Extracellular Vesicle-Derived DNA vs. CfDNA as a Biomarker for the Detection of Colon Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. Extraction of DNA from Tumor Tissue
2.3. Panel Design
2.4. Isolation and Characterization of Extracellular Vesicles
2.5. Extraction of EV-DNA and cfDNA from Plasma
2.6. Library Preparation for Next Generation Sequencing
2.7. Sequencing Data Analysis
2.8. Statistical Analysis
3. Results
3.1. Extra Cellular Vesicles Characterization
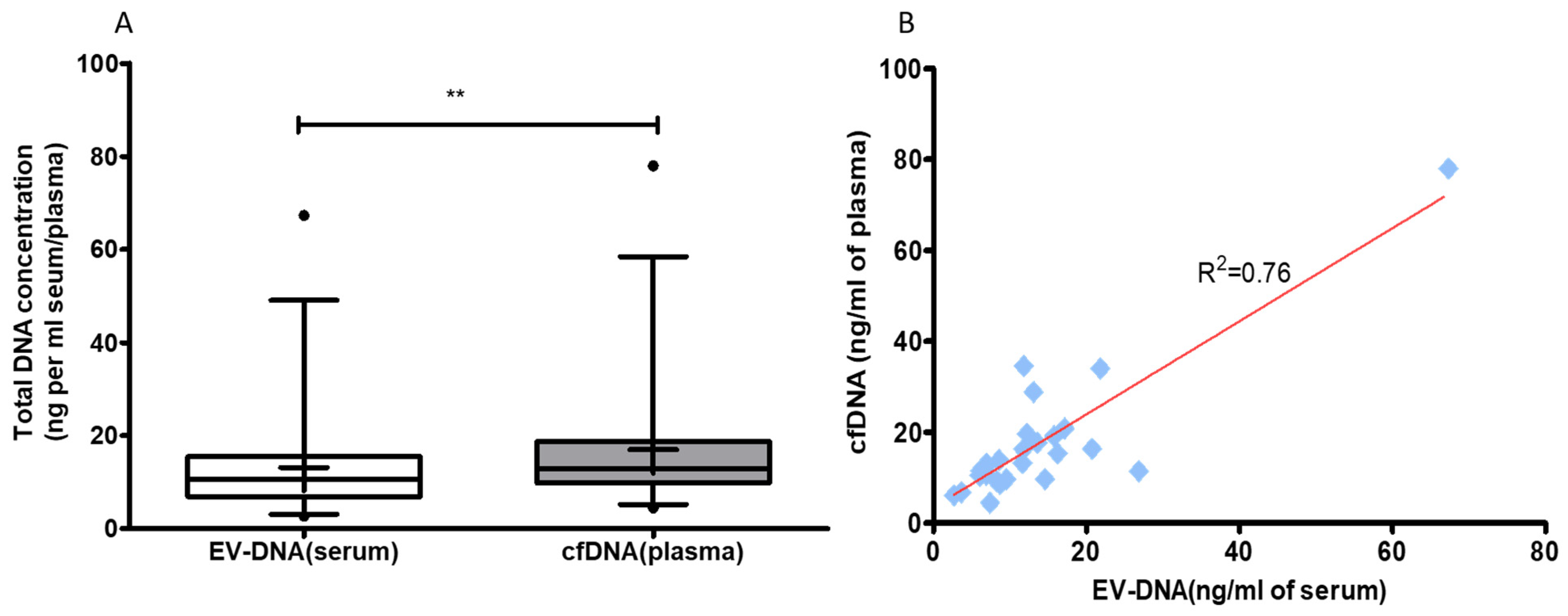
3.2. Blood cfDNA Concentration Is Higher Than EV-DNA
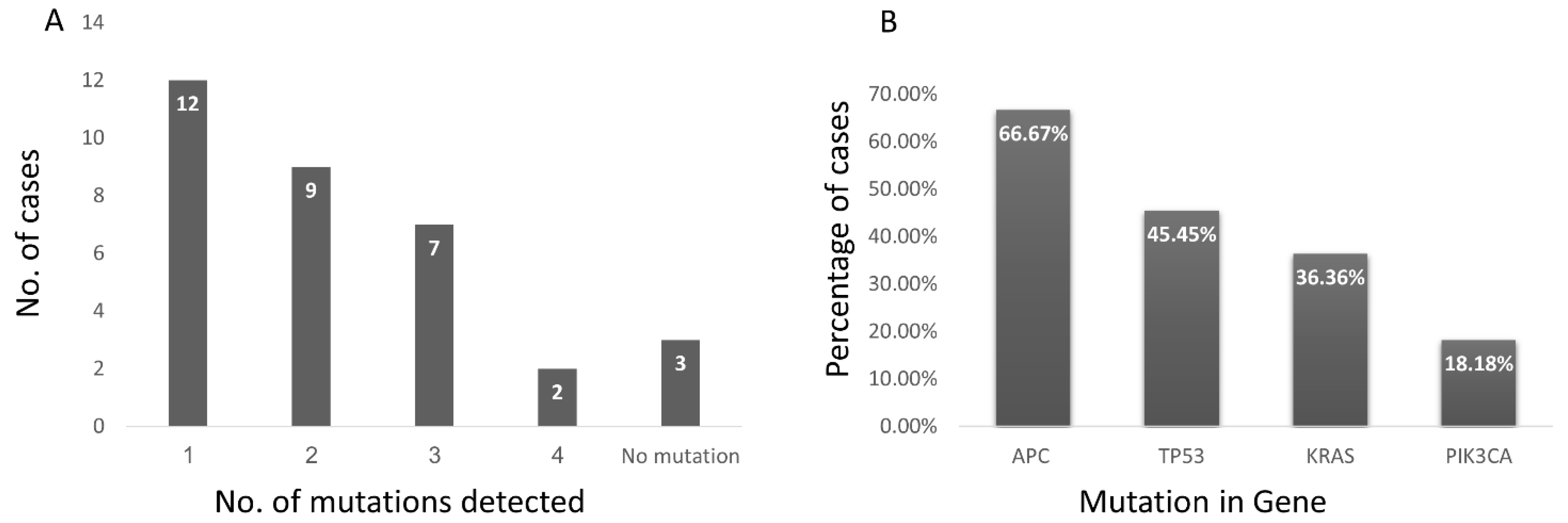
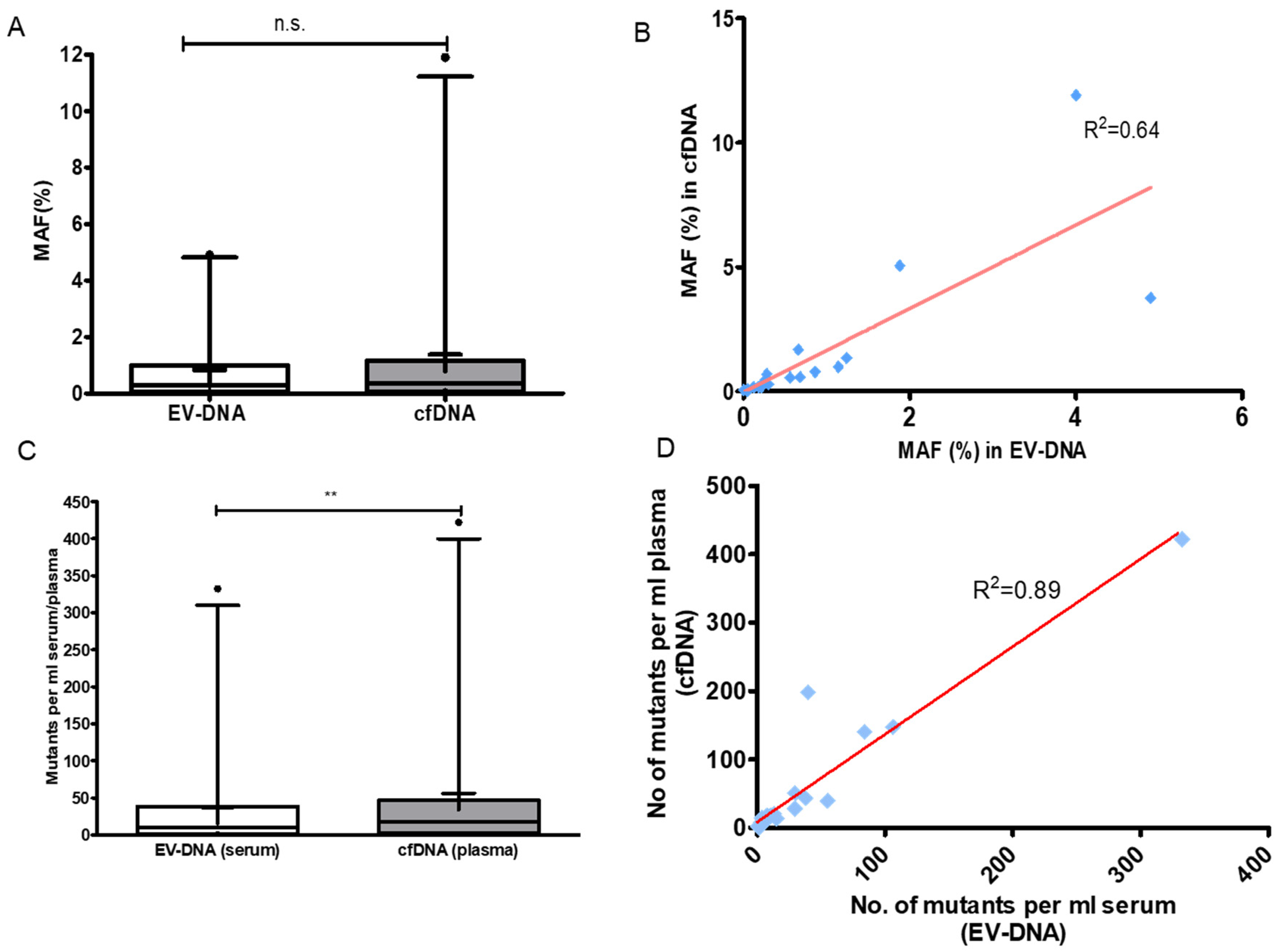
3.3. The Sensitivity of Tumor DNA Detection Was Similar in EV-DNA and cfDNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef] [Green Version]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [Green Version]
- GraçA, R.; Willem, S. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar]
- Elzanowska, J.; Semira, C.; Costa-Silva, B. DNA in extracellular vesicles: Biological and clinical aspects. Mol. Oncol. 2021, 15, 1701–1714. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct. Target. Ther. 2020, 5, 1–14. [Google Scholar] [CrossRef]
- Kahlert, C.; Melo, S.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of Double-stranded Genomic DNA Spanning All Chromosomes with Mutated KRAS and p53 DNA in the Serum Exosomes of Patients with Pancreatic Cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef] [Green Version]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Chen, K.; Wang, Z.; Wang, Y.; Liu, J.; Lin, L.; Shao, Y.; Gao, L.; Yin, H.; Cui, C.; et al. DNA in serum extracellular vesicles is stable under different storage conditions. BMC Cancer 2016, 16, 753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klump, J.; Phillipp, U.; Follo, M.; Eremin, A.; Lehmann, H.; Nestel, S.; von Bubnoff, N.; Nazarenko, I. Extracellular vesicles or free circulating DNA: Where to search for BRAF and cKIT mutations? Nanomed. Nanotechnol. Biol. Med. 2018, 14, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Allenson, K.; Castillo, J.; Lucas, F.A.S.; Scelo, G.; Kim, D.U.; Bernard, V.; Davis, G.; Kumar, T.; Katz, M.; Overman, M.J.; et al. High prevalence of mutantKRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017, 28, 741–747. [Google Scholar] [CrossRef]
- Bernard, V.; Kim, D.U.; Lucas, F.A.S.; Castillo, J.; Allenson, K.; Mulu, F.C.; Stephens, B.M.; Huang, J.; Semaan, A.; Guerrero, P.A.; et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology 2019, 156, 108–118.e4. [Google Scholar] [CrossRef] [Green Version]
- Abe, T.; Nakashima, C.; Sato, A.; Harada, Y.; Sueoka, E.; Kimura, S.; Kawaguchi, A.; Sueoka-Aragane, N. Origin of circulating free DNA in patients with lung cancer. PLoS ONE 2020, 15, e0235611. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [Green Version]
- Biezuner, T.; Spiro, A.; Raz, O.; Amir, S.; Milo, L.; Adar, R.; Chapal-Ilani, N.; Berman, V.; Fried, Y.; Ainbinder, E.; et al. A generic, cost-effective, and scalable cell lineage analysis platform. Genome Res. 2016, 26, 1588–1599. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015, 10, 1556–1566. [Google Scholar] [CrossRef] [PubMed]
- Yohe, S.; Thyagarajan, B. Review of Clinical Next-Generation Sequencing. Arch. Pathol. Lab. Med. 2017, 141, 1544–1557. [Google Scholar] [CrossRef] [Green Version]
- Diaz, L.; Bardelli, A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Alcaide, M.; Cheung, M.; Hillman, J.; Rassekh, S.R.; Deyell, R.; Batist, G.; Karsan, A.; Wyatt, A.W.; Johnson, N.; Scott, D.W.; et al. Evaluating the quantity, quality and size distribution of cell-free DNA by multiplex droplet digital PCR. Sci. Rep. 2020, 10, 12564. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez l’homme. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar] [PubMed]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef] [Green Version]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer man-agement. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-C.; Wang, D.; Jin, L.; Yao, H.