Understanding Resistance Mechanisms to Trifluralin in an Arkansas Palmer Amaranth Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
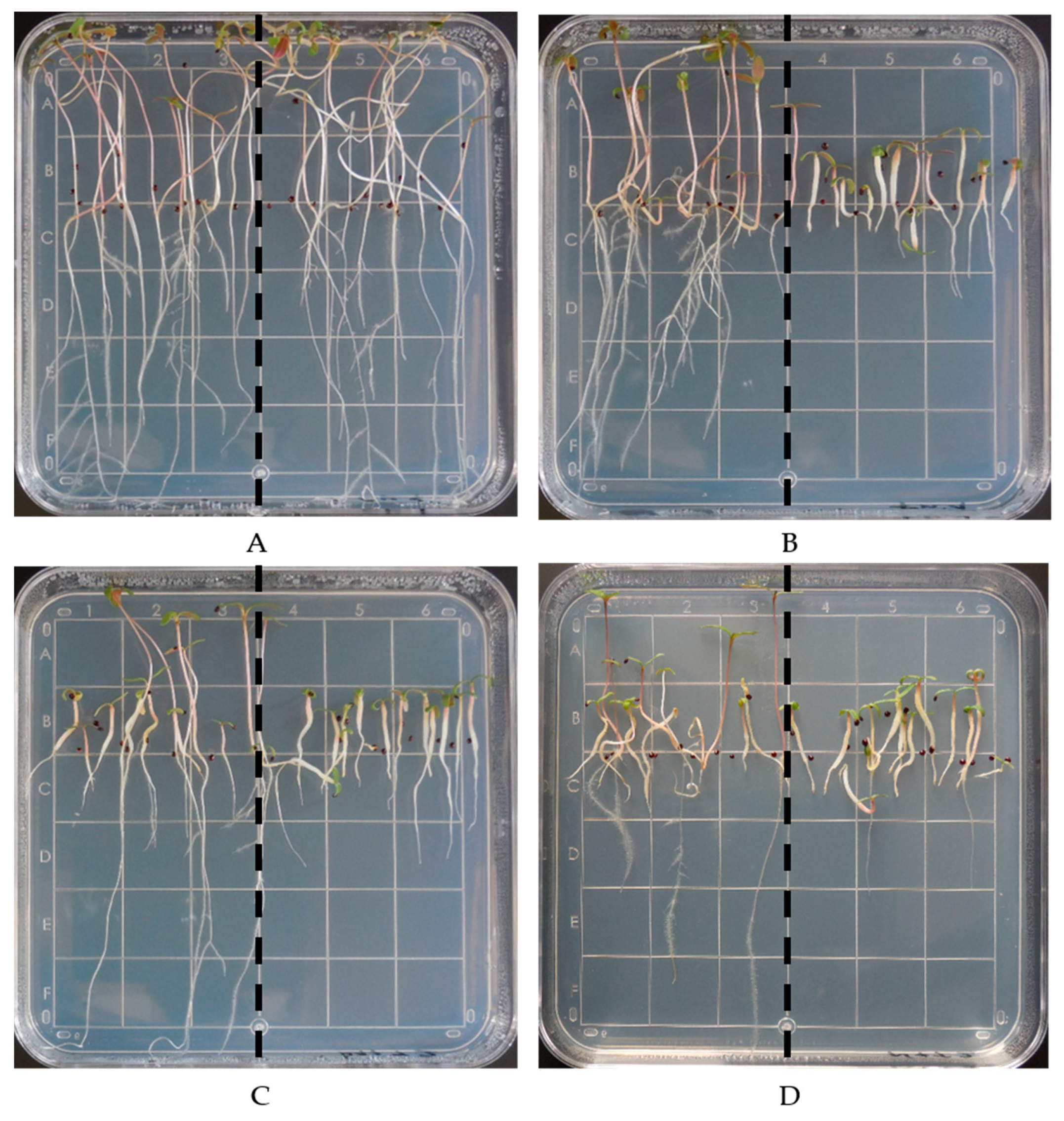
2.2. Dose-Response Experiments on Agar Plates
2.3. Metabolism Assays
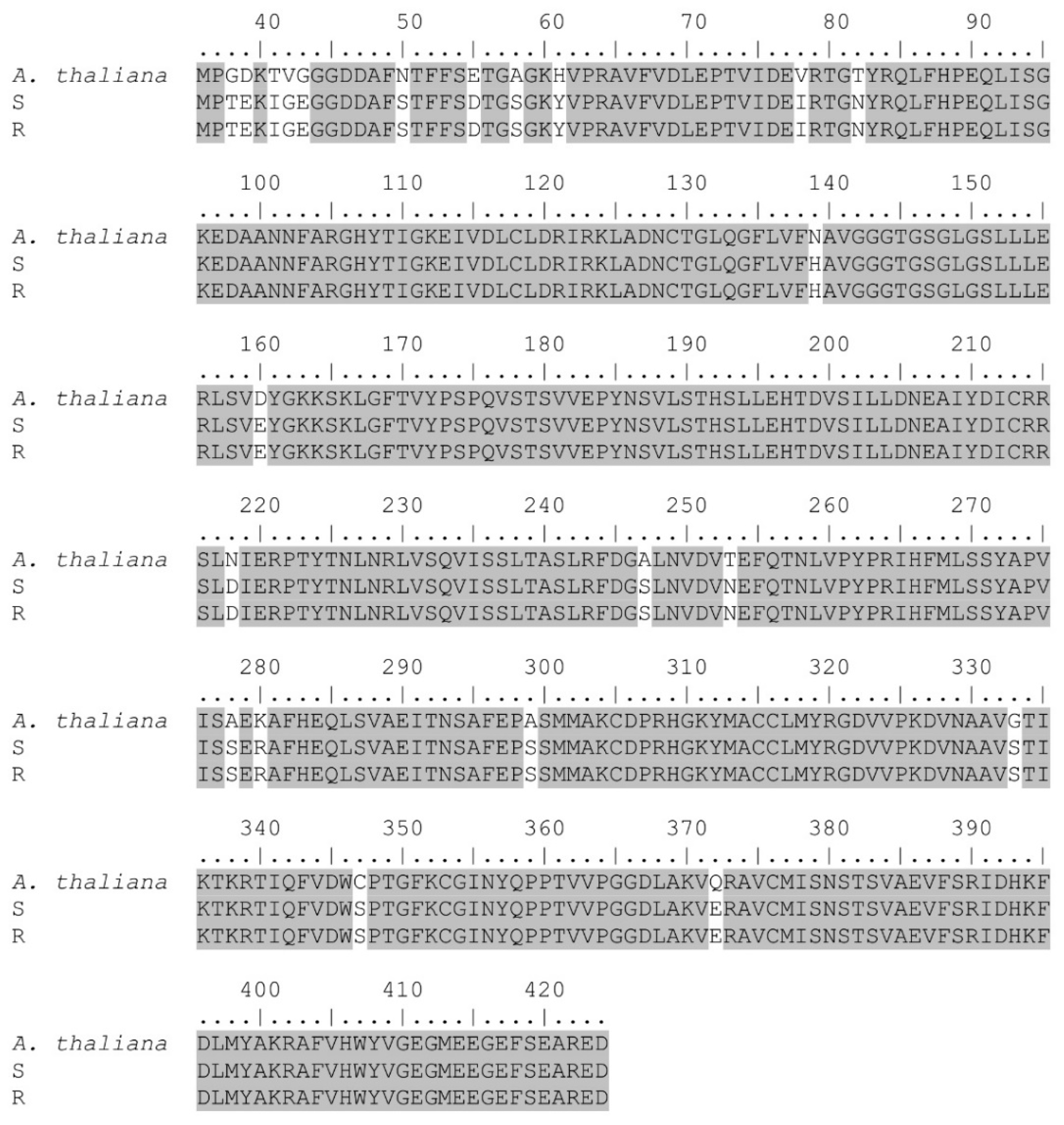
2.4. Tubulin Gene Sequencing
