Advances in Gene Editing of Haploid Tissues in Crops
Abstract
:1. Introduction
2. Application of Doubled Haploid Technology in Modern Breeding and Genetic Analysis
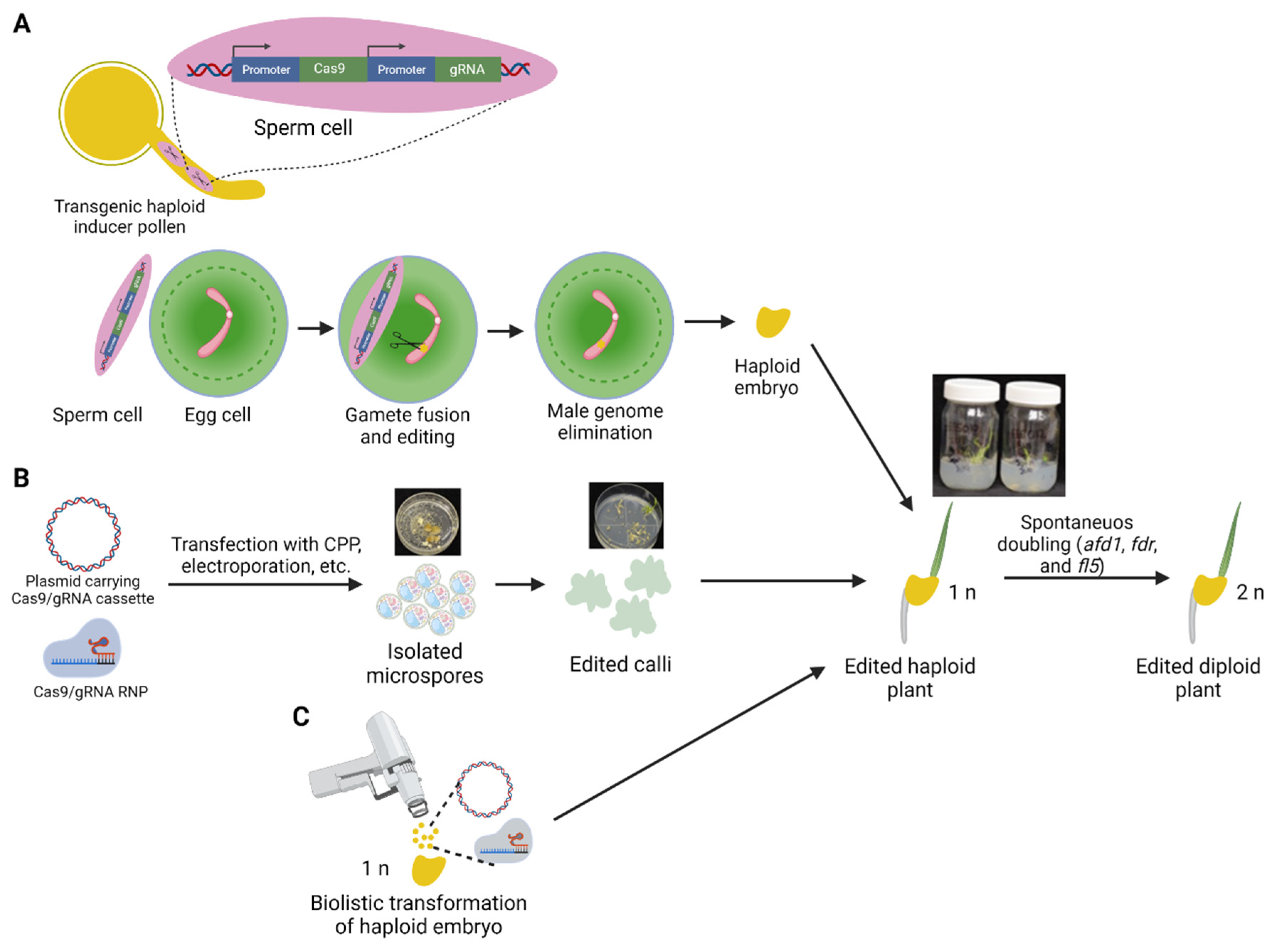
3. Haploid Induction in Crops through Gene Editing
4. Gene Editing in Haploid Cells
5. Haploid Microspores as a Potential Target for Gene Editing
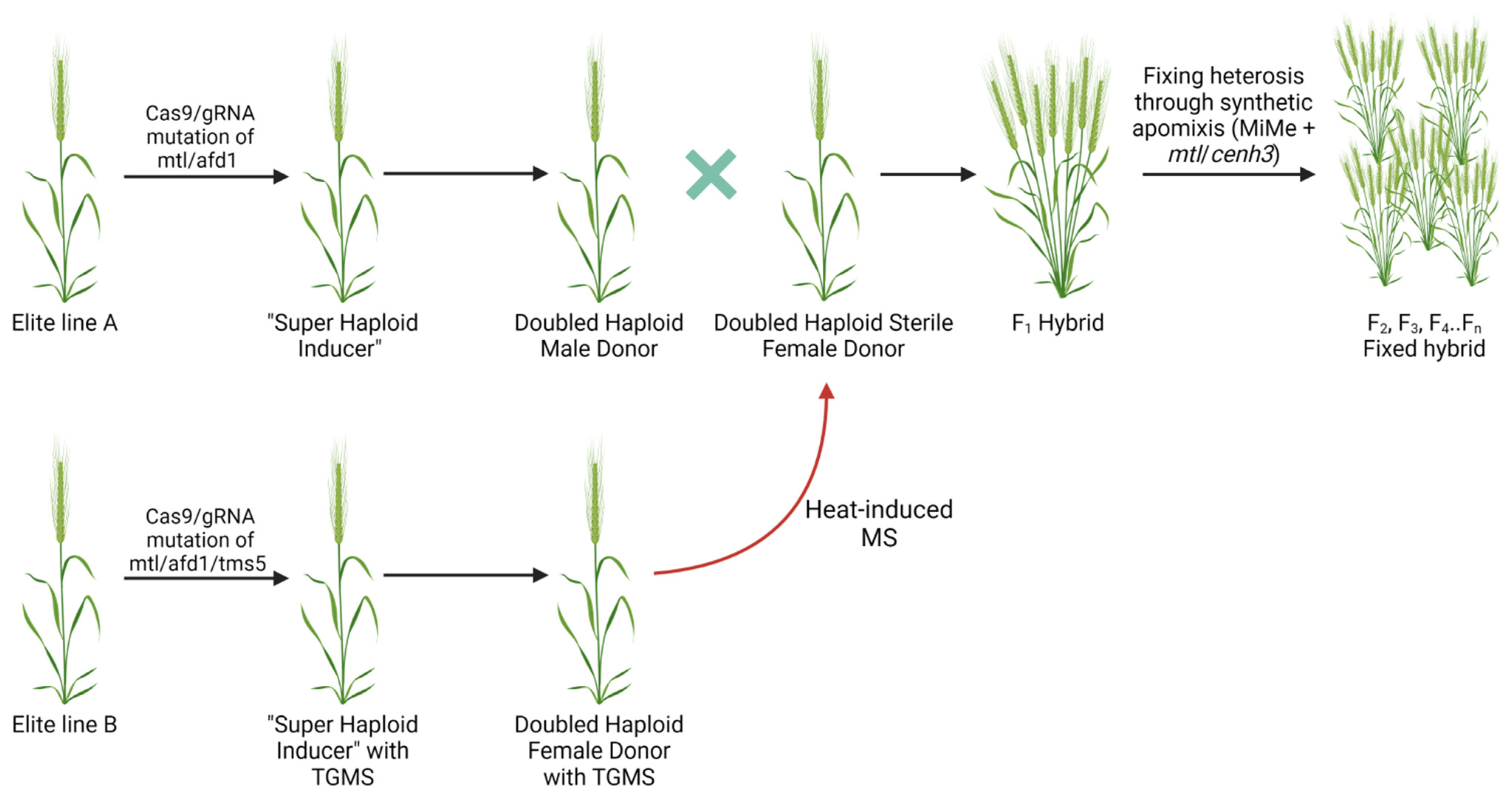
6. Application of Haploid Engineering for Hybrid Crop Production
7. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, H.; Li, C.; Gao, C. Publisher Correction: Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef]
- Schaart, J.G.; van de Wiel, C.C.M.; Smulders, M.J.M. Genome editing of polyploid crops: Prospects, achievements and bottlenecks. Transgenic Res. 2021, 30, 337–351. [Google Scholar] [CrossRef]
- Peng, T.; Sun, X.; Mumm, R.H. Optimized breeding strategies for multiple trait integration: I. Minimizing linkage drag in single event introgression. Mol. Breed. 2014, 33, 89–104. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Liu, C.; Qi, X.; Wu, Y.; Fei, X.; Mao, L.; Cheng, B.; Li, X.; Xie, C. RNA-guided Cas9 as an in vivo desired-target mutator in maize. Plant Biotechnol. J. 2017, 15, 1566–1576. [Google Scholar] [CrossRef] [Green Version]
- Kasha, K.J. Chromosome Doubling and Recovery of Doubled Haploid Plants. In Haploids in Crop Improvement II; Don Palmer, C.E., Keller, W.A., Kasha, K.J., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 123–152. [Google Scholar]
- Ren, J.; Wu, P.; Trampe, B.; Tian, X.; Lübberstedt, T.; Chen, S. Novel technologies in doubled haploid line development. Plant Biotechnol. J. 2017, 15, 1361–1370. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Li, S.; Abdelkhalik, S.; Shahzad, A.; Gu, J.; Yang, Z.; Ding, M.; Liu, K.; Zhao, H.; Yang, M. Development of thermo-photo sensitive genic male sterile lines in wheat using doubled haploid breeding. BMC Plant Biol. 2020, 20, 246. [Google Scholar] [CrossRef]
- Soriano, M.; Li, H.; Boutilier, K. Microspore embryogenesis: Establishment of embryo identity and pattern in culture. Plant Reprod. 2013, 26, 181–196. [Google Scholar] [CrossRef] [Green Version]
- Forster, B.P.; Thomas, W.T.B. Doubled Haploids in Genetics and Plant Breeding. Plant Breed. Rev. 2010, 25, 57–88. [Google Scholar]
- Blakeslee, A.F.; Belling, J.; Farnham, M.E.; Bergner, A.D. A haploid mutant in the jimson weed, “Datura Stramonium”. Science 1922, 55, 646–647. [Google Scholar] [CrossRef] [Green Version]
- Clausen, R.E.; Mann, M.C. Inheritance in Nicotiana Tabacum: V. The Occurrence of Haploid Plants in Interspecific Progenies. Proc. Natl. Acad. Sci. USA 1924, 10, 121–124. [Google Scholar] [CrossRef] [Green Version]
- Gaines, E.F.; Aase, H.C. A Haploid Wheat Plant. Am. J. Bot. 1926, 13, 373–385. [Google Scholar] [CrossRef]
- Comai, L.; Tan, E.H. Haploid Induction and Genome Instability. Trends Genet. 2019, 35, 791–803. [Google Scholar] [CrossRef]
- Kasha, K.J.; Maluszynski, M. Production of doubled haploids in crop plants. An introduction. In Doubled Haploid Production in Crop Plants; Maluszynski, M., Kasha, K., Forster, B.P., Szarejko, I., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 1–4. [Google Scholar]
- Touraev, A.; Forster, B.P.; Jain, S.M. Advances in Haploid Production in Higher Plants; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Vijverberg, K.; Ozias-Akins, P.; Schranz, M.E. Identifying and Engineering Genes for Parthenogenesis in Plants. Front. Plant Sci. 2019, 10, 128. [Google Scholar] [CrossRef] [Green Version]
- Conner, J.; Ozias-Akins, P.; Schmidt, A. Apomixis: Engineering the Ability to Harness Hybrid Vigor in Crop Plants. Methods Mol. Biol. 2017, 1669, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.A.; Mookkan, M.; Huo, H.; Chae, K.; Ozias-Akins, P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. USA 2015, 112, 11205–11210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, K.M.; Jones, G.E. Mingo barley. Can. J. Plant Sci. 1980, 60, 279–280. [Google Scholar] [CrossRef]
