Impact of a Novel W2027L Mutation and Non-Target Site Resistance on Acetyl-CoA Carboxylase-Inhibiting Herbicides in a French Lolium multiflorum Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Seed Samples
2.2. Confirmation of Resistance to Clodinafop-Propargyl
2.3. Mechanism of Resistance Studies
2.3.1. Analysis of ACCase for Target Site Resistance Mutations
2.3.2. Production of Wild WW2027 and Mutant LL2027 Seed Batches to Evaluate the Importance of the W2027L Mutation and Non-Target Site Resistance on ACCase-Herbicide Efficacy
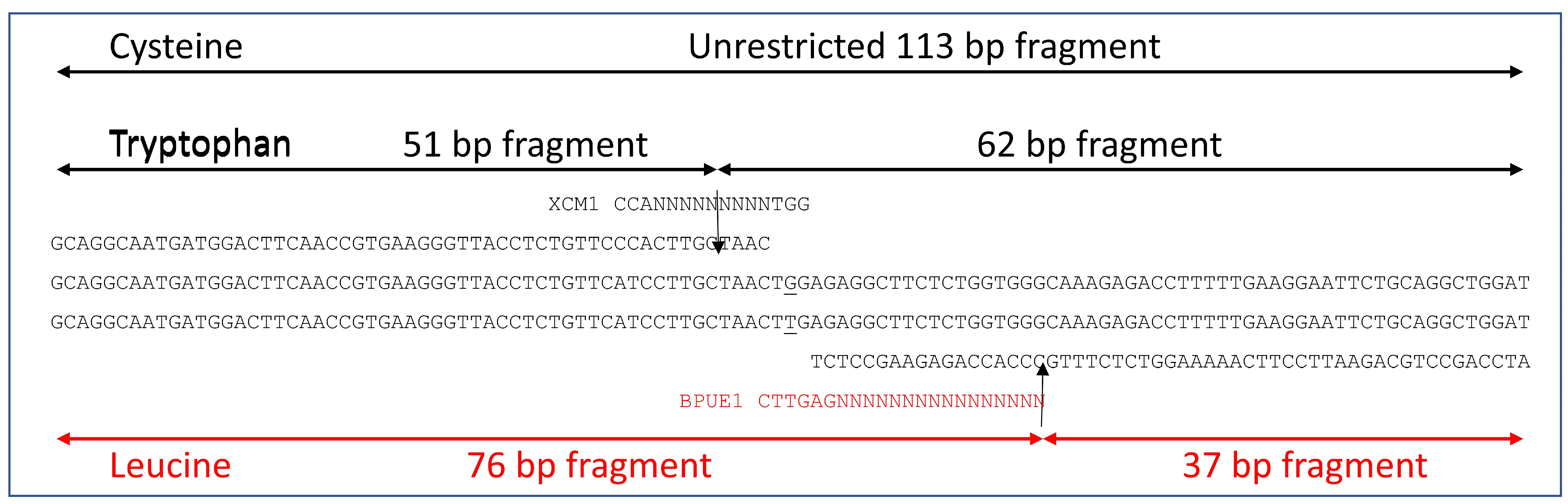
2.3.3. Development of a dPACS Assay for Genotyping the W2027L Mutation
Assay Design
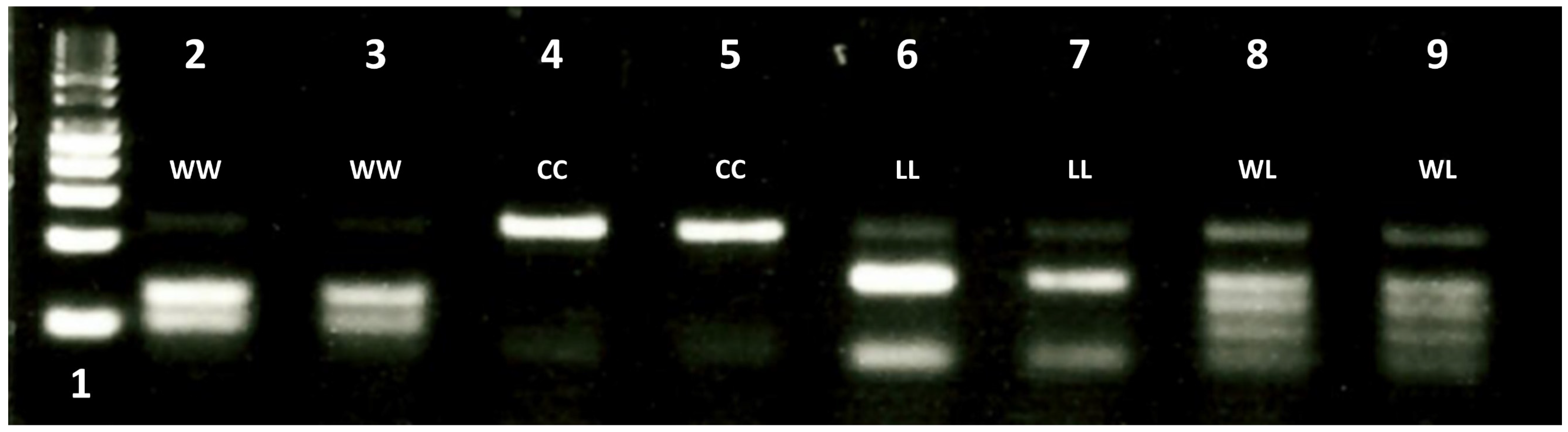
PCR-RFLP and Gel Electrophoresis Procedure
2.3.4. Co-Segregation Studies to Assess the Importance of the W2027L Target Site Mutation and NTSR on the Efficacy of Three ACCase Herbicides
2.3.5. Whole Plant Dose–Response Tests to Assess the Level of Resistance Conferred by the W2027L Mutation and NTSR
3. Results
3.1. Initial Clodinafop-Propargyl Resistance Confirmation Test
3.2. Mechanism of Resistance to Clodinafop-Propargyl
3.2.1. ACCase Analysis for Resistance Mutations
3.2.2. Development of a 2027-ACCase dPACS Assay
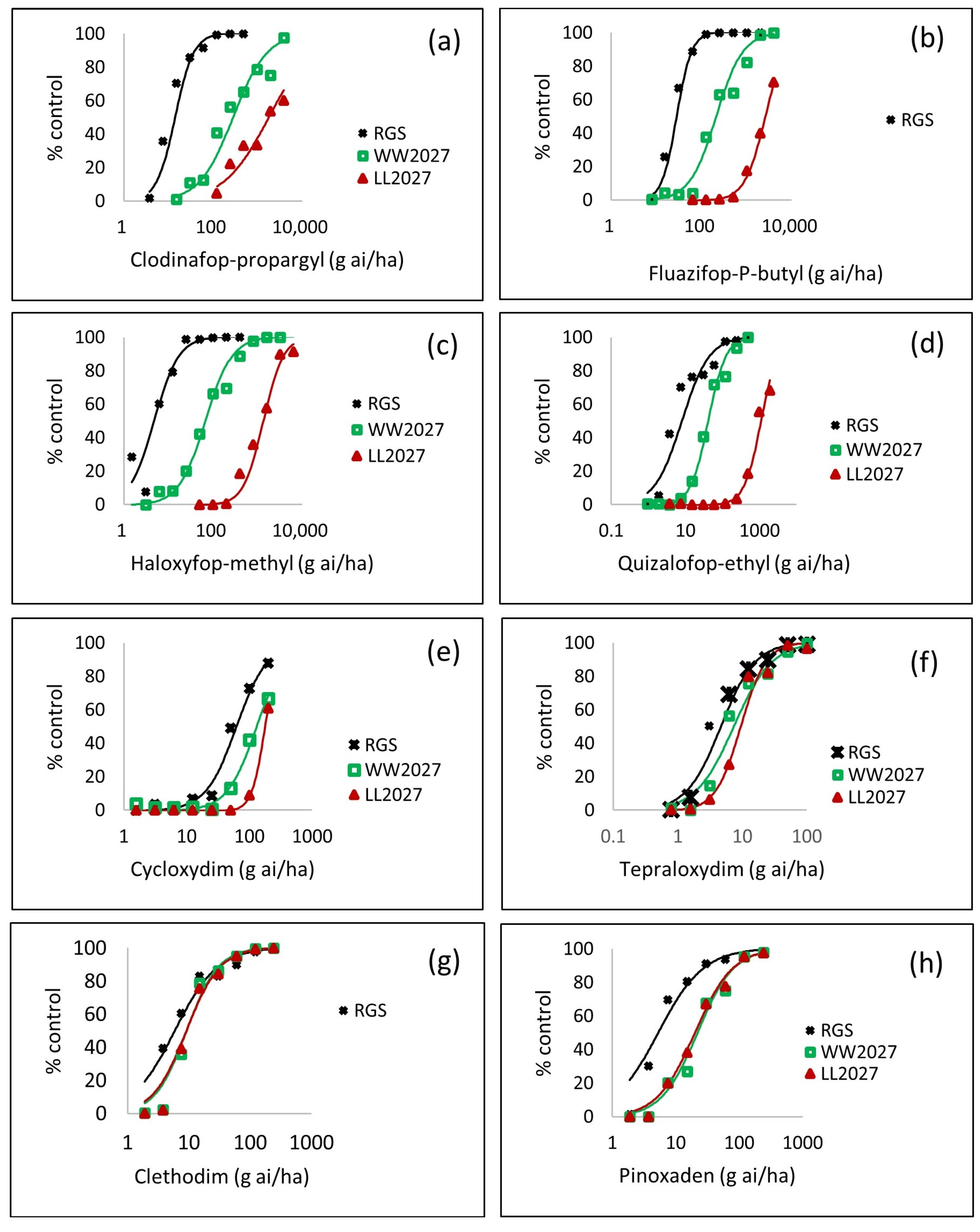
3.2.3. Impact of the W2027L Mutation and NTSR on Representative FOP, DIM, and DEN Herbicides
3.2.4. Level of Resistance Conferred by the W2027L Mutation and Non-Target Site Resistance
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beddows, A.R. Lolium multiflorum Lam. J. Ecol. 1973, 63, 587–600. [Google Scholar] [CrossRef]
- Fearon, C.; Hayward, M.; Lawrence, M. Self-incompatibility in ryegrass V. Genetic control, linkage and seed set in diploid Lolium multiflorum Lam. Heredity 1983, 50, 35–46. [Google Scholar] [CrossRef] [Green Version]
- Stewart, A.; Hayes, R. Ryegrass breeding—Balancing trait priorities. Ir. J. Agric. Food Res. 2011, 50, 31–46. [Google Scholar]
- Humphreys, M.; Feuerstein, U.; Vandewalle, M.; Baert, J. Ryegrasses. In Fodder Crops and Amenity Grasses; Boller, B., Posselt, U.K., Veronesi, F., Eds.; Springer: New York, NY, USA, 2010; pp. 211–260. [Google Scholar]
- Peter-Schmid, M.K.I.; Boller, B.; Kölliker, R. Habitat and management affect genetic structure of Festuca pratensis but not Lolium multiflorum ecotype populations. Plant Breed. 2008, 127, 510–517. [Google Scholar] [CrossRef]
