Gene Expression and Mutational Profile in BAP-1 Inactivated Melanocytic Lesions of Progressive Malignancy from a Patient with Multiple Lesions
Abstract
:1. Introduction
2. Materials and Methods
2.1. RNA Sequencing
2.2. RNA Expression and Pathway Analysis
3. Results
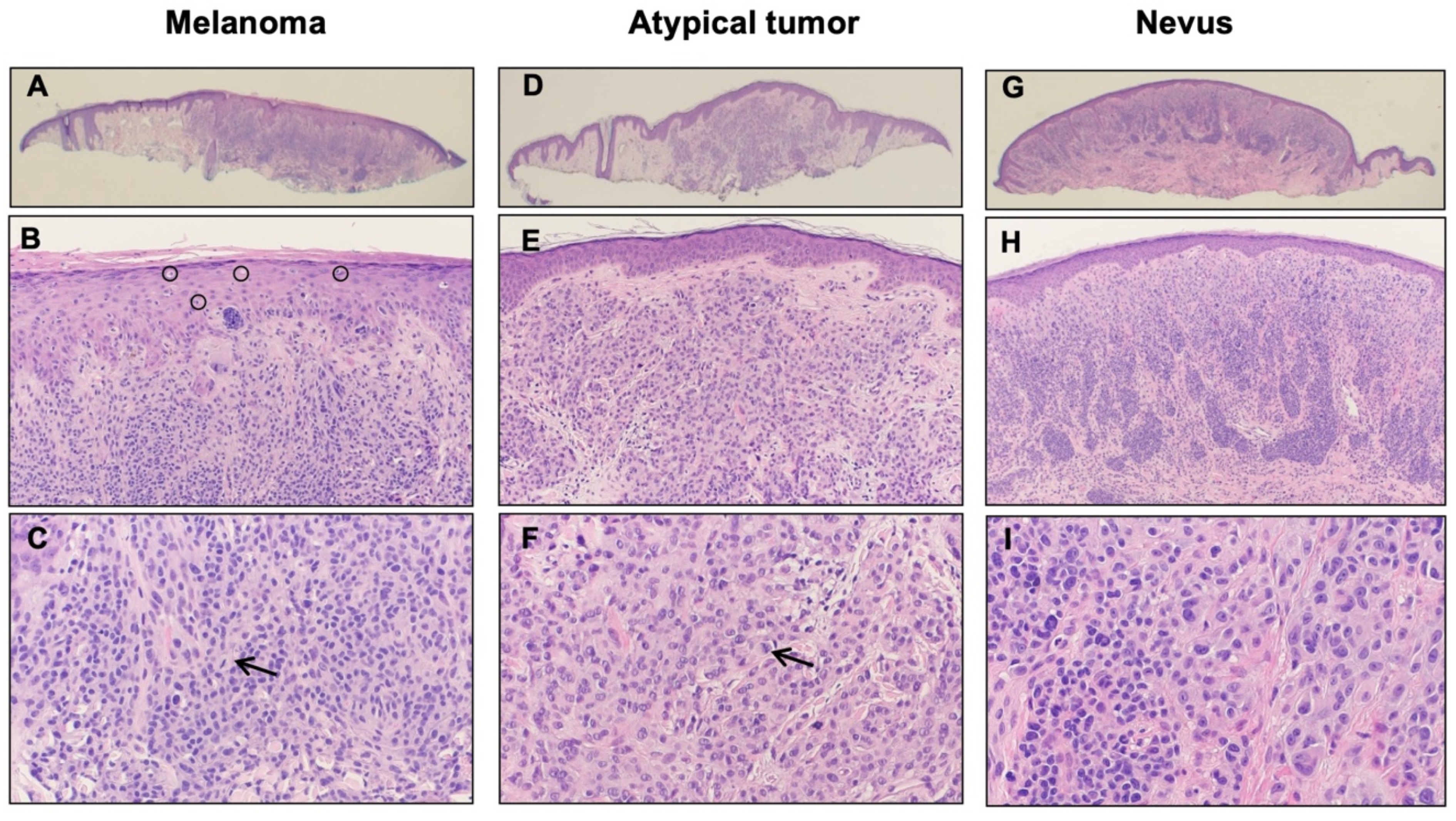
3.1. Case Presentation
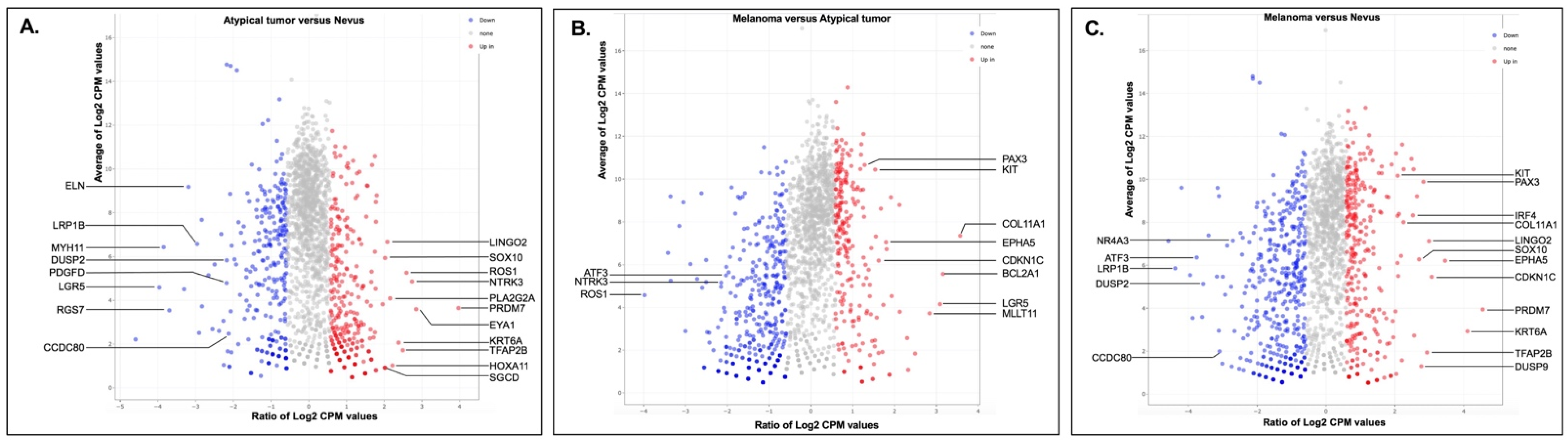
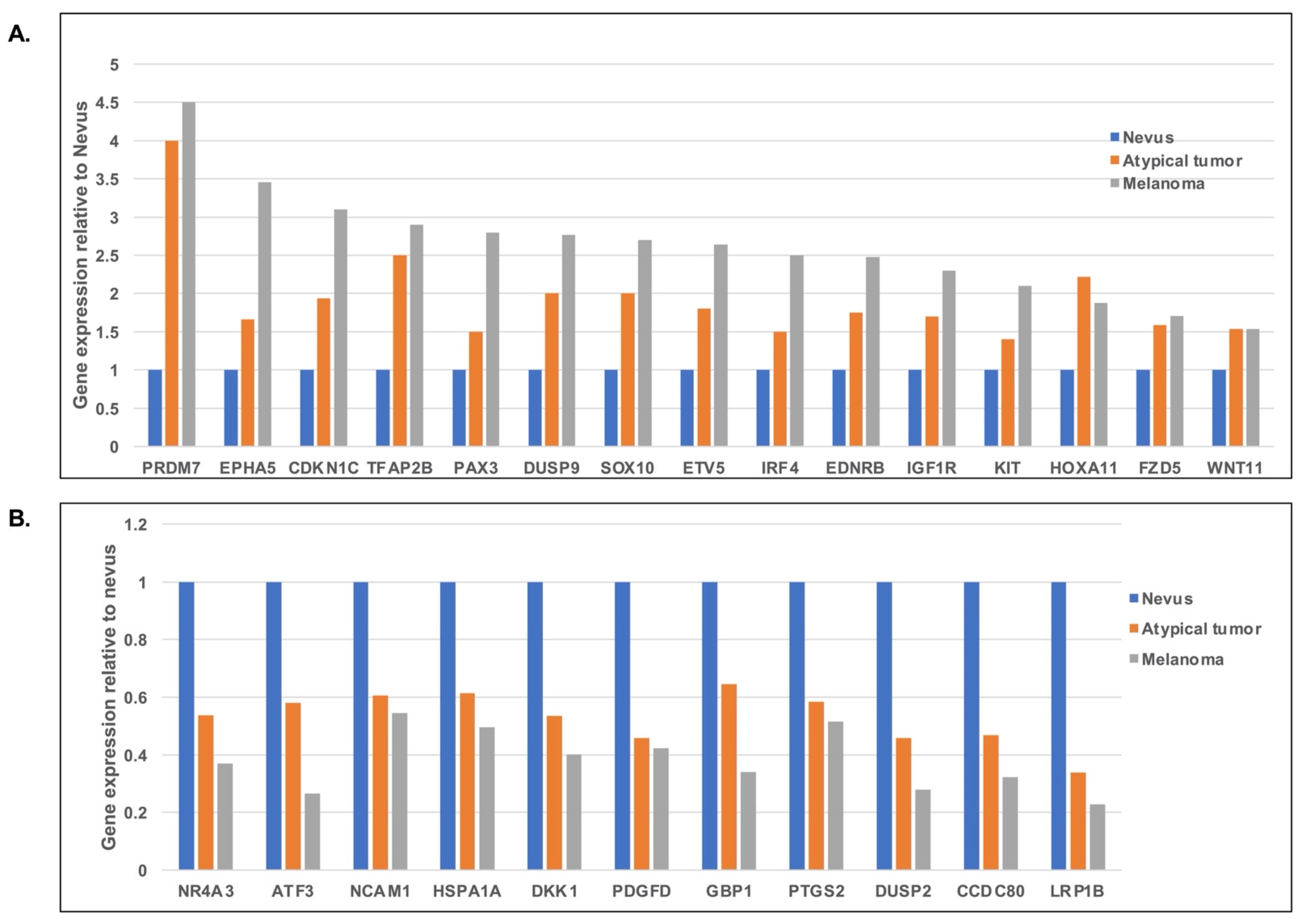
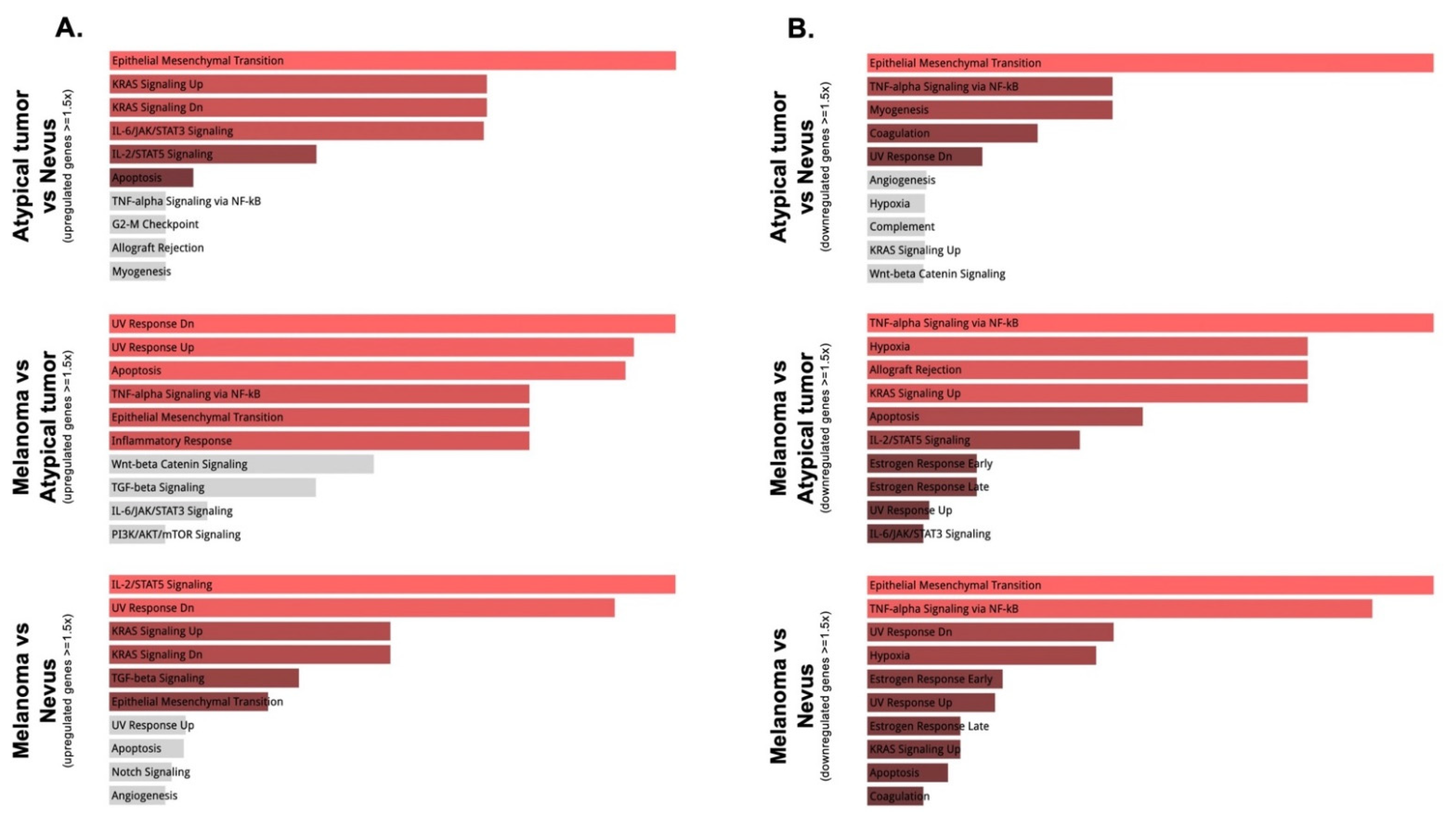
3.2. Gene Expression and Mutation Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carbone, M.; Yang, H.; Pass, H.I.; Krausz, T.; Testa, J.R.; Gaudino, G. BAP1 and cancer. Nat. Rev. Cancer 2013, 13, 153–159. [Google Scholar] [CrossRef]
- Wiesner, T.; Obenauf, A.; Murali, R.; Fried, I.; Griewank, K.; Ulz, P.; Windpassinger, C.; Wackernagel, W.; Loy, S.; Wolf, I.; et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat. Genet. 2011, 43, 1018–1021. [Google Scholar] [CrossRef] [Green Version]
- Wiesner, T.; Fried, I.; Ulz, P.; Stacher, E.; Popper, H.; Murali, R.; Kutzner, H.; Lax, S.; Smolle-Jüttner, F.; Geigl, J.B.; et al. Toward an Improved Definition of the Tumor Spectrum Associated With BAP1 Germline Mutations. J. Clin. Oncol. 2012, 30, e337–e340. [Google Scholar] [CrossRef] [PubMed]
- Walpole, S.; Pritchard, A.L.; Cebulla, C.M.; Pilarski, R.; Stautberg, M.; Davidorf, F.H.; De La Fouchardière, A.; Cabaret, O.; Golmard, L.; Stoppa-Lyonnet, D.; et al. Comprehensive Study of the Clinical Phenotype of Germline BAP1 Variant-Carrying Families Worldwide. J. Natl. Cancer Inst. 2018, 110, 1328–1341. [Google Scholar] [CrossRef]