- Forster, B.P.; Heberle-Bors, E.; Kasha, K.J.; Touraev, A. The resurgence of haploids in higher plants. Trends Plant Sci. 2007, 12, 368–375. [Google Scholar] [CrossRef]
- Kalinowska, K.; Chamas, S.; Unkel, K.; Demidov, D.; Lermontova, I.; Dresselhaus, T.; Kumlehn, J.; Dunemann, F.; Houben, A. State-of-the-art and novel developments of in vivo haploid technologies. Theor. Appl. Genet. 2019, 132, 593–605. [Google Scholar] [CrossRef] [Green Version]
- Maluszynski, M.; Kasha, K.J.; Szarejko, I. Published doubled haploid protocols in plant species. In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 309–335. [Google Scholar]
- Touraev, A.; Pfosser, M.; Heberle-Bors, E. The microspore: A haploid multipurpose cell. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2001; pp. 53–109. [Google Scholar]
- Bilichak, A.; Sastry-Dent, L.; Sriram, S.; Simpson, M.; Samuel, P.; Webb, S.; Jiang, F.; Eudes, F. Genome editing in wheat microspores and haploid embryos mediated by delivery of ZFN proteins and cell-penetrating peptide complexes. Plant Biotechnol. J. 2019, 18, 1307–1316. [Google Scholar] [CrossRef]
- Bhowmik, P.; Ellison, E.; Polley, B.; Bollina, V.; Kulkarni, M.; Ghanbarnia, K.; Song, H.; Gao, C.; Voytas, D.; Kagale, S. Targeted mutagenesis in wheat microspores using CRISPR/Cas9. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Shariatpanahi, M.E.; Bal, U.; Heberle-Bors, E.; Touraev, A. Stresses applied for the re-programming of plant microspores towards in vitro embryogenesis. Physiol. Plant. 2006, 127, 519–534. [Google Scholar] [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.-M.; Van Lammeren, A.A.M.; Miki, B.L.A.; et al. Ectopic Expression of BABY BOOM Triggers a Conversion from Vegetative to Embryonic Growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [Green Version]
- Gordon-Kamm, B.; Sardesai, N.; Arling, M.; Lowe, K.; Hoerster, G.; Betts, S.; Jones, A.T. Using Morphogenic Genes to Improve Recovery and Regeneration of Transgenic Plants. Plants 2019, 8, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidmann, I.; De Lange, B.; Lambalk, J.; Angenent, G.C.; Boutilier, K. Efficient sweet pepper transformation mediated by the BABY BOOM transcription factor. Plant Cell Rep. 2011, 30, 1107–1115. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Y.; Liu, C.; Qi, X.; Jiao, Y.; Wang, D.; Wang, Y.; Liu, Z.; Chen, C.; Chen, B.; Tian, X.; et al. Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 2019, 5, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.M. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development. Plant Cell 2007, 19, 46–62. [Google Scholar] [CrossRef] [Green Version]
- Husbands, A.; Bell, E.M.; Shuai, B.; Smith, H.; Springer, P.S. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007, 35, 6663–6671. [Google Scholar] [CrossRef]
- Coe, E.H. A Line of Maize with High Haploid Frequency. Am. Nat. 1959, 93, 381–382. [Google Scholar] [CrossRef]
- Hu, H.; Schrag, T.A.; Peis, R.; Unterseer, S.; Schipprack, W.; Chen, S.; Lai, J.; Yan, J.; Prasanna, B.M.; Nair, S.; et al. The Genetic Basis of Haploid Induction in Maize Identified with a Novel Genome-Wide Association Method. Genetics 2016, 202, 1267–1276. [Google Scholar] [CrossRef] [Green Version]
- Prigge, V.; Xu, X.; Li, L.; Babu, R.; Chen, S.; Atlin, G.N.; Melchinger, A.E. New Insights into the Genetics of in Vivo Induction of Maternal Haploids, the Backbone of Doubled Haploid Technology in Maize. Genetics 2012, 190, 781–793. [Google Scholar] [CrossRef] [Green Version]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.; McCuiston, J.; Wang, W.; et al. Matrilineal, a sperm-specific phospholipase, triggers maize haploid induction. Nat. Cell Biol. 2017, 542, 105–109. [Google Scholar] [CrossRef]
- Liu, C.; Li, X.; Meng, D.; Zhong, Y.; Chen, C.; Dong, X.; Xu, X.; Chen, B.; Li, W.; Li, L.; et al. A 4-bp Insertion at ZmPLA1 Encoding a Putative Phospholipase A Generates Haploid Induction in Maize. Mol. Plant 2017, 10, 520–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilles, L.M.; Khaled, A.; Laffaire, J.; Chaignon, S.; Gendrot, G.; Laplaige, J.; Bergès, H.; Beydon, G.; Bayle, V.; Barret, P.; et al. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 2017, 36, 707–717. [Google Scholar] [CrossRef] [Green Version]
