Circulatory MicroRNAs in Plasma and Atrial Fibrillation in the General Population: The Rotterdam Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Assessment of Circulatory microRNAs in Plasma
2.3. Assessment of Atrial Fibrillation
2.4. Assessment of Cardiovascular Risk Factors
2.5. Statistical Analyses
2.6. Assessment of Predictive Target Genes
2.7. Literature Review
2.8. In Silico Analyses
3. Results
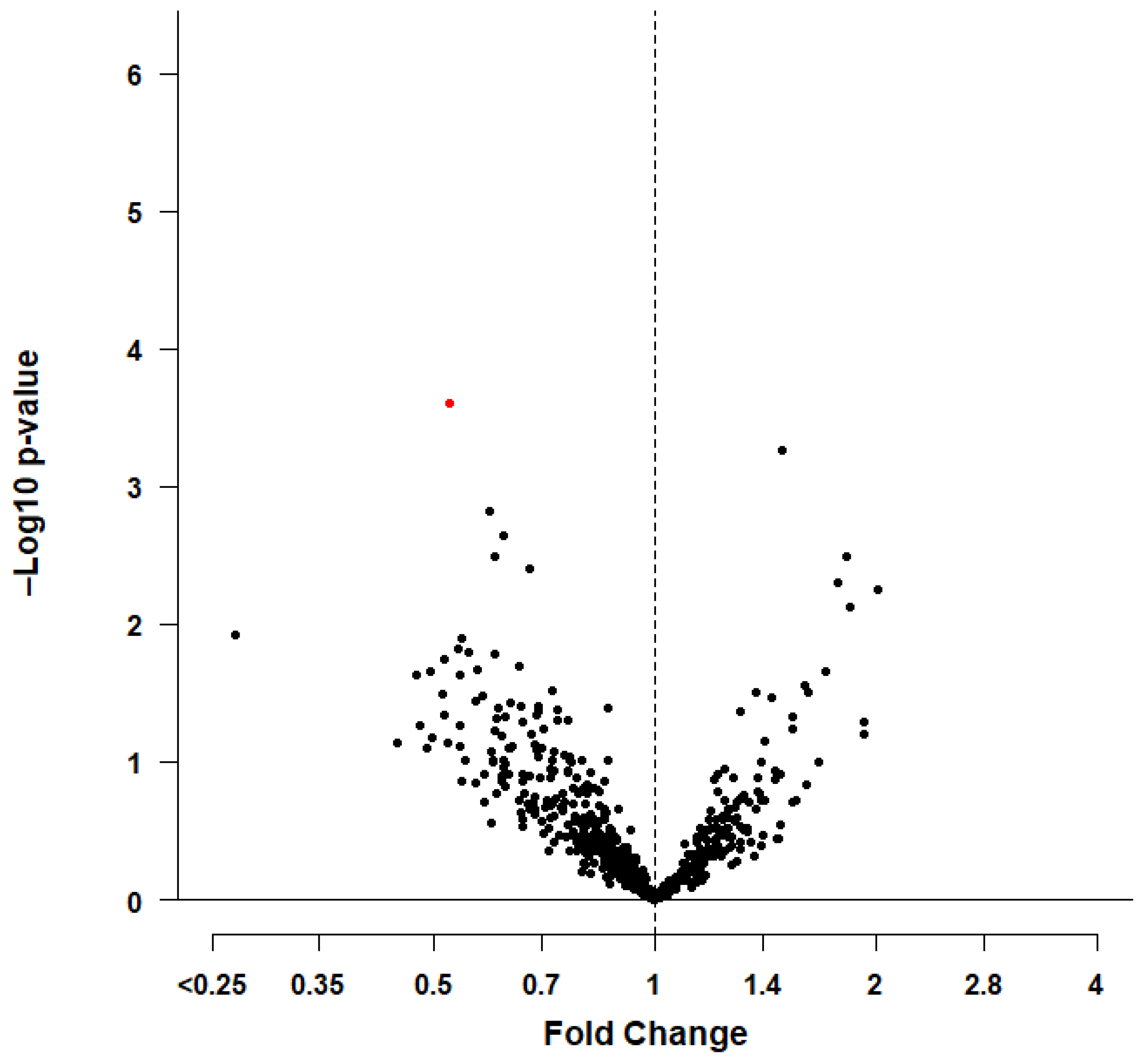
3.1. Statistical Analyses
3.2. Predictive Target Genes
3.3. Literature Review
3.4. In Silico Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chugh, S.S.; Havmoeller, R.; Narayanan, K.; Singh, D.; Rienstra, M.; Benjamin, E.J.; Gillum, R.F.; Kim, Y.H.; McAnulty, J.H., Jr.; Zheng, Z.J.; et al. Worldwide epidemiology of atrial fibrillation: A global burden of disease 2010 study. Circulation 2014, 129, 837–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krijthe, B.P.; Kunst, A.; Benjamin, E.J.; Lip, G.Y.; Franco, O.H.; Hofman, A.; Witteman, J.C.; Stricker, B.H.; Heeringa, J. Projections on the number of individuals with atrial fibrillation in the european union, from 2000 to 2060. Eur. Heart J 2013, 34, 2746–2751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heeringa, J.; van der Kuip, D.A.; Hofman, A.; Kors, J.A.; van Herpen, G.; Stricker, B.H.; Stijnen, T.; Lip, G.Y.; Witteman, J.C. Prevalence, incidence and lifetime risk of atrial fibrillation: The rotterdam study. Eur. Heart J 2006, 27, 949–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandes, A.; Smit, M.D.; Nguyen, B.O.; Rienstra, M.; Van Gelder, I.C. Risk factor management in atrial fibrillation. Arrhythm. Electrophysiol. Rev. 2018, 7, 118–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamin, E.J.; Levy, D.; Vaziri, S.M.; D’Agostino, R.B.; Belanger, A.J.; Wolf, P.A. Independent risk factors for atrial fibrillation in a population-based cohort. The framingham heart study. JAMA 1994, 271, 840–844. [Google Scholar] [CrossRef]
- Schnabel, R.B.; Yin, X.; Gona, P.; Larson, M.G.; Beiser, A.S.; McManus, D.D.; Newton-Cheh, C.; Lubitz, S.A.; Magnani, J.W.; Ellinor, P.T.; et al. 50 year trends in atrial fibrillation prevalence, incidence, risk factors, and mortality in the framingham heart study: A cohort study. Lancet 2015, 386, 154–162. [Google Scholar] [CrossRef] [Green Version]
- Hindricks, G.; Potpara, T.; Dagres, N.; Arbelo, E.; Bax, J.J.; Blomstrom-Lundqvist, C.; Boriani, G.; Castella, M.; Dan, G.A.; Dilaveris, P.E.; et al. 2020 esc guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the european association for cardio-thoracic surgery (eacts). Eur. Heart J. 2021, 42, 373–498. [Google Scholar] [CrossRef]