- Haugh, A.M.; Njauw, C.N.; Bubley, J.A.; Verzì, A.E.; Zhang, B.; Kudalkar, E.; Van den Boom, T.; Walton, K.; Swick, B.L.; Kumar, R.; et al. Genotypic and Phenotypic Features of BAP1 Cancer Syndrome: A Report of 8 New Families and Review of Cases in the Literature. JAMA Dermatol. 2017, 153, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Yeh, I.; Mully, T.W.; Wiesner, T.; Vemula, S.S.; Mirza, S.A.; Sparatta, A.J.; McCalmont, T.H.; Bastian, B.; LeBoit, P.E. Ambiguous Melanocytic Tumors With Loss of 3p21. Am. J. Surg. Pathol. 2014, 38, 1088–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busam, K.J.; Sung, J.; Wiesner, T.; Von Deimling, A.; Jungbluth, A. Combined BRAF(V600E)-positive melanocytic lesions with large epithelioid cells lacking BAP1 expression and conventional nevomelanocytes. Am. J. Surg. Pathol. 2013, 37, 193–199. [Google Scholar] [CrossRef]
- Wiesner, T.; Murali, R.; Fried, I.; Cerroni, L.; Busam, K.; Kutzner, H.; Bastian, B.C. A Distinct Subset of Atypical Spitz Tumors is Characterized by BRAF Mutation and Loss of BAP1 Expression. Am. J. Surg. Pathol. 2012, 36, 818–830. [Google Scholar] [CrossRef] [Green Version]
- Aung, P.P.; Nagarajan, P.; Tetzlaff, M.T.; Curry, J.L.; Tang, G.; Abdullaev, Z.; Pack, S.D.; Ivan, D.; Prieto, V.G.; Torres-Cabala, C.A. Melanoma With Loss of BAP1 Expression in Patients With No Family History of BAP1-Associated Cancer Susceptibility Syndrome: A Case Series. Am. J. Dermatopathol. 2019, 41, 167–179. [Google Scholar] [CrossRef]