- Vallano, D.M.; Selmants, P.C.; Zavaleta, E.S. Simulated nitrogen deposition enhances the performance of an exotic grass relative to native serpentine grassland competitors. Plant Ecol. 2012, 213, 1015–1026. [Google Scholar] [CrossRef]
- Alarcon-Reverte, R. Understanding and Combating the Threat Caused by Lolium multiflorum as a Weed of Arable Crops. Ph.D. Thesis, University of Reading, Reading, UK, 2009. [Google Scholar]
- Moss, S.R.; Hull, R.I.; Perryman, S.A.M.; Cussans, J.W. Lolium multiflorum: Aspects of herbicide resistance, agro-ecology and effects on crop yield in wheat crops. Asp. Appl. Biol. 2017, 134, 151–160. [Google Scholar]
- Schaeffer, A.H.; Schaeffer, O.A.; Silveira, D.C.; Bertol, J.A.G.; Rocha, D.K.; dos Santos, F.M.; Vargas, L.; Lângaro, N.C. Reduction of Ryegrass (Lolium multiflorum Lam.) Natural Re-Sowing with Herbicides and Plant Growth Regulators. Agronomy 2020, 10, 1960. [Google Scholar] [CrossRef]
- Wenger, J.; Niderman, T.; Mathews, C.; Wailes, S. Acetyl-CoA carboxylase inhibitors. In Modern Crop Protection Compounds; Jeschke, P., Witschel, M., Krämer, W., Schirmer, U., Eds.; Wiley-VCH Verlag GmbH & Co: Weinheim, Germany, 2020; pp. 501–528. [Google Scholar]
- Konishi, T.; Sasaki, Y. Compartmentalization of two forms of acetyl-CoA carboxylase in plants and the origin of their tolerance toward herbicides. Proc. Natl. Acad. Sci. USA 1994, 91, 3598–3601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolau, B.J.; Ohlrogge, J.B.; Wurtele, E.S. Plant biotin-containing carboxylases. Arch. Biochem. Biophys 2003, 414, 211–222. [Google Scholar] [CrossRef]
- Herbert, D.; Walker, K.A.; Price, L.J.; Cole, D.J.; Pallett, K.E.; Ridley, S.M.; Harwood, J.L. Acetyl-CoA carboxylase—a graminicide target site. Pestic. Sci. 1997, 50, 67–71. [Google Scholar] [CrossRef]
- Yu, L.P.C.; Kim, Y.S.; Tong, L. Mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by pinoxaden. Proc. Natl. Acad. Sci. USA 2010, 107, 22072–22077. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Tweel, B.; Tong, L. Molecular basis for the inhibition of the carboxyltransferase domain of acetyl-coenzyme-A carboxylase by haloxyfop and diclofop. Proc. Natl. Acad. Sci. USA 2004, 101, 5910–5915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, S.; Callaghan, M.; Watson, K.; Tong, L. A different mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by tepraloxydim. Proc. Natl. Acad. Sci. USA 2009, 106, 20723–20727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofer, U.; Muehlebach, M.; Hole, S.; Zoschke, A. Pinoxaden—For broad spectrum grass weed management in cereal crops. J. Plant Dis. Prot. 2006, 113, 989–995. [Google Scholar]