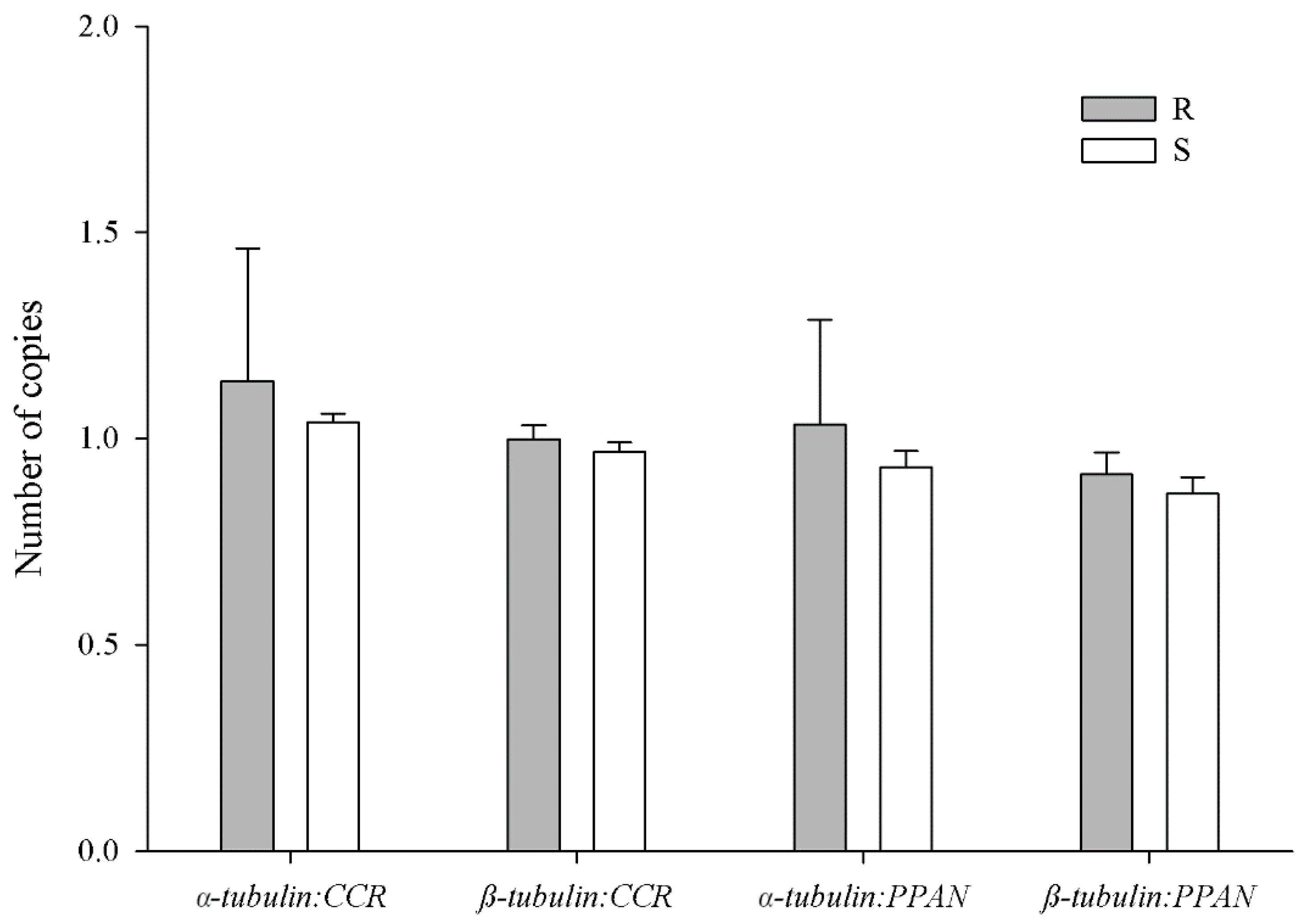
2.5. Tubulin Gene Copy Number Estimation
3. Results and Discussion
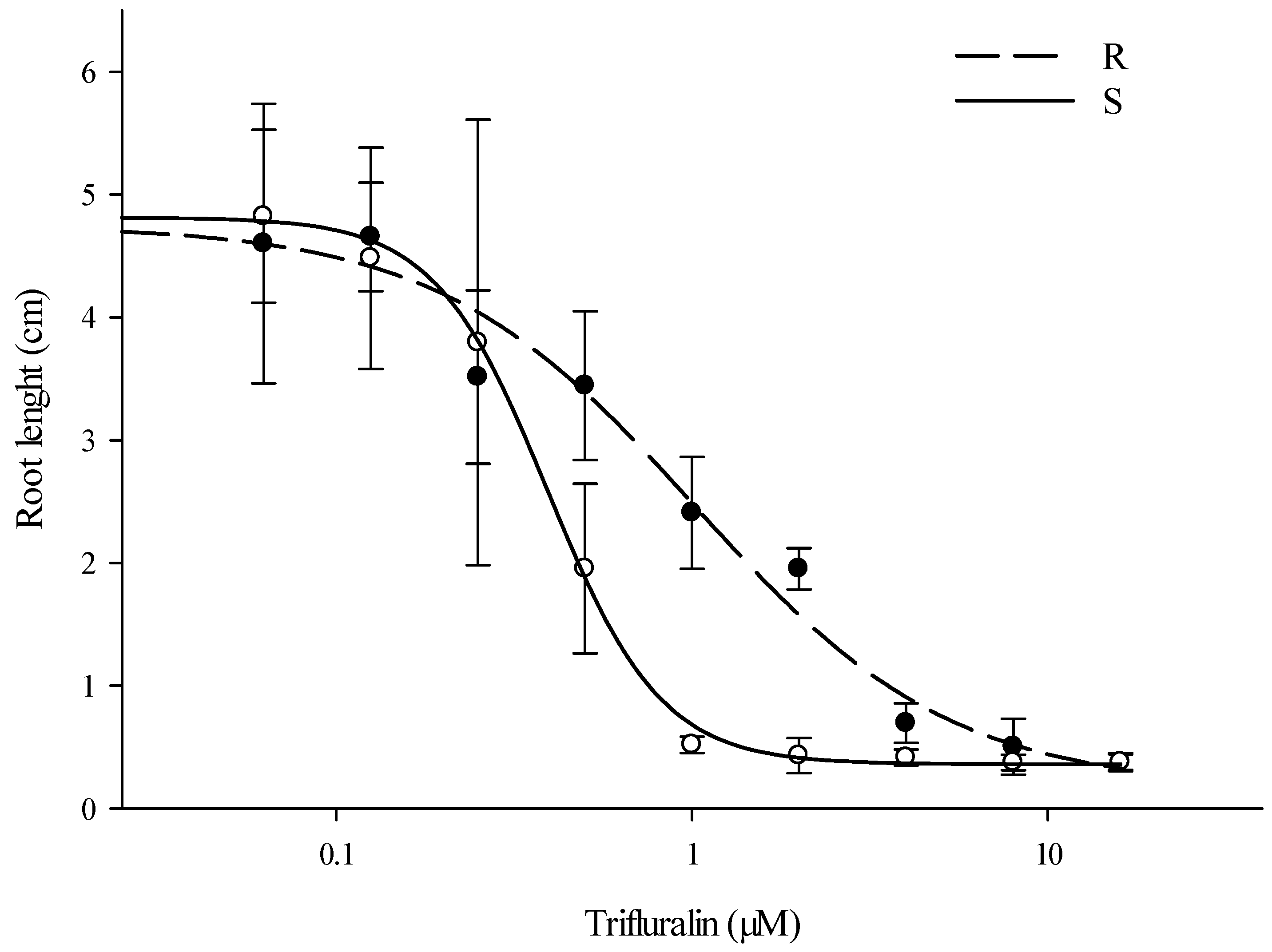
3.1. Dose-Response Experiments on Agar Plates
3.2. Metabolism Assays
3.3. Tubulin Gene Sequencing
3.4. Tubulin Gene Copy Number Estimation
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sauer, J. Recent migration and evolution of the dioecious amaranths. Evolution 1957, 11, 11–31. [Google Scholar] [CrossRef]
- Van Wychen, L. Survey of the Most Common and Troublesome Weeds in Grass Crops, Pasture, and Turf in the United States and Canada. In Weed Science Society of America National Weed Survey Dataset; Weed Science Society of America: Westminster, UK, 2020; Available online: https://wssa.net/wp-content/uploads/2020-Weed-Survey_grass-crops.xlsx (accessed on 4 June 2021).
- Heap, I. The International Herbicide-Resistant Weed Database. Available online: www.weedscience.org (accessed on 28 June 2021).