- Kasha, K.J.; Kao, K.N. High Frequency Haploid Production in Barley (Hordeum vulgare L.). Nat. Cell Biol. 1970, 225, 874–876. [Google Scholar] [CrossRef]
- Devaux, P.; Pickering, R. Haploids in the Improvement of Poaceae. In Haploids in Crop Improvement II; Don Palmer, C.E., Keller, W.A., Kasha, K.J., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 215–242. [Google Scholar]
- Mochida, K.; Tsujimoto, H. Production of Wheat Doubled Haploids by Pollination with Job’s Tears (Coix lachryma-jobi L.). J. Hered. 2001, 92, 81–83. [Google Scholar] [CrossRef]
- Niroula, R.K.; Bimb, H.P. Overview of wheat × maize system of crosses for dihaploid induction in wheat. World Appl. Sci. J. 2009, 7, 1037–1045. [Google Scholar]
- Riera-Lizarazu, O.; Mujeeb-Kazi, A. Polyhaploid production in the Triticeae: Wheat× Tripsacum crosses. Crop Sci. 1993, 33, 973–976. [Google Scholar] [CrossRef]
- Wędzony, M. Protocol for doubled haploid production in hexaploid triticale (x Triticosecale Wittm.) by crosses with maize. In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherland, 2003; pp. 135–140. [Google Scholar]
- Marcińska, I.; Czyczyło-Mysza, I.; Skrzypek, E.; Warchoł, M.; Zieliński, K.; Dubas, E. Obtaining of winter rye (Secale cereale L. ssp. cereale) haploid embryos through hybridization with maize (Zea Mays L.). Cereal Res. Commun. 2018, 46, 521–532. [Google Scholar] [CrossRef] [Green Version]
- Rines, H.W. Oat haploids from wide hybridization. In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherland, 2003; pp. 155–159. [Google Scholar]
- De Maine, M.J. Potato haploid technologies. In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 241–247. [Google Scholar]
- Michalik, B.; Adamus, A.; Nowak, E. Gynogenesis in Polish Onion Cultivars. J. Plant Physiol. 2000, 156, 211–216. [Google Scholar] [CrossRef]
- Weich, E.W.; Levall, M.W. Doubled haploid production of sugar beet (Beta vulgaris L.). In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 255–263. [Google Scholar]
- Von Aderkas, P.; Dawkins, M.D. Haploid embryogenesis in trees. In Biosafety of Forest Transgenic Trees; Springer Science and Business Media LLC: Heidelberg, Germany, 1993; Volume 41, pp. 57–65. [Google Scholar]
- Kumlehn, J. Embryogenesis and Plant Regeneration from Isolated Wheat Zygotes. In In Vitro Embryogenesis in Higher Plants; Germana, M.A., Lambardi, M., Eds.; Springer: New York, NY, USA, 2016; pp. 503–514. [Google Scholar]
- Mayor, P.J.; Bernardo, R. Genomewide Selection and Marker-Assisted Recurrent Selection in Doubled Haploid versus F2 Populations. Crop. Sci. 2009, 49, 1719–1725. [Google Scholar] [CrossRef]
- Chen, G.; Zhou, Y.; Kishchenko, O.; Stepanenko, A.; Jatayev, S.; Zhang, D.; Borisjuk, N. Gene editing to facilitate hybrid crop production. Biotechnol. Adv. 2021, 46, 107676. [Google Scholar] [CrossRef]
- Dirks, R.; Van Dun, K.; De Snoo, C.B.; Berg, M.V.D.; Lelivelt, C.L.C.; Voermans, W.; Woudenberg, L.; De Wit, J.P.C.; Reinink, K.; Schut, J.W.; et al. Reverse breeding: A novel breeding approach based on engineered meiosis. Plant Biotechnol. J. 2009, 7, 837–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijnker, E.; Van Dun, K.; De Snoo, C.B.; Lelivelt, C.L.C.; Keurentjes, J.; Naharudin, N.S.; Ravi, M.; Chan, S.W.L.; De Jong, H.; Dirks, R. Reverse breeding in Arabidopsis thaliana generates homozygous parental lines from a heterozygous plant. Nat. Genet. 2012, 44, 467–470. [Google Scholar] [CrossRef]
- Wang, N.; Gent, J.I.; Dawe, R.K. Haploid induction by a maize cenh3 null mutant. Sci. Adv. 2021, 7, eabe2299. [Google Scholar] [CrossRef]
- Liu, C.; Zhong, Y.; Qi, X.; Chen, M.; Liu, Z.; Chen, C.; Tian, X.; Li, J.; Jiao, Y.; Wang, D.; et al. Extension of the in vivo haploid induction system from diploid maize to hexaploid wheat. Plant Biotechnol. J. 2019, 18, 316–318. [Google Scholar] [CrossRef] [Green Version]
