H.; Cado, I.; Weber, A.; Tiebe, M.; Grummt, I.; Voit, R. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc. Natl. Acad. Sci. USA 2009, 106, 17781–17786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murayama, A.; Ohmori, K.; Fujimura, A.; Minami, H.; Yasuzawa-Tanaka, K.; Kuroda, T.; Oie, S.; Daitoku, H.; Okuwaki, M.; Nagata, K.; et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell 2008, 133, 627–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumazawa, T.; Nishimura, K.; Kuroda, T.; Ono, W.; Yamaguchi, C.; Katagiri, N.; Tsuchiya, M.; Masumoto, H.; Nakajima, Y.; Murayama, A.; et al. Novel nucleolar pathway connecting intracellular energy status with p53 activation. J. Biol. Chem. 2011, 286, 20861–20869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Song, T.; Chen, L.; Kabra, N.; Zheng, H.; Koomen, J.; Seto, E.; Chen, J. Regulation of SirT1-nucleomethylin binding by rRNA coordinates ribosome biogenesis with nutrient availability. Mol. Cell. Biol. 2013, 33, 3835–3848. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Song, T.; Chen, L.; Soliman, H.; Chen, J. Nucleolar repression facilitates initiation and maintenance of senescence. Cell Cycle 2015, 14, 3613–3623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waku, T.; Nakajima, Y.; Yokoyama, W.; Nomura, N.; Kako, K.; Kobayashi, A.; Shimizu, T.; Fukamizu, A. NML-mediated rRNA base methylation links ribosomal subunit formation to cell proliferation in a p53-dependent manner. J. Cell Sci. 2016, 129, 2382–2393. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Dammert, M.A.; Grummt, I.; Bierhoff, H. lncRNA-Induced Nucleosome Repositioning Reinforces Transcriptional Repression of rRNA Genes upon Hypotonic Stress. Cell Rep. 2016, 14, 1876–1882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Dammert, M.A.; Hoppe, S.; Bierhoff, H.; Grummt, I. Heat shock represses rRNA synthesis by inactivation of TIF-IA and lncRNA-dependent changes in nucleosome positioning. Nucleic Acids Res. 2016, 44, 8144–8152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Sentürk, N.; Song, C.; Grummt, I. lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev. 2018, 32, 836–848. [Google Scholar] [CrossRef]
- Mayer, C.; Bierhoff, H.; Grummt, I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005, 19, 933–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audas, T.E.; Jacob, M.D.; Lee, S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell 2012, 45, 147–157. [Google Scholar] [CrossRef]
- Audas, T.E.; Jacob, M.D.; Lee, S. The nucleolar detention pathway: A cellular strategy for regulating molecular networks. Cell Cycle 2012, 11, 2059–2062. [Google Scholar] [CrossRef] [Green Version]
- Yap, K.; Mukhina, S.; Zhang, G.; Tan, J.S.C.; Ong, H.S.; Makeyev, E.V. A Short Tandem Repeat-Enriched RNA Assembles a Nuclear Compartment to Control Alternative Splicing and Promote Cell Survival. Mol. Cell 2018, 72, 525–540. [Google Scholar] [CrossRef]
- Jacob, M.D.; Audas, T.E.; Uniacke, J.; Trinkle-Mulcahy, L.; Lee, S. Environmental cues induce a long noncoding RNA-dependent remodeling of the nucleolus. Mol. Biol. Cell 2013, 24, 2943–2953. [Google Scholar] [CrossRef]
- Wang, M.; Tao, X.; Jacob, M.D.; Bennett, C.A.; Ho, J.J.D.; Gonzalgo, M.L.; Audas, T.E.; Lee, S. Stress-Induced Low Complexity RNA Activates Physiological Amyloidogenesis. Cell Rep. 2018, 24, 1713–1721. [Google Scholar] [CrossRef] [Green Version]
- Theodoridis, P.R.; Bokros, M.; Marijan, D.; Balukoff, N.C.; Wang, D.; Kirk, C.C.; Budine, T.D.; Goldsmith, H.D.; Wang, M.; Audas, T.E.; et al. Local translation in nuclear condensate amyloid bodies. Proc. Natl. Acad. Sci. USA 2021, 118, e2014457118. [Google Scholar] [CrossRef]
- Abraham, K.J.; Khosraviani, N.; Chan, J.N.Y.; Gorthi, A.; Samman, A.; Zhao, D.Y.; Wang, M.; Bokros, M.; Vidya, E.; Ostrowski, L.A.; et al. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature 2020, 585, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Nadel, J.; Athanasiadou, R.; Lemetre, C.; Wijetunga, N.A.; Ó Broin, P.; Sato, H.; Zhang, Z.; Jeddeloh, J.; Montagna, C.; Golden, A.; et al. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenet. Chromatin. 2015, 8, 46. [Google Scholar] [CrossRef] [Green Version]