- Horak, M.J.; Peterson, D.E. Biotypes of palmer amaranth (Amaranthus palmeri) and common waterhemp (Amaranthus rudis) are resistant to imazethapyr and thifensulfuron. Weed Technol. 1995, 9, 192–195. [Google Scholar] [CrossRef]
- Gossett, B.J.; Murdock, E.C.; Toler, J.E. Resistance of palmer amaranth (Amaranthus palmeri) to the dinitroaniline herbicides. Weed Technol. 1992, 6, 587–591. [Google Scholar] [CrossRef]
- Peterson, D.; Jugulam, M.; Shyam, C.; Borgato, E. Palmer Amaranth Resistance to 2,4-D and Dicamba Confirmed in Kansas. Available online: https://webapp.agron.ksu.edu/agr_social/eu_article.throck?article_id=2110 (accessed on 24 June 2020).
- Chahal, P.S.; Jugulam, M.; Jhala, A.J. Mechanism of atrazine resistance in atrazine-and HPPD inhibitor-resistant palmer amaranth (Amaranthus palmeri S. Wats.) from Nebraska. Can. J. Plant Sci. 2019, 99, 815–823. [Google Scholar] [CrossRef]
- Culpepper, A.S.; Grey, T.L.; Vencill, W.K.; Kichler, J.M.; Webster, T.M.; Brown, S.M.; York, A.C.; Davis, J.W.; Hanna, W.W. Glyphosate-resistant palmer amaranth (Amaranthus palmeri) confirmed in Georgia. Weed Sci. 2006, 54, 620–626. [Google Scholar] [CrossRef]
- Varanasi, V.K.; Brabham, C.; Norsworthy, J.K. Confirmation and characterization of non–target site resistance to fomesafen in palmer amaranth (Amaranthus palmeri). Weed Sci. 2018, 66, 702–709. [Google Scholar] [CrossRef]
- Brabham, C.; Norsworthy, J.K.; Houston, M.M.; Varanasi, V.K.; Barber, T. Confirmation of S-metolachlor resistance in palmer Amaranth (Amaranthus palmeri). Weed Technol. 2019, 33, 720–726. [Google Scholar] [CrossRef] [Green Version]
- Nakka, S.; Godar, A.S.; Wani, P.S.; Thompson, C.R.; Peterson, D.E.; Roelofs, J.; Jugulam, M. Physiological and molecular characterization of hydroxyphenylpyruvate dioxygenase (HPPD)-inhibitor resistance in palmer amaranth (Amaranthus palmeri S. Wats.). Front. Plant Sci. 2017, 8, 555. [Google Scholar] [CrossRef] [Green Version]
- Epp, J.B.; Schmitzer, P.R.; Crouse, G.D. Fifty years of herbicide research: Comparing the discovery of trifluralin and halauxifen-methyl. Pest Manag. Sci. 2018, 74, 9–16. [Google Scholar] [CrossRef] [PubMed]
- USDA-NASS US Department of Agriculture (2021) National Agricultural Statistics Service: Quick Stats. Available online: https://quickstats.nass.usda.gov/ (accessed on 3 June 2021).
- Nogales, E.; Wolf, S.G.; Downing, K.H. Structure of the αβ tubulin dimer by electron crystallography. Nature 1998, 391, 199–203. [Google Scholar] [CrossRef]
- Anthony, R.G.; Hussey, P.J. Dinitroaniline herbicide resistance and the microtubule cytoskeleton. Trends Plant Sci. 1999, 4, 112–116. [Google Scholar] [CrossRef]
- Senseman, S.A. Herbicide Handbook, 9th ed.; Weed Science Society of America: Champaign, IL, USA, 2007. [Google Scholar]
- Busi, R.; Goggin, D.E.; Onofri, A.; Boutsalis, P.; Preston, C.; Powles, S.B.; Beckie, H.J. Loss of trifluralin metabolic resistance in Lolium rigidum plants exposed to prosulfocarb recurrent selection. Pest Manag. Sci. 2020, 76, 3926–3934. [Google Scholar] [CrossRef]