- Yao, L.; Zhang, Y.; Liu, C.; Liu, Y.; Wang, Y.; Liang, D.; Liu, J.; Sahoo, G.; Kelliher, T. OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants 2018, 4, 530–533. [Google Scholar] [CrossRef]
- Tuvesson, S.; Dayteg, C.; Hagberg, P.; Manninen, O.; Tanhuanpää, P.; Tenhola-Roininen, T.; Kiviharju, E.; Weyen, J.; Förster, J.; Schondelmaier, J.; et al. Molecular markers and doubled haploids in European plant breeding programmes. Euphytica 2007, 158, 305–312. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef] [Green Version]
- McCallum, C.M.; Comai, L.; Greene, E.A.; Henikoff, S. Targeted screening for induced mutations. Nat. Biotechnol. 2000, 18, 455–457. [Google Scholar] [CrossRef] [PubMed]
- Szarejko, I.; Forster, B.P. Doubled haploidy and induced mutation. Euphytica 2006, 158, 359–370. [Google Scholar] [CrossRef]
- Sanei, M.; Pickering, R.; Kumke, K.; Nasuda, S.; Houben, A. Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc. Natl. Acad. Sci. USA 2011, 108, E498–E505. [Google Scholar] [CrossRef] [Green Version]
- Ravi, M.; Chan, S.W.L. Haploid plants produced by centromere-mediated genome elimination. Nat. Cell Biol. 2010, 464, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Kuppu, S.; Ron, M.; Marimuthu, M.P.; Li, G.; Huddleson, A.; Siddeek, M.H.; Terry, J.; Buchner, R.; Shabek, N.; Comai, L.; et al. A variety of changes, including CRISPR/Cas9-mediated deletions, in CENH3 lead to haploid induction on outcrossing. Plant Biotechnol. J. 2020, 18, 2068–2080. [Google Scholar] [CrossRef]
- Wang, N.; Dawe, R.K. Centromere Size and Its Relationship to Haploid Formation in Plants. Mol. Plant 2018, 11, 398–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Wu, Y.; Zhang, W.; Dawe, R.K.; Jiang, J. Maize centromeres expand and adopt a uniform size in the genetic background of oat. Genome Res. 2014, 24, 107–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, J.; Yu, K.; Wei, J.; Gui, H.; Liu, C.; Liang, D.; Wang, Y.; Zhou, H.; Carlin, R.; Rich, R.; et al. Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3. Nat. Biotechnol. 2020, 38, 1397–1401. [Google Scholar] [CrossRef]
- Liu, C.; Li, W.; Zhong, Y.; Dong, X.; Hu, H.; Tian, X.; Wang, L.; Chen, B.; Chen, C.; Melchinger, A.E.; et al. Fine mapping of qhir8 affecting in vivo haploid induction in maize. Theor. Appl. Genet. 2015, 128, 2507–2515. [Google Scholar] [CrossRef]
- Li, X.; Meng, D.; Chen, S.; Luo, H.; Zhang, Q.; Jin, W.; Yan, J. Single nucleus sequencing reveals spermatid chromosome fragmentation as a possible cause of maize haploid induction. Nat. Commun. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Zhao, X.; Xu, X.; Xie, H.; Chen, S.; Jin, W. Fertilization and Uniparental Chromosome Elimination during Crosses with Maize Haploid Inducers. Plant Physiol. 2013, 163, 721–731. [Google Scholar] [CrossRef] [Green Version]
- Qiu, F.; Liang, Y.; Li, Y.; Liu, Y.; Wang, L.; Zheng, Y. Morphological, cellular and molecular evidences of chromosome random elimination in vivo upon haploid induction in maize. Curr. Plant Biol. 2014, 1, 83–90. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.; Li, L.; Liu, C.; Liu, C.; Geng, S.; Li, X.; Huang, C.; Mao, L.; Chen, S.; Xie, C. Genome Editing and Double-Fluorescence Proteins Enable Robust Maternal Haploid Induction and Identification in Maize. Mol. Plant 2018, 11, 1214–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Wang, K.; Jia, Z.; Gong, Q.; Lin, Z.; Du, L.; Pei, X.; Ye, X. Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 2020, 71, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Chen, B.; Li, M.; Wang, D.; Jiao, Y.; Qi, X.; Wang, M.; Liu, Z.; Chen, C.; Wang, Y.; et al. A DMP-triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis. Nat. Plants 2020, 6, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Mori, T.; Ueda, K.; Yamada, L.; Nagahara, S.; Higashiyama, T.; Sawada, H.; Igawa, T. The male gamete membrane protein DMP9/DAU2 is required for double fertilization in flowering plants. Development 2018, 145, dev170076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cyprys, P.; Lindemeier, M.; Sprunck, S. Gamete fusion is facilitated by two sperm cell-expressed DUF679 membrane proteins. Nat. Plants 2019, 5, 253–257. [Google Scholar] [CrossRef]