- Ko, D.; Rahman, F.; Schnabel, R.B.; Yin, X.; Benjamin, E.J.; Christophersen, I.E. Atrial fibrillation in women: Epidemiology, pathophysiology, presentation, and prognosis. Nat. Rev. Cardiol. 2016, 13, 321–332. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mrnas are conserved targets of micrornas. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Grasedieck, S.; Scholer, N.; Bommer, M.; Niess, J.H.; Tumani, H.; Rouhi, A.; Bloehdorn, J.; Liebisch, P.; Mertens, D.; Dohner, H.; et al. Impact of serum storage conditions on microrna stability. Leukemia 2012, 26, 2414–2416. [Google Scholar] [CrossRef]
- Small, E.M.; Olson, E.N. Pervasive roles of micrornas in cardiovascular biology. Nature 2011, 469, 336–342. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Cui, G.; Esmailian, F.; Plunkett, M.; Marelli, D.; Ardehali, A.; Odim, J.; Laks, H.; Sen, L. Atrial extracellular matrix remodeling and the maintenance of atrial fibrillation. Circulation 2004, 109, 363–368. [Google Scholar] [CrossRef] [Green Version]
- Wei, J.; Zhang, Y.; Li, Z.; Wang, X.; Chen, L.; Du, J.; Liu, J.; Liu, J.; Hou, Y. Gch1 attenuates cardiac autonomic nervous remodeling in canines with atrial-tachypacing via tetrahydrobiopterin pathway regulated by microrna-206. Pacing Clin. Electrophysiol. 2018, 41, 459–471. [Google Scholar] [CrossRef]
- Bingen, B.O.; Askar, S.F.; Neshati, Z.; Feola, I.; Panfilov, A.V.; de Vries, A.A.; Pijnappels, D.A. Constitutively active acetylcholine-dependent potassium current increases atrial defibrillation threshold by favoring post-shock re-initiation. Sci. Rep. 2015, 5, 15187. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Liu, L.; Hu, J.; Song, L. Microrna regulatory network revealing the mechanism of inflammation in atrial fibrillation. Med. Sci. Monit. 2015, 21, 3505–3513. [Google Scholar] [CrossRef] [Green Version]
- Dawson, K.; Wakili, R.; Ordog, B.; Clauss, S.; Chen, Y.; Iwasaki, Y.; Voigt, N.; Qi, X.Y.; Sinner, M.F.; Dobrev, D.; et al. Microrna29: A mechanistic contributor and potential biomarker in atrial fibrillation. Circulation 2013, 127, 1466–1475, 1475e1461-1428. [Google Scholar] [CrossRef] [Green Version]
- Harling, L.; Lambert, J.; Ashrafian, H.; Darzi, A.; Gooderham, N.J.; Athanasiou, T. Elevated serum microrna 483-5p levels may predict patients at risk of post-operative atrial fibrillation. Eur. J. Cardiothorac. Surg. 2017, 51, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Komal, S.; Yin, J.J.; Wang, S.H.; Huang, C.Z.; Tao, H.L.; Dong, J.Z.; Han, S.N.; Zhang, L.R. Micrornas: Emerging biomarkers for atrial fibrillation. J. Cardiol. 2019, 74, 475–482. [Google Scholar] [CrossRef]
- Liu, T.; Zhong, S.; Rao, F.; Xue, Y.; Qi, Z.; Wu, S. Catheter ablation restores decreased plasma mir-409-3p and mir-432 in atrial fibrillation patients. Europace 2016, 18, 92–99. [Google Scholar] [CrossRef]
- McManus, D.D.; Lin, H.; Tanriverdi, K.; Quercio, M.; Yin, X.; Larson, M.G.; Ellinor, P.T.; Levy, D.; Freedman, J.E.; Benjamin, E.J. Relations between circulating micrornas and atrial fibrillation: Data from the framingham offspring study. Heart Rhythm 2014, 11, 663–669. [Google Scholar] [CrossRef] [Green Version]