- Schultz, D.P.; Funderburk, H.H.; Negi, N.S. Effect of trifluralin on growth, morphology, and nucleic acid synthesis. Plant Physiol. 1968, 43, 265–273. [Google Scholar] [CrossRef] [Green Version]
- Morrison, I.N.; Todd, B.G.; Nawolsky, K.M. Confirmation of trifluralin-resistant green foxtail (Setaria viridis) in manitoba. Weed Technol. 1989, 3, 544–551. [Google Scholar] [CrossRef]
- Délye, C.; Menchari, Y.; Michel, S.; Darmency, H. Molecular bases for sensitivity to tubulin-binding herbicides in green foxtail. Plant Physiol. 2004, 136, 3920–3932. [Google Scholar] [CrossRef] [Green Version]
- McAlister, F.M.; Holtum, J.A.M.; Powles, S.B. Dintroaniline herbicide resistance in rigid ryegrass (Lolium rigidum). Weed Sci. 1995, 43, 55–62. [Google Scholar] [CrossRef]
- Chen, J.; Chu, Z.; Han, H.; Goggin, D.E.; Yu, Q.; Sayer, C.; Powles, S.B. A Val-202-Phe α-tubulin mutation and enhanced metabolism confer dinitroaniline resistance in a single Lolium rigidum population. Pest Manag. Sci. 2020, 76, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.; Chen, J.; Nyporko, A.; Han, H.; Yu, Q.; Powles, S. Novel α-tubulin mutations conferring resistance to dinitroaniline herbicides in Lolium rigidum. Front. Plant Sci. 2018, 9, 97. [Google Scholar] [CrossRef]
- Fleet, B.; Malone, J.; Preston, C.; Gill, G. Target-site point mutation conferring resistance to trifluralin in rigid ryegrass (Lolium rigidum). Weed Sci. 2018, 66, 246–253. [Google Scholar] [CrossRef]
- Chen, J.; Goggin, D.; Han, H.; Busi, R.; Yu, Q.; Powles, S. Enhanced trifluralin metabolism can confer resistance in Lolium rigidum. J. Agric. Food Chem. 2018, 66, 7589–7596. [Google Scholar] [CrossRef]
- Busi, R.; Gaines, T.A.; Powles, S. Phorate can reverse P450 metabolism-based herbicide resistance in Lolium rigidum. Pest Manag. Sci. 2017, 73, 410–417. [Google Scholar] [CrossRef] [PubMed]
- González-Torralva, F.; Norsworthy, J.K.; Piveta, L.B.; Varanasi, V.K.; Barber, T.; Brabham, C. Susceptibility of Arkansas palmer amaranth accessions to common herbicide sites of action. Weed Technol. 2020, 34, 770–775. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Seefeldt, S.; Jensen, J.E.; Fuerst, P. Log-logistic analysis of herbicide dose-response relationships. Weed Technol. 1995, 9, 218–227. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Q.; Patterson, E.; Sayer, C.; Powles, S. Dinitroaniline herbicide resistance and mechanisms in weeds. Front. Plant Sci. 2021, 12, 634018. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McInnes, R.; Lidgett, A.; Lynch, D.; Huxley, H.; Jones, E.; Mahoney, N.; Spangenberg, G. Isolation and characterization of a cinnamoyl-CoA reductase gene from perennial ryegrass (Lolium perenne). J. Plant Physiol. 2002, 159, 415–422. [Google Scholar] [CrossRef]
- Salas, R.A.; Dayan, F.E.; Pan, Z.; Watson, S.B.; Dickson, J.W.; Scott, R.C.; Burgos, N.R. EPSPS gene amplification in glyphosate-resistant italian ryegrass (Lolium perenne ssp. multiflorum) from Arkansas. Pest Manag. Sci. 2012, 68, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Information Resource (TAIR). Available online: https://www.arabidopsis.org (accessed on 27 March 2021).