- Stajič, E.; Kiełkowska, A.; Murovec, J.; Bohanec, B. Deep sequencing analysis of CRISPR/Cas9 induced mutations by two delivery methods in target model genes and the CENH3 region of red cabbage (Brassica oleracea var. capitata f. rubra). Plant Cell Tissue Organ Cult. 2019, 139, 227–235. [Google Scholar] [CrossRef]
- Van Dun, C.M.P.; Lelivelt, C.L.C.; Movahedi, S. Non-Transgenic Haploid Inducer Lines in Cucurbits. 2018. Patent WO/2017/081009, 18 May 2017. [Google Scholar]
- Dunemann, F.; Unkel, K.; Sprink, T. Using CRISPR/Cas9 to Produce Haploid Inducers of Carrot through Targeted Mutations of Centromerichistone H3 (CENH3). Acta Hortic. 2019, 1264, 211–220. [Google Scholar] [CrossRef]
- Karimi-Ashtiyani, R.; Ishii, T.; Niessen, M.; Stein, N.; Heckmann, S.; Gurushidze, M.; Banaei-Moghaddam, A.M.; Fuchs, J.; Schubert, V.; Koch, K.; et al. Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc. Natl. Acad. Sci. USA 2015, 112, 11211–11216. [Google Scholar] [CrossRef] [Green Version]
- Den Camp, R.H.M.O.; van Dijk, P.J.; Gallard, A. Method for the Production of Haploid and Subsequent Doubled Haploid Plants. 2020. Patent WO2017058023A1, 6 April 2017. [Google Scholar]
- Che, P.; Anand, A.; Wu, E.; Sander, J.D.; Simon, M.K.; Zhu, W.; Sigmund, A.L.; Zastrow-Hayes, G.; Miller, M.; Liu, D.; et al. Developing a flexible, high-efficiency Agrobacterium -mediated sorghum transformation system with broad application. Plant Biotechnol. J. 2018, 16, 1388–1395. [Google Scholar] [CrossRef] [Green Version]
- Kelliher, T.; Starr, D.; Wang, W.; McCuiston, J.; Zhong, H.; Nuccio, M.L.; Martin, B. Maternal Haploids Are Preferentially Induced by CENH3-tailswap Transgenic Complementation in Maize. Front. Plant Sci. 2016, 7, 414. [Google Scholar] [CrossRef] [Green Version]
- Kermicle, J.L. Androgenesis Conditioned by a Mutation in Maize. Science 1969, 166, 1422–1424. [Google Scholar] [CrossRef]
- Zong, Y.; Song, Q.; Li, C.; Jin, S.; Zhang, D.; Wang, Y.; Qiu, J.-L.; Gao, C. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 2018, 36, 950–953. [Google Scholar] [CrossRef]
- Abe, F.; Haque, E.; Hisano, H.; Tanaka, T.; Kamiya, Y.; Mikami, M.; Kawaura, K.; Endo, M.; Onishi, K.; Hayashi, T.; et al. Genome-Edited Triple-Recessive Mutation Alters Seed Dormancy in Wheat. Cell Rep. 2019, 28, 1362–1369.e4. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Hua, L.; Gupta, A.; Tricoli, D.; Edwards, K.J.; Yang, B.; Li, W. Development of an Agrobacterium -delivered CRISPR /Cas9 system for wheat genome editing. Plant Biotechnol. J. 2019, 17, 1623–1635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, C.; Su, H.; Bai, H.; Wang, R.; Liu, Y.; Guo, X.; Liu, C.; Zhang, J.; Yuan, J.; Birchler, J.A.; et al. High-efficiency genome editing using a dmc1 promoter-controlled CRISPR/Cas9 system in maize. Plant Biotechnol. J. 2018, 16, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Mao, Y.; Xu, N.; Zhang, B.; Wei, P.; Yang, D.; Wang, Z.; Zhang, Z.; Zheng, R.; Yang, L.; et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modi-fications in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4632–4637. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.-L.; Dong, L.; Wang, Z.-P.; Zhang, H.-Y.; Han, C.-Y.; Liu, B.; Wang, X.-C.; Chen, Q.-J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-P.; Xing, H.-L.; Dong, L.; Zhang, H.-Y.; Han, C.-Y.; Wang, X.-C.; Chen, Q.-J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015, 16, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Zhu, L.; Zhao, B.; Zhao, Y.; Xie, Y.; Zheng, Z.; Li, Y.; Sun, J.; Wang, H. Development of a Haploid-Inducer Mediated Genome Editing System for Accelerating Maize Breeding. Mol. Plant 2019, 12, 597–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelliher, T.; Starr, D.; Su, X.; Tang, G.; Chen, Z.; Carter, J.; Wittich, P.E.; Dong, S.; Green, J.; Burch, E.; et al. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 2019, 37, 287–292. [Google Scholar] [CrossRef]