- Heap, I.M. International Survey of Herbicide-Resistant Weeds. 2021. Available online: http://www.weedscience.com (accessed on 18 November 2020).
- Powles, S.; Yu, Q. Evolution in action: Plants reistant to herbicides. Annu. Rev. Plant Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef] [Green Version]
- Gaines, T.; Duke, S.O.; Morran, S.; Rigon, C.A.G.; Tranel, P.J.; Küpper, A.; Dayan, F.E. Mechanisms of evolved herbicide resistance. J. Biol. Chem. 2020, 295, 10307–10330. [Google Scholar] [CrossRef] [PubMed]
- Kaundun, S.S. Resistance to acetyl-CoA carboxylase-inhibiting herbicides. Pest. Manag. Sci. 2014, 70, 1405–1417. [Google Scholar] [CrossRef]
- Jang, S.; Marjanovic, J.; Gornicki, P. Resistance to herbicides caused by single amino acid mutations in acetyl-CoA carboxylase in resistant populations of grassy weeds. New Phytol. 2013, 197, 1110–1116. [Google Scholar] [CrossRef]
- Kaundun, S.S.; Bailly, G.; Dale, R.; Hutchings, S.-J.; Mcindoe, E. A novel W1999S mutation and non-target site resistance impact on acetyl-CoA carboxylase inhibiting herbicides to varying degrees in a UK Lolium multiflorum population. PLoS ONE 2013, 8, e58012. [Google Scholar] [CrossRef] [Green Version]
- Jugulam, M.; Shyam, C. Non-target-site resistance to herbicides: Recent developments. Plants 2019, 8, 417. [Google Scholar] [CrossRef] [Green Version]
- Busi, R.; Vila-Aiub, M.M.; Powles, S.B. Genetic control of a cytochrome P450 metabolism-based herbicide resistance mechanism in Lolium rigidum. Heredity 2011, 106, 817–824. [Google Scholar] [CrossRef] [Green Version]
- Manalil, S.; Busi, R.; Renton, M.; Powles, S.B. Rapid evolution of herbicide resistance by low herbicide dosages. Weed Sci. 2011, 59, 210–217. [Google Scholar] [CrossRef]
- Yu, Q.; Han, H.; Cawthray, G.R.; Wang, S.F.; Powles, S.B. Enhanced rates of herbicide metabolism in low herbicide-dose selected resistant Lolium rigidum. Plant Cell Environ. 2013, 36, 818–827. [Google Scholar] [CrossRef] [PubMed]
- Iwakami, S.; Kamidate, Y.; Yamaguchi, T.; Ishizaka, M.; Endo, M.; Suda, H.; Nagai, K.; Sunohara, Y.; Toki, S.; Uchino, A.; et al. CYP81A P450s are involved in concomitant cross-resistance to acetolactate synthase and acetyl-CoA carboxylase herbicides in Echinochloa phyllopogon. New Phytol. 2019, 221, 2112–2122. [Google Scholar] [CrossRef] [PubMed]