- Yan, Q.; Zhu, C.; Guang, S.; Feng, X. The Functions of Non-coding RNAs in rRNA Regulation. Front. Genet. 2019, 10, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCann, K.L.; Baserga, S.J. Driving nucleolar assembly. Genes Dev. 2014, 28, 211–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Sluis, M.; McStay, B. Nucleolar reorganization in response to rDNA damage. Curr. Opin. Cell Biol. 2017, 46, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Van Sluis, M.; McStay, B. Nucleolar DNA Double-Strand Break Responses Underpinning rDNA Genomic Stability. Trends Genet. 2019, 35, 743–753. [Google Scholar] [CrossRef]
- Caudron-Herger, M.; Pankert, T.; Rippe, K. Regulation of nucleolus assembly by non-coding RNA polymerase II transcripts. Nucleus 2016, 7, 308–318. [Google Scholar] [CrossRef] [Green Version]
- Mars, J.C.; Tremblay, M.G.; Valere, M.; Sibai, D.S.; Sabourin-Felix, M.; Lessard, F.; Moss, T. The chemotherapeutic agent CX-5461 irreversibly blocks RNA polymerase I initiation and promoter release to cause nucleolar disruption, DNA damage and cell inviability. NAR Cancer 2020, 2, zcaa032. [Google Scholar] [CrossRef]
- Hirai, H. Chromosome Dynamics Regulating Genomic Dispersion and Alteration of Nucleolus Organizer Regions (NORs). Cells 2020, 9, 971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harding, S.M.; Boiarsky, J.A.; Greenberg, R.A. ATM Dependent Silencing Links Nucleolar Chromatin Reorganization to DNA Damage Recognition. Cell Rep. 2015, 13, 251–259. [Google Scholar] [CrossRef] [Green Version]
- Van Sluis, M.; McStay, B. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev. 2015, 29, 1151–1163. [Google Scholar] [CrossRef] [Green Version]
- Floutsakou, I.; Agrawal, S.; Nguyen, T.T.; Seoighe, C.; Ganley, A.R.; McStay, B. The shared genomic architecture of human nucleolar organizer regions. Genome Res. 2013, 23, 2003–2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caudron-Herger, M.; Pankert, T.; Seiler, J.; Németh, A.; Voit, R.; Grummt, I.; Rippe, K. Alu element-containing RNAs maintain nucleolar structure and function. EMBO J. 2015, 34, 2758–2774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, Y.-H.; Yao, R.-W.; Zhang, Y.; Guo, C.-J.; Jiang, S.; Xu, G.; Dong, R.; Yang, L.; Chen, L.-L. SLERT Regulates DDX21 Rings Associated with Pol I Transcription. Cell 2017, 169, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, J.; Wang, M.; Li, X.; Gong, H.; Tang, H.; Chen, L.; Wan, L.; Liu, Q. Activity dependent LoNA regulates translation by coordinating rRNA transcription and methylation. Nat. Commun. 2018, 9, 1726. [Google Scholar] [CrossRef]
- Voit, R.; Hoffmann, M.; Grummt, I. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 1999, 18, 1891–1899. [Google Scholar] [CrossRef] [Green Version]
- Voit, R.; Grummt, I. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc. Natl. Acad. Sci. USA 2001, 98, 13631–13636. [Google Scholar] [CrossRef] [Green Version]
- Stefanovsky, V.Y.; Pelletier, G.; Hannan, R.; GagnonKugler, T.; Rothblum, L.I.; Moss, T. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 2001, 8, 1063–1073. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, X.; Fro, M.; Grummt, I. ERK-Dependent Phosphorylation of the Transcription Initiation Factor TIF-IA Is Required for RNA Polymerase, I. Transcr. Cell Growth. Mol. Cell 2003, 11, 405–413. [Google Scholar] [CrossRef]