- Budhagatapalli, N.; Halbach, T.; Hiekel, S.; Büchner, H.; Müller, A.E.; Kumlehn, J. Site-directed mutagenesis in bread and durum wheat via pollination by cas9 /guide RNA-transgenic maize used as haploidy inducer. Plant Biotechnol. J. 2020, 18, 2376–2378. [Google Scholar] [CrossRef]
- Maheshwari, S.; Tan, E.H.; West, A.; Franklin, F.C.H.; Comai, L.; Chan, S.W.L. Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids. PLoS Genet. 2015, 11, e1004970. [Google Scholar] [CrossRef] [Green Version]
- Hamada, H.; Liu, Y.; Nagira, Y.; Miki, R.; Taoka, N.; Imai, R. Biolistic-delivery-based transient CRISPR/Cas9 expression enables in planta genome editing in wheat. Sci. Rep. 2018, 8, 14422. [Google Scholar] [CrossRef]
- Liu, Y.; Luo, W.; Linghu, Q.; Abe, F.; Hisano, H.; Sato, K.; Kamiya, Y.; Kawaura, K.; Onishi, K.; Endo, M.; et al. In planta Genome Editing in Commercial Wheat Varieties. Front. Plant Sci. 2021, 12, 648841. [Google Scholar] [CrossRef]
- Bilichak, A.; Luu, J.; Eudes, F. Intracellular delivery of fluorescent protein into viable wheat microspores using cationic peptides. Front. Plant Sci. 2015, 6, 666. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.-J.; Zhang, D. Molecular Control of Male Fertility for Crop Hybrid Breeding. Trends Plant Sci. 2018, 23, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K.; Balyan, H.S.; Gahlaut, V.; Saripalli, G.; Pal, B.; Basnet, B.; Joshi, A.K. Hybrid wheat: Past, present and future. Theor. Appl. Genet. 2019, 132, 2463–2483. [Google Scholar] [CrossRef]
- Singh, S.P.; Srivastava, R.; Kumar, J. Male sterility systems in wheat and opportunities for hybrid wheat development. Acta Physiol. Plant. 2014, 37, 1713. [Google Scholar] [CrossRef]
- Chang, Z.; Chen, Z.; Wang, N.; Xie, G.; Lu, J.; Yan, W.; Zhou, J.; Tang, X.; Deng, X.W. Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc. Natl. Acad. Sci. USA 2016, 113, 14145–14150. [Google Scholar] [CrossRef] [Green Version]
- Skibbe, D.; Schnable, P.J.M. Male Sterility in Maize. Maydica 2005, 50, 367–376. [Google Scholar]
- Pallotta, M.A.; Warner, P.; Kouidri, A.; Tucker, E.J.; Baes, M.; Suchecki, R.; Watson-Haigh, N.; Okada, T.; Garcia, M.; Sandhu, A.; et al. Wheat ms5 male-sterility is induced by recessive homoeologous A and D genome non-specific lipid transfer proteins. Plant J. 2019, 99, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kumar, M.; Thilges, K.; Cho, M.-J.; Cigan, A.M. MS26/CYP704B is required for anther and pollen wall development in bread wheat (Triticum aestivum L.) and combining mutations in all three homeologs causes male sterility. PLoS ONE 2017, 12, e0177632. [Google Scholar] [CrossRef]
- Djukanovic, V.; Smith, J.; Lowe, K.; Yang, M.; Gao, H.; Jones, S.; Nicholson, M.G.; West, A.; Lape, J.; Bidney, D.; et al. Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J. 2013, 76, 888–899. [Google Scholar] [CrossRef]
- Cigan, A.M.; Singh, M.; Benn, G.; Feigenbutz, L.; Kumar, M.; Cho, M.-J.; Svitashev, S.; Young, J. Targeted mutagenesis of a conserved anther-expressed P450 gene confers male sterility in monocots. Plant Biotechnol. J. 2017, 15, 379–389. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, M.; Albertsen, M.C.; Young, J.K.; Cigan, A.M. Concurrent modifications in the three homeologs of Ms45 gene with CRISPR-Cas9 lead to rapid generation of male sterile bread wheat (Triticum aestivum L.). Plant Mol. Biol. 2018, 97, 371–383. [Google Scholar] [CrossRef]
- Okada, A.; Arndell, T.; Borisjuk, N.; Sharma, N.; Watson-Haigh, N.; Tucker, E.J.; Baumann, U.; Langridge, P.; Whitford, R. CRISPR /Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 2019, 17, 1905–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.-H.; Dend, J.-Y. A Dominant Gene for Male Sterility in Wheat. Plant Breed. 1986, 97, 204–209. [Google Scholar] [CrossRef]