- Kaundun, S.S. An aspartate to glycine change in the carboxyl transferase domain of acetyl CoA carboxylase is in part associated with resistance to ACCase inhibitor herbicides in a Lolium multiflorum population. Pest. Manag. Sci. 2010, 66, 1249–1256. [Google Scholar] [CrossRef]
- Kaundun, S.S.; Marchegiani, E.; Hutchings, S.-J.; Baker, K. Derived Polymorphic Amplified Cleaved Sequence (dPACS): A novel PCR-RFLP procedure for detecting known single nucleotide and deletion-insertion polymorphisms. Int. J. Mol. Sci. 2019, 20, 3193. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Zhang, L.; Wang, H.; Li, Q.; Liu, W.; Wang, J. A Rare Ile-2041-Thr mutation in the ACCase gene confers resistance to ACCase-inhibiting herbicides in shortawn foxtail (Alopecurus aequalis). Weed Sci. 2017, 65, 239–246. [Google Scholar] [CrossRef]
- Délye, C.; Zhang, X.Q.; Michel, S.; Matéjicek, A.; Powles, S.B. Molecular bases for sensitivity to acetyl-coenzyme A carboxylase inhibitors in black-grass. Plant Physiol. 2005, 137, 794–806. [Google Scholar] [CrossRef] [Green Version]
- Gherekhloo, J.; Osuna, M.D.; De Prado, R. Biochemical and molecular basis of resistance to ACCase-inhibiting herbicides in Iranian Phalaris minor populations. Weed Res. 2012, 52, 367–372. [Google Scholar] [CrossRef]
- Li, L.; Du, L.; Liu, W.; Yuan, G.; Wang, J. Target-site mechanism of ACCase-inhibitors resistance in American sloughgrass (Beckmannia syzigachne Steud.) from China. Pestic. Biochem. Physiol. 2014, 110, 57–62. [Google Scholar] [CrossRef]
- Osuna, M.D.; Goulart, I.C.G.R.; Vidal, R.A.; Kalsing, A.; Ruiz Santaella, J.P.; De Prado, R. Resistance to ACCase inhibitors in Eleusine indica from Brazil involves a target site mutation. Planta Daninha. 2012, 30, 675–681. [Google Scholar] [CrossRef] [Green Version]
- Menchari, Y.; Chauvel, B.; Darmency, H.; Delye, C. Fitness costs associated with three mutant acetyl-coenzyme A carboxylase alleles endowing herbicide resistance in black-grass Alopecurus myosuroides. J. Appl. Ecol. 2008, 45, 939–947. [Google Scholar] [CrossRef]
- Alarcon-Reverte, R.; Hanley, S.; Kaundun, S.S.; Karp, A.; Moss, S.R. A SNaPshot assay for the rapid and simple detection of known point mutations conferring resistance to ACCase-inhibiting herbicides in Lolium spp. Weed Res. 2013, 53, 12–20. [Google Scholar] [CrossRef]