-W.; Zhang, J.-H.; Wang, J.; Zhao, X.-M.; Shen, C.-Y.; Chen, W.; Wang, X.-L.; et al. Circulating tumor DNA detectable in early- and late-stage colorectal cancer patients. Biosci. Rep. 2018, 38, BSR20180322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, J.D.; Javed, A.A.; Thoburn, C.; Wong, F.; Tie, J.; Gibbs, P.; Schmidt, C.M.; Yip-Schneider, M.T.; Allen, P.J.; Schattner, M.; et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 10202–10207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.H.; Yoon, H.; Park, S.; Kim, J.S.; Ahn, Y.-H.; Kwon, K.; Lee, D.; Kim, K.H. Urinary Exosomal and cell-free DNA Detects Somatic Mutation and Copy Number Alteration in Urothelial Carcinoma of Bladder. Sci. Rep. 2018, 8, 14707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Y.; Liu, B.; Lei, H.; Zhang, B.; Huang, H.; Chen, S.; Feng, Y.; Zhu, L.; Gu, Y.; Zhang, Q.; et al. Nanoscale extracellular vesicle-derived DNA is superior to circulating cell-free DNA for mutation detection in early-stage non-small-cell lung cancer. Ann. Oncol. 2018, 29, 2379–2383. [Google Scholar] [CrossRef]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef] [Green Version]
- Vagner, T.; Spinelli, C.; Minciacchi, V.R.; Balaj, L.; Zandian, M.; Conley, A.; Zijlstra, A.; Freeman, M.R.; Demichelis, F.; De, S.; et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. Vesicles 2018, 7, 1505403. [Google Scholar] [CrossRef] [Green Version]
- Malkin, E.Z.; Bratman, S.V. Bioactive DNA from extracellular vesicles and particles. Cell Death Dis. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Johansson, G.; Andersson, D.; Filges, S.; Li, J.; Muth, A.; Godfrey, T.E.; Ståhlberg, A. Considerations and quality controls when analyzing cell-free tumor DNA. Biomol. Detect. Quantif. 2019, 17, 100078. [Google Scholar] [CrossRef]
- Gansauge, M.-T.; Gerber, T.; Glocke, I.; Korlević, P.; Lippik, L.; Nagel, S.; Riehl, L.M.; Schmidt, A.; Meyer, M. Single-stranded DNA library preparation from highly degraded DNA usingT4DNA ligase. Nucleic Acids Res. 2017, 45, e79. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Age (Years) | Mean ± SD | 69 ± 14.29 | |