- Radstrom, P.; Lofstrom, C.; Lovenklev, M. Strategies for overcoming PCR inhibition. In PCR Primer: A Laboratory Manual; Dieffenbach, C., Dveksler, G., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2003; pp. 149–161. [Google Scholar]
- Gaines, T.A.; Zhang, W.; Wang, D.; Bukun, B.; Chisholm, S.T.; Shaner, D.L.; Nissen, S.J.; Patzoldt, W.L.; Tranel, P.J.; Culpepper, A.S.; et al. Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Natl. Acad. Sci. USA 2010, 107, 1029–1034. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Salas-Perez, R.A.; Saski, C.A.; Noorai, R.E.; Srivastava, S.K.; Lawton-Rauh, A.L.; Nichols, R.L.; Roma-Burgos, N. RNA-seq transcriptome analysis of Amaranthus palmeri with differential tolerance to glufosinate herbicide. PLoS ONE 2018, 13, e0195488. [Google Scholar] [CrossRef] [PubMed]
- Strom, S.A.; Hager, A.G.; Seiter, N.J.; Davis, A.S.; Riechers, D.E. Metabolic resistance to S-metolachlor in two waterhemp (Amaranthus tuberculatus) populations from Illinois, USA. Pest Manag. Sci. 2020, 76, 3139–3148. [Google Scholar] [CrossRef]
- Wang, T.; Fleury, A.; Ma, J.; Darmency, H. Genetic control of dinitroaniline resistance in foxtail millet (Setaria italica). J. Hered. 1996, 87, 423–426. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Lu, H.; Han, H.; Yu, Q.; Sayer, C.; Powles, S. Genetic inheritance of dinitroaniline resistance in an annual ryegrass population. Plant Sci. 2019, 283, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Jasieniuk, M.; Brûlé-Babel, A.L.; Morrison, I.N. Inheritance of trifluralin resistance in green foxtail (Setaria viridis). Weed Sci. 1994, 42, 123–127. [Google Scholar] [CrossRef]
- Franco-Ortega, S.; Goldberg-Cavalleri, A.; Walker, A.; Brazier-Hicks, M.; Onkokesung, N.; Edwards, R. Non-target site herbicide resistance is conferred by two distinct mechanisms in black-grass (Alopecurus myosuroides). Front. Plant Sci. 2021, 12, 636652. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Powles, S. Metabolism-based herbicide resistance and cross-resistance in crop weeds: A threat to herbicide sustainability and global crop production. Plant Physiol. 2014, 166, 1106–1118. [Google Scholar] [CrossRef] [Green Version]
- Shyam, C.; Borgato, E.A.; Peterson, D.E.; Dille, J.A.; Jugulam, M. Predominance of metabolic resistance in a six-way-resistant palmer amaranth (Amaranthus palmeri) population. Front. Plant Sci. 2021, 11, 614618. [Google Scholar] [CrossRef] [PubMed]
- Iwakami, S.; Endo, M.; Saika, H.; Okuno, J.; Nakamura, N.; Yokoyama, M.; Watanabe, H.; Toki, S.; Uchino, A.; Inamura, T. Cytochrome P450 CYP81A12 and CYP81A21 are associated with resistance to two acetolactate synthase inhibitors in Echinochloa phyllopogon. Plant Physiol. 2014, 165, 618–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwakami, S.; Kamidate, Y.; Yamaguchi, T.; Ishizaka, M.; Endo, M.; Suda, H.; Nagai, K.; Sunohara, Y.; Toki, S.; Uchino, A.; et al. CYP81A P450s are involved in concomitant cross-resistance to acetolactate synthase and Acetyl-CoA carboxylase herbicides in Echinochloa phyllopogon. New Phytol. 2019, 221, 2112–2122. [Google Scholar] [CrossRef] [PubMed]
- Duhoux, A.; Carrère, S.; Gouzy, J.; Bonin, L.; Délye, C. RNA-seq analysis of rye-grass transcriptomic response to an herbicide inhibiting acetolactate-synthase identifies transcripts linked to non-target-site-based resistance. Plant Mol. Biol. 2015, 87, 473–487. [Google Scholar] [CrossRef]
- Duhoux, A.; Carrère, S.; Duhoux, A.; Délye, C. Transcriptional markers enable identification of rye-grass (Lolium sp.) plants with non-target-site-based resistance to herbicides inhibiting acetolactate-synthase. Plant Sci. 2017, 257, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Gaines, T.A.; Lorentz, L.; Figge, A.; Herrmann, J.; Maiwald, F.; Ott, M.; Han, H.; Busi, R.; Yu, Q.; Powles, S.B.; et al. RNA-seq transcriptome analysis to identify genes involved in metabolism-based diclofop resistance in Lolium rigidum. Plant J. 2014, 78, 865–876. [Google Scholar] [CrossRef]