- Yu, Q.; Collavo, A.; Zheng, M.-Q.; Owen, M.; Sattin, M.; Powles, S.B. Diversity of acetyl-coenzyme A carboxylase mutations in resistant Lolium populations: Evaluation using clethodim. Plant Physiol. 2007, 145, 547–558. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Devine, D. A possible point mutation in the plastidic ACCase gene conferring resistance to sethoxydim in green foxtail (Setaria viridis). In Proceedings of the Weed Science Society of America Meeting, Toronto, ON, Canada, 9 February 2000. Abstract 81. [Google Scholar]
- Délye, C.; Matéjicek, A.; Gasquez, J. PCR-based detection of resistance to acetyl-CoA carboxylase-inhibiting herbicides in black-grass (Alopecurus myosuroides Huds) and ryegrass (Lolium rigidum Gaud). Pest. Manag. Sci. 2002, 58, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Scarabel, L.; Panozzo, S.; Loddo, D.; Mathiassen, S.K.; Kristensen, M.; Kudsk, P.; Gitsopoulos, T.; Travlos, I.; Tani, E.; Chachalis, D.; et al. Diversified resistance mechanisms in multi-resistant Lolium spp. in three European countries. Front. Plant Sci. 2020, 11, 608845. [Google Scholar] [CrossRef]
- Beckie, H.J.; Warwick, S.I.; Sauder, C.A. Basis for herbicide resistance in Canadian populations of wild oat (Avena fatua). Weed Sci. 2012, 60, 10–18. [Google Scholar] [CrossRef]
- Cruz-Hipolito, H.; Fernandez, P.; Alcantara, R.; Gherekhloo, J.; Osuna, M.D.; De Prado, R. Ile-1781-Leu and Asp-2078-Gly mutations in ACCase gene, endow cross-resistance to APP, CHD, and PPZ in Phalaris minor from Mexico. Int. J. Mol. Sci. 2015, 16, 21363–21377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, W.; Cai, J.; Zhang, J.; Chen, Y.; Chen, Y.; Di, Y.; Yuan, S. Molecular basis of resistance to ACCase-inhibiting herbicide cyhalofop-butyl in Chinese sprangletop (Leptochloa chinensis (L.) Nees) from China. Pestic. Biochem. Phys. 2019, 158, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Huan, Z.-B.; Jin, T.; Zhang, S.-Y.; Wang, J.-X. Cloning and sequence analysis of plastid acetyl-CoA carboxylase cDNA from two Echinochloa crus-galli biotypes. J. Pestic. Sci. 2011, 36, 461–466. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; Qu, M.; Jiang, X.; Li, X.; Ju, Q.; Lu, X.; Wang, J. Fitness costs associated with acetyl-coenzyme A carboxylase mutations endowing herbicide resistance in American sloughgrass (Beckmannia syzigachne Steud.). Ecol. Evol. 2019, 9, 2220–2230. [Google Scholar] [CrossRef] [Green Version]