- Busam, K.J.; Wanna, M.; Wiesner, T. Multiple epithelioid Spitz nevi or tumors with loss of BAP1 expression: A clue to a hereditary tumor syndrome. JAMA Dermatol. 2013, 149, 335–339. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.A.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Brown, L.C.; Tucker, M.D.; Sedhom, R.; Schwartz, E.B.; Zhu, J.; Kao, C.; Labriola, M.K.; Gupta, R.T.; Marin, D.; Wu, Y.; et al. LRP1B mutations are associated with favorable outcomes to immune checkpoint inhibitors across multiple cancer types. J. Immunother. Cancer 2021, 9, e001792. [Google Scholar] [CrossRef]
- Jenkins, R.W.; Fisher, D.E. Treatment of Advanced Melanoma in 2020 and Beyond. J. Investig. Dermatol. 2020, 141, 23–31. [Google Scholar] [CrossRef]
- Sunshine, J.C.; Kim, D.; Zhang, B.; Compres, E.V.; Khan, A.U.; Busam, K.J.; Gerami, P. Melanocytic Neoplasms With MAP2K1 in Frame Deletions and Spitz Morphology. Am. J. Dermatopathol. 2020, 42, 923–931. [Google Scholar] [CrossRef]
- Capoluongo, E. Insulin-Like Growth Factor System and Sporadic Malignant Melanoma. Am. J. Pathol. 2011, 178, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Quan, V.L.; Panah, E.; Zhang, B.; Shi, K.; Mohan, L.S.; Gerami, P. The role of gene fusions in melanocytic neoplasms. J. Cutan. Pathol. 2019, 46, 878–887. [Google Scholar] [CrossRef] [Green Version]