- Rangani, G.; Noguera, M.; Salas-Perez, R.; Benedetti, L.; Roma-Burgos, N. Mechanism of resistance to S-metolachlor in palmer amaranth. Front. Plant Sci. 2021, 12, 652581. [Google Scholar] [CrossRef]
- Iwakami, S.; Shimono, Y.; Manabe, Y.; Endo, M.; Shibaike, H.; Uchino, A.; Tominaga, T. Copy number variation in acetolactate synthase genes of thifensulfuron-methyl resistant Alopecurus aequalis (shortawn foxtail) accessions in Japan. Front. Plant Sci. 2017, 8, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Küpper, A.; Peter, F.; Zöllner, P.; Lorentz, L.; Tranel, P.J.; Beffa, R.; Gaines, T.A. Tembotrione detoxification in 4-hydroxyphenylpyruvate dioxygenase (HPPD) inhibitor-resistant palmer amaranth (Amaranthus palmeri S. Wats.). Pest Manag. Sci. 2018, 74, 2325–2334. [Google Scholar] [CrossRef]
- Gaines, T.A.; Barker, A.L.; Patterson, E.L.; Westra, E.P.; Wilson, R.G. EPSPS gene copy number and whole-plant glyphosate resistance level in Kochia scoparia. PLoS ONE 2016, 11, e0168295. [Google Scholar] [CrossRef] [Green Version]
- Godar, A.S.; Stahlman, P.W.; Jugulam, M.; Dille, J.A. Glyphosate-resistant kochia (Kochia scoparia) in Kansas: EPSPS gene copy number in relation to resistance levels. Weed Sci. 2015, 63, 587–595. [Google Scholar] [CrossRef]
- Ngo, T.D.; Malone, J.M.; Boutsalis, P.; Gill, G.; Preston, C. EPSPS gene amplification conferring resistance to glyphosate in windmill grass (Chloris truncata) in Australia. Pest Manag. Sci. 2018, 74, 1101–1108. [Google Scholar] [CrossRef]
- Malone, J.M.; Morran, S.; Shirley, N.; Boutsalis, P.; Preston, C. EPSPS gene amplification in glyphosate-resistant Bromus diandrus. Pest Manag. Sci. 2016, 72, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, H.; Zhang, C.; Wei, S.; Huang, Z.; Chen, J.; Wang, X. Mutations and amplification of EPSPS gene confer resistance to glyphosate in goosegrass (Eleusine indica). Planta 2015, 242, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Laforest, M.; Soufiane, B.; Simard, M.-J.; Obeid, K.; Page, E.; Nurse, R.E. Acetyl-CoA carboxylase overexpression in herbicide-resistant large crabgrass (Digitaria sanguinalis). Pest Manag. Sci. 2017, 73, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Adu-Yeboah, P.; Malone, J.M.; Fleet, B.; Gill, G.; Preston, C. EPSPS gene amplification confers resistance to glyphosate resistant populations of Hordeum glaucum stued (northern barley grass) in South Australia. Pest Manag. Sci. 2020, 76, 1214–1221. [Google Scholar] [CrossRef]

| Gene 1 | Sequence (5′ → 3′) 2 | Amplicon (bp) | Slope | Efficiency (%) 3 |
|---|---|---|---|---|
| α-tubulin | F GAGAAGGTGGAGACGATGCAT R AATTCCCGGTCCGAATTTCGT | 123 | −3.352 | 98.8 |
| β-tubulin | F GCTGTCTTAGGTTCCCAGGTC R GCCATGAGAGGTTAGAGGAGC | 119 | −3.421 | 96.0 |
| CCR | F CGACGGAAAATAGCAACAAAGTG R GTCTTTGACGGTGGCGTTAAC | 116 | −3.355 | 98.6 |
| PPAN | F TGCTCCATTTTTGAGGGTTGC R GACATCGAGGCCTCAACTGTG | 113 | −3.3033 | 100.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Torralva, F.; Norsworthy, J.K. Understanding Resistance Mechanisms to Trifluralin in an Arkansas Palmer Amaranth Population. Genes 2021, 12, 1225. https://doi.org/10.3390/genes12081225
González-Torralva F, Norsworthy JK. Understanding Resistance Mechanisms to Trifluralin in an Arkansas Palmer Amaranth Population. Genes. 2021; 12(8):1225. https://doi.org/10.3390/genes12081225
Chicago/Turabian StyleGonzález-Torralva, Fidel, and Jason K. Norsworthy. 2021. "Understanding Resistance Mechanisms to Trifluralin in an Arkansas Palmer Amaranth Population" Genes 12, no. 8: 1225. https://doi.org/10.3390/genes12081225