- Délye, C.; Michel, S. ‘Universal’ primers for PCR-sequencing of grass chloroplastic acetyl-CoA carboxylase domains involved in resistance to herbicides. Weed Res. 2005, 45, 323–330. [Google Scholar] [CrossRef]
- Herbert, D.; Cole, D.J.; Pallett, K.E.; Harwood, J.L. Susceptibilities of different test systems from maize (Zea mays), Poa annua, and Festuca rubra to herbicides that inhibit the enzyme acetyl-coenzyme A carboxylase. Pestic. Biochem. Phys. 1996, 55, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.-L.; Yang, W.-C.; Yu, N.-X.; Yang, S.-G.; Yang, G.-F. Computational simulations of structural role of the active-site W374C mutation of acetyl-coenzyme-A carboxylase: Multi-drug resistance mechanism. J. Mol. Model. 2011, 17, 495–503. [Google Scholar] [CrossRef]
- Yu, J.; Gao, H.; Pan, L.; Yao, Z.; Dong, L. Mechanism of resistance to cyhalofop-butyl in Chinese sprangletop (Leptochloa chinensis (L.) Nees). Pestic. Biochem. Phys. 2017, 143, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Délye, C.; Matéjicek, A.; Michel, S. Cross-resistance patterns to ACCase-inhibiting herbicides conferred by mutant ACCase isoforms in Alopecurus myosuroides Huds. (black-grass), re-examined at the recommended herbicide field rate. Pest. Manag. Sci. 2008, 64, 1179–1186. [Google Scholar] [CrossRef]
- Du, L.; Liu, W.; Yuan, G.; Guo, W.; Li, Q.; Wang, J. Cross-resistance patterns to ACCase-inhibitors in American sloughgrass (Beckmannia syzigachne Steud.) homozygous for specific ACCase mutations. Pestic. Biochem. Phys. 2016, 126, 42–48. [Google Scholar] [CrossRef]
- Yuan, G.; Tian, Z.; Li, T.; Qian, Z.; Guo, W.; Shen, G. Cross-resistance pattern to ACCase-inhibiting herbicides in a rare Trp-2027-Ser mutation Chinese sprangletop (Leptochloa chinensis) population. Chil. J. Agric. Res. 2021, 81, 62–69. [Google Scholar] [CrossRef]
- Delye, C.; Pernin, F.; Michel, S. ‘Universal’ PCR assays detecting mutations in acetyl-coenzyme A carboxylase or acetolactate synthase that endow herbicide resistance in grass weeds. Weed Res. 2011, 51, 353–362. [Google Scholar] [CrossRef]
- Murphy, B.P.; Tranel, P.J. Target-site mutations conferring herbicide resistance. Plants 2019, 8, 382. [Google Scholar] [CrossRef] [Green Version]