- McManus, D.D.; Tanriverdi, K.; Lin, H.; Esa, N.; Kinno, M.; Mandapati, D.; Tam, S.; Okike, O.N.; Ellinor, P.T.; Keaney, J.F., Jr.; et al. Plasma micrornas are associated with atrial fibrillation and change after catheter ablation (the mirhythm study). Heart Rhythm 2015, 12, 3–10. [Google Scholar] [CrossRef] [Green Version]
- Soeki, T.; Matsuura, T.; Bando, S.; Tobiume, T.; Uematsu, E.; Ise, T.; Kusunose, K.; Yamaguchi, K.; Yagi, S.; Fukuda, D.; et al. Relationship between local production of microrna-328 and atrial substrate remodeling in atrial fibrillation. J. Cardiol. 2016, 68, 472–477. [Google Scholar] [CrossRef] [Green Version]
- Tsoporis, J.N.; Fazio, A.; Rizos, I.K.; Izhar, S.; Proteau, G.; Salpeas, V.; Rigopoulos, A.; Sakadakis, E.; Toumpoulis, I.K.; Parker, T.G. Increased right atrial appendage apoptosis is associated with differential regulation of candidate micrornas 1 and 133a in patients who developed atrial fibrillation after cardiac surgery. J. Mol. Cell Cardiol. 2018, 121, 25–32. [Google Scholar] [CrossRef]
- Vaze, A.; Tran, K.V.; Tanriverdi, K.; Sardana, M.; Lessard, D.; Donahue, J.K.; Barton, B.; Aurigemma, G.; Lubitz, S.A.; Lin, H.; et al. Relations between plasma micrornas, echocardiographic markers of atrial remodeling, and atrial fibrillation: Data from the framingham offspring study. PLoS ONE 2020, 15, e0236960. [Google Scholar] [CrossRef]
- Wei, X.J.; Han, M.; Yang, F.Y.; Wei, G.C.; Liang, Z.G.; Yao, H.; Ji, C.W.; Xie, R.S.; Gong, C.L.; Tian, Y. Biological significance of mir-126 expression in atrial fibrillation and heart failure. Braz. J. Med. Biol. Res. 2015, 48, 983–989. [Google Scholar] [CrossRef]
- Leening, M.J.; Kavousi, M.; Heeringa, J.; van Rooij, F.J.; Verkroost-van Heemst, J.; Deckers, J.W.; Mattace-Raso, F.U.; Ziere, G.; Hofman, A.; Stricker, B.H.; et al. Methods of data collection and definitions of cardiac outcomes in the rotterdam study. Eur. J. Epidemiol. 2012, 27, 173–185. [Google Scholar] [CrossRef] [Green Version]
- Ikram, M.A.; Brusselle, G.; Ghanbari, M.; Goedegebure, A.; Ikram, M.K.; Kavousi, M.; Kieboom, B.C.T.; Klaver, C.C.W.; de Knegt, R.J.; Luik, A.I.; et al. Objectives, design and main findings until 2020 from the Rotterdam study. Eur. J. Epidemiol. 2020, 35, 483–517. [Google Scholar] [CrossRef]
- Zhang, X.; Mens, M.M.J.; Abozaid, Y.J.; Bos, D.; Darwish Murad, S.; de Knegt, R.J.; Ikram, M.A.; Pan, Q.; Ghanbari, M. Circulatory micrornas as potential biomarkers for fatty liver disease: The rotterdam study. Aliment. Pharmacol. Ther. 2021, 53, 432–442. [Google Scholar]
- van Bemmel, J.H.; Kors, J.A.; van Herpen, G. Methodology of the modular ecg analysis system means. Methods Inf. Med. 1990, 29, 346–353. [Google Scholar]