| Sex | Male | 15 (45.45%) | |
| Female | 18 (54.54%) | ||
| Diameter (cm) | Mean ± SD | 4.59 ± 1.65 | |
| Grade (Differentiation) | Well | 7 (21.21%) | |
| Moderate | 20 (60.60%) | ||
| Poor | 3 (9.09%) | ||
| Mucinous | 3 (9.09%) | ||
| Stage | T | T1 | 1 (3.03%) |
| T2 | 11 (33.33%) | ||
| T3 | 18 (54.54%) | ||
| T4 | 3 (9.09%) | ||
| N | N0 | 22 (66.66%) | |
| N1 | 4 (12.12%) | ||
| N2 | 7 (21.12%) | ||
| I | 10 (30.30%) | ||
| II | 10 (30.30%) | ||
| III | 9 (27.27%) | ||
| IV | 4 (12.12%) | ||
| Metastasis | Yes | 4 (12.12%) | |
| No | 29 (87.87%) | ||
| Vascular Invasion | Yes | 7 (21.12%) | |
| No | 16 (48.48%) | ||
| Serial No | Gene | Mutation | Stage | Tumor Size (cm) | MAF in EV-DNA (%) | No. of Mutant Reads from Each Duplicate | MAF in cfDNA (Plasma) (%) | No. of Mutant Reads from Each Duplicate | Noise in Wild Type DNA (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | TP53 | p.R175H (c.524G>A) | II | 2.5 | 0.30% | ≥10 | 0.29% | ≥10 | 0% |
| 2 | KRAS | p.G12V (c.35G>T) | II | 2.5 | 0.05% | <10 | 0.04% | <10 | 0.008% |
| 3 | APC | p.R1450X (c.4348C>T) | III | 4 | 1.14% | ≥10 | 0.99% | ≥10 | 0% |
| 4 | TP53 | p.R248W (c.742C>T) | IV | 4.5 | 4.00% | ≥10 | 11.90% | ≥10 | 0% |
| 5 | TP53 | p.R273H (c.818G>A) | IV | 6 | 1.24% | ≥10 | 1.34% | ≥10 | 0.026% |
| 6 | KRAS | p.G12S (c.34G>A) | IV | 5.5 | 0.24% | ≥10 | 0.37% | ≥10 | 0.012% |
| 7 | APC | p.E1309X (c.3925G>T) | IV | 2.5 | 0.04% | <10 | 0.02% | <10 | 0.00% |
| 8 | PIK3CA | p.E545K (c.1633G>A) | II | 6 | 0.68% | ≥10 | 0.57% | ≥10 | 0% |
| 9 | PIK3CA | p.H1047L (c.3140A>T) | II | 6 | 4.90% | 10 | 3.75% | ≥10 | 0% |
| 10 | TP53 | p.C242R (c.724T>C) | I | 6 | NA | NA | NA | <10 | 0.05% |
| 11 | KRAS | p.G12D (c.35G>A) | II | 2.5 | 0% | <10 | 0.05% | <10 | 0.014% |
| 12 | KRAS | p.G12V (c.35G>T) | II | 2 | 0.05% | <10 | 0.07% | <10 | 0.008% |
| 13 | TP53 | p.R175H (c.524G>A) | III | 8 | 0.66% | ≥10 | 1.68% | ≥10 | 0% |