- Kaundun, S.S.; Hutchings, S.-J.; Dale, R.P.; McIndoe, E. Broad resistance to ACCase inhibiting herbicides in a ryegrass population is due only to a cysteine to arginine mutation in the target enzyme. PLoS ONE 2012, 7, e39759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petit, C.; Bay, G.; Pernin, F.; Délye, C. Prevalence of cross- or multiple resistance to the acetyl-coenzyme A carboxylase inhibitors fenoxaprop, clodinafop and pinoxaden in black-grass (Alopecurus myosuroides Huds.) in France. Pest. Manag. Sci. 2010, 66, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Hull, R.; Tatnell, L.; Cook, S.; Beffa, R.; Moss, S. Current status of herbicide-resistant weeds in the UK. In Crop Production in Southern Britain, Precision Decisions for Profitable Cropping; Association of Applied Biologists (AAB): Wellesbourne Warwick, UK, 2014; Volume 127, pp. 261–272. [Google Scholar]
- Han, H.; Yu, Q.; Beffa, R.; González, S.; Maiwald, F.; Wang, J.; Powles, S.B. Cytochrome P450 CYP81A10v7 in Lolium rigidum confers metabolic resistance to herbicides across at least five modes of action. Plant J. 2021, 105, 79–92. [Google Scholar] [CrossRef] [PubMed]
- White, G.M.; Moss, S.R.; Karp, A. Differences in the molecular basis of resistance to the cyclohexanedione herbicide sethoxydim in Lolium multiflorum. Weed Res. 2005, 45, 440–448. [Google Scholar] [CrossRef]
- Comont, D.; Lowe, C.; Hull, R.; Crook, L.; Hicks, H.L.; Onkokesung, N.; Beffa, R.; Childs, D.Z.; Edwards, R.; Freckleton, R.P.; et al. Evolution of generalist resistance to herbicide mixtures reveals a trade-off in resistance management. Nat. Commun. 2020, 11, 3086. [Google Scholar] [CrossRef] [PubMed]

| Herbicide | Rates Applied in g ai/ha |
|---|---|
| Clodinafop-propargyl | 3.8, 7.5, 15, 30, 60, 120, 240, 480 |
| Diclofop-methyl | 15, 31.25, 62.5, 125, 250, 500, 1000, 2000, 4000, 8000, 16,000 |
| Fluazifop-P-butyl | 7.8, 15, 31,25, 62.5, 125, 250, 500, 1000 |
| Haloxyfop-methyl | 1.5, 3.125, 6.25, 12.5, 25, 50, 100, 200, 400 |
| Quizalofop-ethyl | 0.93, 1.875, 3.75, 7.5, 15, 30, 60, 120 |
| Cycloxydim | 3.125, 6.25, 12.5, 25, 50, 100, 200, 400 |
| Tepraloxydim | 6.25, 12.5, 25, 50, 100, 200, 400, 800 |
| Clethodim | 1.9, 3.8, 7.5, 15, 30, 60, 120, 240 |
| Pinoxaden | 1.875, 3.75, 7.5, 15, 30, 60, 120, 240 |
| Herbicide | Rate (g/ha) | Comparison | No. of Survivors/Total | p-Value (2-Sided) | |
|---|---|---|---|---|---|
| Genotype 1 | Genotype 2 | ||||
| Fluazifop-P-butyl | 200 | WW2027 v WL2027 | 18/24 | 51/53 | 0.0097 |
| WW2027 v LL2027 | 18/24 | 19/19 | 0.0265 | ||
| WL2027 v LL2027 | 51/53 | 19/19 | 1 | ||
| RGS v WW2027 | 0/24 | 18/24 | <0.00001 | ||