- Ni, F.; Qi, J.; Hao, Q.; Lyu, B.; Luo, M.-C.; Wang, Y.; Chen, F.; Wang, S.; Zhang, C.; Epstein, L.; et al. Wheat Ms2 encodes for an orphan protein that confers male sterility in grass species. Nat. Commun. 2017, 8, 15121. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Zhang, L.; Zou, C.; Gu, Y.; Duan, J.; Zhao, G.; Wu, J.; Liu, Y.; Fang, X.; Gao, L.; et al. A TRIM insertion in the promoter of Ms2 causes male sterility in wheat. Nat. Commun. 2017, 8, 15407. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Liu, H.; Zhou, Y.; Liu, H.; Du, L.; Wang, K.; Ye, X. Fertility recovery of wheat male sterility controlled by Ms2 using CRISPR/Cas9. Plant Biotechnol. J. 2021, 19, 224–226. [Google Scholar] [CrossRef]
- Zhou, H.; He, M.; Li, J.; Chen, L.; Huang, Z.; Zheng, S.; Zhu, L.; Ni, E.; Jiang, D.; Zhao, B.; et al. Development of Commercial Thermo-sensitive Genic Male Sterile Rice Accelerates Hybrid Rice Breeding Using the CRISPR/Cas9-mediated TMS5 Editing System. Sci. Rep. 2016, 6, 37395. [Google Scholar] [CrossRef] [Green Version]
- Sasakuma, T.; Ohtsuka, I. Cytoplasmic effects of Aegilops species having D genome in wheat. I. Cytoplasmic differentiation among five species regarding pistillody induction. Seiken Jiho Rep. Kihara Inst. Biol. Res. 1979. Available online: https://agris.fao.org/agris-search/search.do?recordID=US201301374634 (accessed on 21 August 2021).
- Mura, K. F1 seed production efficiency by using photoperiod-sensitive cytoplasmic male sterility and performance of F1 hybrid lines in wheat. Jpn. J. Breed. 1998, 48, 35–40. [Google Scholar] [CrossRef] [Green Version]
- Murai, K. Factors responsible for levels of male sterility in photoperiod-sensitive cytoplasmic male sterile (PCMS) wheat lines. Euphytica 2001, 117, 111–116. [Google Scholar] [CrossRef]
- Van Dijk, P.J.; Rigola, D.; Schauer, S.E. Plant Breeding: Surprisingly, Less Sex Is Better. Curr. Biol. 2016, 26, R122–R124. [Google Scholar] [CrossRef] [Green Version]
- Hojsgaard, D. Apomixis Technology: Separating the Wheat from the Chaff. Genes 2020, 11, 411. [Google Scholar] [CrossRef] [Green Version]
- D’Erfurth, I.; Jolivet, S.; Froger, N.; Catrice, O.; Novatchkova, M.; Mercier, R. Turning Meiosis into Mitosis. PLoS Biol. 2009, 7, e1000124. [Google Scholar] [CrossRef] [Green Version]
- Mieulet, D.; Jolivet, S.; Rivard, M.; Cromer, L.; Vernet, A.; Mayonove, P.; Pereira, L.; Droc, G.; Courtois, B.; Guiderdoni, E.; et al. Turning rice meiosis into mitosis. Cell Res. 2016, 26, 1242–1254. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Q.; Shen, Y.; Hua, Y.; Wang, J.; Lin, J.; Wu, M.; Sun, T.; Cheng, Z.; Mercier, R.; et al. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 2019, 37, 283–286. [Google Scholar] [CrossRef]
- Melchinger, A.E.; Molenaar, W.S.; Mirdita, V.; Schipprack, W. Colchicine Alternatives for Chromosome Doubling in Maize Haploids for Doubled-Haploid Production. Crop. Sci. 2016, 56, 559–569. [Google Scholar] [CrossRef]
- De La Fuente, G.N. Improvements to the Maize (Zea mays L.) In Vivo Maternal Doubled Haploid System. Ph.D. Thesis, Iowa State University, Ames, IA, USA, 2015. [Google Scholar]
- Seguí-Simarro, J.M.; Nuez, F. Pathways to doubled haploidy: Chromosome doubling during androgenesis. Cytogenet. Genome Res. 2008, 120, 358–369. [Google Scholar] [CrossRef]
- Castillo, A.M.; Cistué, L.; Vallés, M.P.; Soriano, M. Chromosome Doubling in Monocots. In Advances in Haploid Production in Higher Plants; Touraev, A., Forster, B.P., Jain, S.M., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 329–338. [Google Scholar]
- Ahmadi, B.; Ebrahimzadeh, H. In vitro androgenesis: Spontaneous vs. artificial genome doubling and characterization of regenerants. Plant Cell Rep. 2020, 39, 299–316. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wu, P.; Tian, X.; Lubberstedt, T.; Chen, S. QTL mapping for haploid male fertility by a segregation distortion method and fine mapping of a key QTL qhmf4 in maize. Theor. Appl. Genet. 2017, 130, 1349–1359. [Google Scholar] [CrossRef]
- Chang, M.-T.; Coe, E.H. Doubled Haploids. In Molecular Genetic Approaches to Maize Improvement; Kriz, A.L., Larkins, B.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 127–142. [Google Scholar]