- Tetzlaff, M.T.; Reuben, A.; Billings, S.D.; Prieto, V.G.; Curry, J.L. Toward a Molecular-Genetic Classification of Spitzoid Neoplasms. Clin. Lab. Med. 2017, 37, 431–448. [Google Scholar] [CrossRef]
- Shakhova, O.; Zingg, D.; Schaefer, S.M.; Hari, L.; Civenni, G.; Blunschi, J.; Claudinot, S.; Okoniewski, M.; Beermann, F.; Mihic-Probst, D.; et al. Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nature 2012, 14, 882–890. [Google Scholar] [CrossRef] [PubMed]
- Cronin, J.C.; Watkins-Chow, D.; Incao, A.; Hasskamp, J.H.; Schönewolf, N.; Aoude, L.; Hayward, N.; Bastian, B.; Dummer, R.; Loftus, S.K.; et al. SOX10 Ablation Arrests Cell Cycle, Induces Senescence, and Suppresses Melanomagenesis. Cancer Res. 2013, 73, 5709–5718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graf, S.A.; Busch, C.; Bosserhoff, A.-K.; Besch, R.; Berking, C. SOX10 Promotes Melanoma Cell Invasion by Regulating Melanoma Inhibitory Activity. J. Investig. Dermatol. 2014, 134, 2212–2220. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, S.; Takahashi, A.; Kikuchi, R.; Nishibu, S.; Lo, J.A.; Hejna, M.; Moon, W.M.; Kato, S.; Zhou, Y.; Hodi, F.S.; et al. SOX10 regulates melanoma immunogenicity through an IRF4-IRF1 axis. Cancer Res. 2021. [Google Scholar] [CrossRef]
- Gibbs, D.C.; Ward, S.V.; Orlow, I.; Cadby, G.; Kanetsky, P.A.; Luo, L.; Busam, K.J.; Kricker, A.; Armstrong, B.K.; Cust, A.E.; et al. Functional melanoma-risk variant IRF 4 rs12203592 associated with Breslow thickness: A pooled international study of primary melanomas. Br. J. Dermatol. 2017, 177, e180–e182. [Google Scholar] [CrossRef]