| Cycloxydim | 200 | WW2027 v WL2027 | 4/19 | 13/55 | 1 |
| WW2027 v LL2027 | 4/19 | 8/22 | 0.3246 | ||
| WL2027 v LL2027 | 13/55 | 8/22 | 0.2712 | ||
| RGS v WW2027 | 0/24 | 4/19 | 0.0314 | ||
| Pinoxaden | 60 | WW2027 v WL2027 | 8/24 | 25/48 | 0.2093 |
| WW2027 v LL2027 | 8/24 | 4/24 | 0.3177 | ||
| WL2027 v LL2027 | 25/48 | 4/24 | 0.0049 | ||
| RGS v WW2027 | 0/24 | 8/24 | 0.0039 | ||
| Herbicide | WW2027 vs. RGS | LL2027 vs. RGS | LL2027 vs. WW2027 |
|---|---|---|---|
| Clodinafop-propargyl | 19.9 (13.6–29.3) | 125.7 (62.8–251.6) | 6.3 (3.0–13.1) |
| Diclofop-methyl | 37.6 (22.6–62.5) | >121.2 | >3.2 |
| Fluazifop-P-butyl | 7.5 (5.6–10.1) | 92.7 (63.0–136.6) | 12.3 (8.1–18.6) |
| Haloxyfop-methyl | 14.0 (9.5–20.6) | 279.6 (191.3–408.5) | 20.0 (14.5–27.5) |
| Quizalofop-ethyl | 5.13 (3.30–7.97) | 137.4 (83.5–226.0) | 26.8 (18.0–39.9) |
| Cycloxydim | 2.09 (1.31–3.33) | 2.84 (2.00–4.03) | 1.36 (0.88–2.08) |
| Tepraloxydim | 1.58 (1.00–2.51) | 2.03 (1.41–2.92) | 1.28 (0.85–1.95) |
| Clethodim | 1.56 (0.82–2.95) | 1.54 (0.80–2.97) | 0.99 (0.54–1.81) |
| Pinoxaden | 4.17 (2.20–7.91) | 3.80 (1.98–7.31) | 0.91 (0.57–1.46) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaundun, S.S.; Downes, J.; Jackson, L.V.; Hutchings, S.-J.; Mcindoe, E. Impact of a Novel W2027L Mutation and Non-Target Site Resistance on Acetyl-CoA Carboxylase-Inhibiting Herbicides in a French Lolium multiflorum Population. Genes 2021, 12, 1838. https://doi.org/10.3390/genes12111838
Kaundun SS, Downes J, Jackson LV, Hutchings S-J, Mcindoe E. Impact of a Novel W2027L Mutation and Non-Target Site Resistance on Acetyl-CoA Carboxylase-Inhibiting Herbicides in a French Lolium multiflorum Population. Genes. 2021; 12(11):1838. https://doi.org/10.3390/genes12111838
Chicago/Turabian StyleKaundun, Shiv Shankhar, Joe Downes, Lucy Victoria Jackson, Sarah-Jane Hutchings, and Eddie Mcindoe. 2021. "Impact of a Novel W2027L Mutation and Non-Target Site Resistance on Acetyl-CoA Carboxylase-Inhibiting Herbicides in a French Lolium multiflorum Population" Genes 12, no. 11: 1838. https://doi.org/10.3390/genes12111838
APA StyleKaundun, S. S., Downes, J., Jackson, L. V., Hutchings, S.-J., & Mcindoe, E. (2021). Impact of a Novel W2027L Mutation and Non-Target Site Resistance on Acetyl-CoA Carboxylase-Inhibiting Herbicides in a French Lolium multiflorum Population. Genes, 12(11), 1838. https://doi.org/10.3390/genes12111838