| 14 | APC | p.Q1406X (c.4216C>T) | III | 5.2 | 0.86% | ≥10 | 0.78% | ≥10 | 0.09% |
| 15 | TP53 | p.G245S (c.733G>A) | I | 5 | 0.56% | ≥10 | 0.56% | ≥10 | 0.02% |
| 16 | TP53 | p.A161T (c.481G>A) | I | 1.2 | 0.06% | <10 | 0.06% | <10 | 0% |
| 17 | TP53 | p.M237V (c.709A>G) | I | 6 | 0.04% | <10 | 0.06% | <10 | 0% |
| 18 | TP53 | p.R175H (c.524G>A) | III | 5.5 | 0.18% | <10 | 0.18% | <10 | 0% |
| 19 | TP53 | p.R306X (c.916C>T) | I | 1 | NA | NA | NA | NA | 0.06% |
| 20 | APC | p.R232X (c.694C>T) | III | 6 | 0.28% | ≥10 | 0.69% | ≥10 | 0.07% |
| 21 | APC | p. P1432Hfs*4 | I | 4.5 | 1.88% | ≥10 | 5.05% | ≥10 | 0.24% |
| 22 | TP53 | p.R175H (c.524G>A) | I | 3 | NA | NA | NA | NA | 0% |
| 23 | APC | p.R1450X (c.4348C>T) | II | 4 | 0.12% | <10 | 0.17% | ≥10 | 0% |
| 24 | TP53 | p.R175H (c.524G>A) | II | 6 | NA | NA | NA | NA | 0% |
| 25 | KRAS | p.G12V (c.35G>T) | I | 5 | 0.21% | ≥10 | 0.17% | ≥10 | 0.008% |

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thakur, K.; Singh, M.S.; Feldstein-Davydova, S.; Hannes, V.; Hershkovitz, D.; Tsuriel, S. Extracellular Vesicle-Derived DNA vs. CfDNA as a Biomarker for the Detection of Colon Cancer. Genes 2021, 12, 1171. https://doi.org/10.3390/genes12081171
Thakur K, Singh MS, Feldstein-Davydova S, Hannes V, Hershkovitz D, Tsuriel S. Extracellular Vesicle-Derived DNA vs. CfDNA as a Biomarker for the Detection of Colon Cancer. Genes. 2021; 12(8):1171. https://doi.org/10.3390/genes12081171
Chicago/Turabian StyleThakur, Kavita, Manu Smriti Singh, Sara Feldstein-Davydova, Victoria Hannes, Dov Hershkovitz, and Shlomo Tsuriel. 2021. "Extracellular Vesicle-Derived DNA vs. CfDNA as a Biomarker for the Detection of Colon Cancer" Genes 12, no. 8: 1171. https://doi.org/10.3390/genes12081171
APA StyleThakur, K., Singh, M. S., Feldstein-Davydova, S., Hannes, V., Hershkovitz, D., & Tsuriel, S. (2021). Extracellular Vesicle-Derived DNA vs. CfDNA as a Biomarker for the Detection of Colon Cancer. Genes, 12(8), 1171. https://doi.org/10.3390/genes12081171