- Kubic, J.D.; Little, E.C.; Lui, J.W.; Iizuka, T.; Lang, D. PAX3 and ETS1 synergistically activate MET expression in melanoma cells. Oncogene 2014, 34, 4964–4974. [Google Scholar] [CrossRef] [Green Version]
- Cohen-Solal, K.A.; Kaufman, H.L.; Lasfar, A. Transcription factors as critical players in melanoma invasiveness, drug resistance, and opportunities for therapeutic drug development. Pigment. Cell Melanoma Res. 2017, 31, 241–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Lesion | Gene | Mutation | Type |
|---|---|---|---|
| Nevus | NIPBL | p.Q338X | stopgain |
| AKAP9 | p.Q2911X | stopgain | |
| EP400 | p.E2200X | stopgain | |
| CREBBP | p.Q1075X | stopgain | |
| COL1A1 | p.P817fs | frameshift deletion | |
| ZNF687 | p.Q474X | stopgain | |
| PDGFRB | p.Q412X | stopgain | |
| ASPH | p.R659X | stopgain | |
| NT5C2 | p.Q173X | stopgain | |
| NIN | p.Q1291X | stopgain | |
| HIF1A | p.Q379X | stopgain | |
| CHD2 | p.Q366X | stopgain | |
| USP7 | p.Q822X | stopgain | |
| MYO18A | p.Q830X | stopgain | |
| KPNB1 | p.Q111X | stopgain | |
| RPS6KA3 | p.R383W | nonsynonymous SNV | |
| TBL1XR1 | p.S332F | nonsynonymous SNV | |
| MAP2K1 | p.A172V | nonsynonymous SNV | |
| Atypical tumor | NIPBL | p.Q1567X | stopgain |
| AKAP9 | p.Q2480X | stopgain | |
| EP400 | p.Q148X | stopgain | |
| CAD | p.Q30X | stopgain | |
| PPP1CB | p.Q293X | stopgain | |
| BIRC6 | p.Q1072X | stopgain | |