- Chalyk, S.T. Properties of maternal haploid maize plants and potential application to maize breeding. Euphytica 1994, 79, 13–18. [Google Scholar] [CrossRef]
- Kleiber, D.; Prigge, V.; Melchinger, A.E.; Burkard, F.; Vicente, F.S.; Palomino, G.; Gordillo, G.A. Haploid Fertility in Temperate and Tropical Maize Germplasm. Crop. Sci. 2012, 52, 623–630. [Google Scholar] [CrossRef]
- Ren, J.; Boerman, N.; Liu, R.; Wu, P.; Trampe, B.; Vanous, K.; Frei, U.K.; Chen, S.; Lübberstedt, T. Mapping of QTL and identification of candidate genes conferring spontaneous haploid genome doubling in maize (Zea mays L.). Plant Sci. 2020, 293, 110337. [Google Scholar] [CrossRef]
- Sugihara, N.; Higashigawa, T.; Aramoto, D.; Kato, A. Haploid plants carrying a sodium azide-induced mutation (fdr1) produce fertile pollen grains due to first division restitution (FDR) in maize (Zea mays L.). Theor. Appl. Genet. 2013, 126, 2931–2941. [Google Scholar] [CrossRef]
- Trampe, B.; Dos Santos, I.G.; Frei, U.K.; Ren, J.; Chen, S.; Lübberstedt, T. QTL mapping of spontaneous haploid genome doubling using genotyping-by-sequencing in maize (Zea mays L.). Theor. Appl. Genet. 2020, 133, 2131–2140. [Google Scholar] [CrossRef]
- Chaikam, V.; Gowda, M.; Nair, S.K.; Melchinger, A.E.; Boddupalli, P.M. Genome-wide association study to identify genomic regions influencing spontaneous fertility in maize haploids. Euphytica 2019, 215, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Cifuentes, M.; Rivard, M.; Pereira, L.; Chelysheva, L.; Mercier, R. Haploid Meiosis in Arabidopsis: Double-Strand Breaks Are Formed and Repaired but Without Synapsis and Crossovers. PLoS ONE 2013, 8, e72431. [Google Scholar] [CrossRef] [Green Version]

| Gene Name | Gene Description | Plant | Haploid Induction Rate | Hi Engineering through GE | References |
|---|---|---|---|---|---|
| Centromere histone H3 (CENH3) | Centromere-specific histone variant | Arabidopsis thaliana | Up to 34% | Yes | [65] |
| Up to 44% | [66] | ||||
| Brassica oleracea var. capitata | Not tested | Yes | [79] | ||
| Cucumis melo | 1.50% | Not tested | [80] | ||
| Cucumis sativus | 1% | Not tested | [80] | ||
| Daucus carota | Not tested | Yes | [81] | ||
| Hordeum vulgare | 0 | Not tested | [82] | ||
| Oryza sativa | 1% | Not tested | [83] | ||
| Solanum lycopersicum | 2.30% | Not tested | [83] | ||
| Sorghum bicolor | Not tested | Yes | [84] | ||
| Triticum aestivum | ~7% | Yes | [69] | ||
| Zea mays | Up to 3.6% | Yes | [57,85] | ||
| DOMAIN OF UNKNOWN FUNCTION 679 membrane protein (DMP) | DMPs are involved in gamete fusion during double fertilization | Arabidopsis thaliana | ~2.1% in Atdmp8dmp9 double mutant | Yes | [76] |
| Zea mays | 0.30% | Yes | [31] | ||
| indeterminate gametophyte1 (ig1)/LATERAL ORGAN BOUNDARIES (LOB)-domain protein | Involved in the lateral organ development in higher plants | Zea mays | 3% | Not tested | [86] |
| MATRILINEAL (MTL)/Patatin-Like Phospholipase A (ZmPLA1)/NOT LIKE DAD (NLD) | Pollen-specific phospholipase | Oryza sativa | ~6% in Osmatl background | Yes | [59] |
| Triticum aestivum | Up to 15.7% in tapla-a and tapla-d double mutant | Yes | [58] | ||
| 31.6% in TaMTL triple mutant | Yes | [75] | |||
| Not tested | C–to–T base editing | [87] | |||
| Zea mays | 3% | Yes | [37] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhowmik, P.; Bilichak, A. Advances in Gene Editing of Haploid Tissues in Crops. Genes 2021, 12, 1410. https://doi.org/10.3390/genes12091410
Bhowmik P, Bilichak A. Advances in Gene Editing of Haploid Tissues in Crops. Genes. 2021; 12(9):1410. https://doi.org/10.3390/genes12091410
Chicago/Turabian StyleBhowmik, Pankaj, and Andriy Bilichak. 2021. "Advances in Gene Editing of Haploid Tissues in Crops" Genes 12, no. 9: 1410. https://doi.org/10.3390/genes12091410
APA StyleBhowmik, P., & Bilichak, A. (2021). Advances in Gene Editing of Haploid Tissues in Crops. Genes, 12(9), 1410. https://doi.org/10.3390/genes12091410