- Zhang, C.; Comai, L.; Johnson, D.L. PTEN represses RNA Polymerase I transcription by disrupting the SL1 complex. Mol. Cell. Biol. 2005, 25, 6899–6911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, H.; Chen, X.; Yin, Q.; Ruan, D.; Zhao, X.; Zhang, C.; McNutt, M.A.; Yin, Y. PTENβ is an alternatively translated isoform of PTEN that regulates rDNA transcription. Nat. Commun. 2017, 8, 14771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lessard, F.; Morin, F.; Ivanchuk, S.; Langlois, F.; Stefanovsky, V.; Rutka, J.; Moss, T. The ARF tumor suppressor controls ribosome biogenesis by regulating the RNA polymerase I transcription factor TTF-I. Mol. Cell 2010, 38, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Pilch, P.F. PTRF/Cavin-1 promotes efficient ribosomal RNA transcription in response to metabolic challenges. eLife 2016, 5, e17508. [Google Scholar] [CrossRef] [PubMed]
- Liu, L. Lessons from cavin-1 deficiency. Biochem. Soc. Trans. 2020, 48, 147–154. [Google Scholar] [CrossRef]
- Shiue, C.-N.; Berkson, R.G.; Wright, A.P.H. c-Myc induces changes in higher order rDNA structure on stimulation of quiescent cells. Oncogene 2009, 28, 1833–1842. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Schmitz, K.M.; Mayer, C.; Yuan, X.; Akhtar, A.; Grummt, I. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat. Cell Biol. 2009, 11, 1010–1016. [Google Scholar] [CrossRef]
- Santoro, R.; Schmitz, K.M.; Sandoval, J.; Grummt, I. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep. 2010, 11, 52–58. [Google Scholar] [CrossRef] [Green Version]
- Guetg, C.; Lienemann, P.; Sirri, V.; Grummt, I.; Hernandez-Verdun, D.; Hottiger, M.O.; Fussenegger, M.; Santoro, R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010, 29, 2135–2146. [Google Scholar] [CrossRef]
- Lee, D.; An, J.; Park, Y.U. SHPRH regulates rRNA transcription by recognizing the histone code in an mTOR-dependent manner. Proc. Natl. Acad. Sci. USA 2017, 114, E3424–E3433. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.; Park, J.H.; Kim, S.; Lee, S.G.; Myung, K. SHPRH as a new player in ribosomal RNA transcription and its potential role in homeostasis of ribosomal DNA repeats. Transcription 2018, 9, 190–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarshad, A.; Sadeghifar, F.; Louvet, E.; Mori, R.; Böhm, S.; Al-Muzzaini, B.; Vintermist, A.; Fomproix, N.; Östlund, A.K.; Percipalle, P. Nuclear myosin 1c facilitates the chromatin modifications required to activate rRNA gene transcription and cell cycle progression. PLoS Genet. 2013, 9, e1003397. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.C.; Greco, T.M.; Boonmee, A.; Miteva, Y.; Cristea, I.M. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol. Cell. Proteom. 2012, 11, 60–76. [Google Scholar] [CrossRef] [Green Version]
- Kiran, S.; Anwar, T.; Kiran, M.; Ramakrishna, G. Sirtuin 7 in cell proliferation, stress and disease: Rise of the Seventh Sirtuin! Cell Signal. 2015, 27, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Barnett, C.; Yazgan, O.; Kuo, H.C.; Malakar, S.; Thomas, T.; Fitzgerald, A.; Harbour, W.; Henry, J.J.; Krebs, J.E. Williams Syndrome Transcription Factor is critical for neural crest cell function in Xenopus laevis. Mech. Dev. 2012, 129, 324–338. [Google Scholar] [CrossRef]
- Franek, M.; Kovaříková, A.; Bártová, E.; Kozubek, S. Nucleolar Reorganization Upon Site-Specific Double-Strand Break Induction. J. Histochem. Cytochem. 2016, 64, 669–686. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tariq, K.; Östlund Farrants, A.-K. Antagonising Chromatin Remodelling Activities in the Regulation of Mammalian Ribosomal Transcription. Genes 2021, 12, 961. https://doi.org/10.3390/genes12070961
Tariq K, Östlund Farrants A-K. Antagonising Chromatin Remodelling Activities in the Regulation of Mammalian Ribosomal Transcription. Genes. 2021; 12(7):961. https://doi.org/10.3390/genes12070961
Chicago/Turabian StyleTariq, Kanwal, and Ann-Kristin Östlund Farrants. 2021. "Antagonising Chromatin Remodelling Activities in the Regulation of Mammalian Ribosomal Transcription" Genes 12, no. 7: 961. https://doi.org/10.3390/genes12070961
APA StyleTariq, K., & Östlund Farrants, A.-K. (2021). Antagonising Chromatin Remodelling Activities in the Regulation of Mammalian Ribosomal Transcription. Genes, 12(7), 961. https://doi.org/10.3390/genes12070961