| COL6A3 | p.Q2366X | stopgain | |
| TFG | p.Q266X | stopgain | |
| EIF4A2 | p.Q209X | stopgain | |
| AFF4 | p.Q537X | stopgain | |
| ARHGAP26 | p.R120X | stopgain | |
| MAML1 | p.Q683X | stopgain | |
| DST | p.Q1890X | stopgain | |
| HDAC2 | p.Q128X | stopgain | |
| PTK2 | p.Q734X | stopgain | |
| SYK | p.Q239X | stopgain | |
| NUMA1 | p.Q832X | stopgain | |
| PICALM | p.Q256X | stopgain | |
| EED | p.Q302X | stopgain | |
| SIK3 | p.Q675X | stopgain | |
| KDM5A | p.R266X | stopgain | |
| PRICKLE1 | p.Q348X | stopgain | |
| ARID2 | p.Q720X | stopgain | |
| NUP107 | p.Q236X | stopgain | |
| BRCA2 | p.Q2943X | stopgain | |
| TCF12 | p.Q605X | stopgain | |
| TOP2A | p.Q517X | stopgain | |
| ITGB3 | p.Q616X | stopgain | |
| SMARCA4 | p.R978X | stopgain | |
| NOTCH3 | p.Q646X | stopgain | |
| CSNK2A1 | p.Q71X | stopgain | |
| NCOA3 | p.Q478X | stopgain | |
| ETS2 | p.Q234X | stopgain | |
| EP300 | p.Q523X | stopgain | |
| OFD1 | p.Q39X | stopgain | |
| AR | p.Q825X | stopgain | |
| ZMYM3 | p.Q763X | stopgain | |
| BMPR1A | p.R244X | stopgain | |
| NOTCH2 | p.Q1814X | stopgain | |
| TPR | p.Q2344X | stopgain | |
| BRAF | p.V600E | nonsynonymous SNV | |
| CSF1R | p.A781V | nonsynonymous SNV | |
| PTEN | p.T277I | nonsynonymous SNV | |
| Melanoma | LRPPRC | p.G1050fs | frameshift insertion |
| EXT2 | p.R215X | stopgain | |
| SPEN | p.Q3373X | stopgain | |
| BAP1 | p.Y223X | stopgain | |
| AHR | p.Q705X | stopgain | |
| KMT2A | p.Q1207X | stopgain | |
| CDH1 | p.Q388X | stopgain | |
| RABEP1 | p.Q225X | stopgain | |
| CLTC | p.Q1358X | stopgain | |
| SUGP2 | p.R831X | stopgain | |
| TIAM1 | p.Q714X | stopgain | |
| BRAF | p.V600E | nonsynonymous SNV |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Nelson, A.C.; He, Y.; Munro, S.A.; Song, K.Y.; Domingo-Musibay, E.; Giubellino, A. Gene Expression and Mutational Profile in BAP-1 Inactivated Melanocytic Lesions of Progressive Malignancy from a Patient with Multiple Lesions. Genes 2022, 13, 10. https://doi.org/10.3390/genes13010010
Zhou Y, Nelson AC, He Y, Munro SA, Song KY, Domingo-Musibay E, Giubellino A. Gene Expression and Mutational Profile in BAP-1 Inactivated Melanocytic Lesions of Progressive Malignancy from a Patient with Multiple Lesions. Genes. 2022; 13(1):10. https://doi.org/10.3390/genes13010010
Chicago/Turabian StyleZhou, Yan, Andrew C. Nelson, Yuyu He, Sarah A. Munro, Kyu Young Song, Evidio Domingo-Musibay, and Alessio Giubellino. 2022. "Gene Expression and Mutational Profile in BAP-1 Inactivated Melanocytic Lesions of Progressive Malignancy from a Patient with Multiple Lesions" Genes 13, no. 1: 10. https://doi.org/10.3390/genes13010010
APA StyleZhou, Y., Nelson, A. C., He, Y., Munro, S. A., Song, K. Y., Domingo-Musibay, E., & Giubellino, A. (2022). Gene Expression and Mutational Profile in BAP-1 Inactivated Melanocytic Lesions of Progressive Malignancy from a Patient with Multiple Lesions. Genes, 13(1), 10. https://doi.org/10.3390/genes13010010