- Li, J.; Ji, L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 2005, 95, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Cheverud, J.M. A simple correction for multiple comparisons in interval mapping genome scans. Heredity 2001, 87, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, J.; Zhang, H.; Lu, J. Micrornas in the same clusters evolve to coordinately regulate functionally related genes. Mol. Biol. Evol. 2016, 33, 2232–2247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Team R core. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, 2020. Available online: https://www.R-project.Org/ (accessed on 24 August 2020).
- Liu, W.; Wang, X. Prediction of functional microrna targets by integrative modeling of microrna binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microrna target sites in mammalian mrnas. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. Mirdb: An online database for prediction of functional microrna targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.Y.; Lin, Y.C.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.G.; Jin, C.N.; Yu, Y.; et al. Mirtarbase 2020: Updates to the experimentally validated microrna-target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, J.B.; Thorolfsdottir, R.B.; Fritsche, L.G.; Zhou, W.; Skov, M.W.; Graham, S.E.; Herron, T.J.; McCarthy, S.; Schmidt, E.M.; Sveinbjornsson, G.; et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 2018, 50, 1234–1239. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Lutter, D.; Marr, C.; Krumsiek, J.; Lang, E.W.; Theis, F.J. Intronic micrornas support their host genes by mediating synergistic and antagonistic regulatory effects. BMC Genom. 2010, 11, 224. [Google Scholar] [CrossRef] [Green Version]
- Hinske, L.C.; Franca, G.S.; Torres, H.A.; Ohara, D.T.; Lopes-Ramos, C.M.; Heyn, J.; Reis, L.F.; Ohno-Machado, L.; Kreth, S.; Galante, P.A. Miriad-integrating microrna inter- and intragenic data. Database 2014, 2014, bau099. [Google Scholar] [CrossRef]
- Shen, N.N.; Zhang, C.; Li, Z.; Kong, L.C.; Wang, X.H.; Gu, Z.C.; Wang, J.L. Microrna expression signatures of atrial fibrillation: The critical systematic review and bioinformatics analysis. Exp. Biol. Med. 2020, 245, 42–53. [Google Scholar] [CrossRef] [Green Version]
- Ling, T.Y.; Wang, X.L.; Chai, Q.; Lu, T.; Stulak, J.M.; Joyce, L.D.; Daly, R.C.; Greason, K.L.; Wu, L.Q.; Shen, W.K.; et al. Regulation of cardiac cacnb2 by microrna-499: Potential role in atrial fibrillation. BBA Clin. 2017, 7, 78–84. [Google Scholar] [CrossRef]
- Ling, T.Y.; Wang, X.L.; Chai, Q.; Lau, T.W.; Koestler, C.M.; Park, S.J.; Daly, R.C.; Greason, K.L.; Jen, J.; Wu, L.Q.; et al. Regulation of the sk3 channel by microrna-499—Potential role in atrial fibrillation. Heart Rhythm 2013, 10, 1001–1009. [Google Scholar] [CrossRef] [Green Version]
- Yamac, A.H.; Kucukbuzcu, S.; Ozansoy, M.; Gok, O.; Oz, K.; Erturk, M.; Yilmaz, E.; Ersoy, B.; Zeybek, R.; Goktekin, O.; et al. Altered expression of micro-rna 199a and increased levels of cardiac sirt1 protein are associated with the occurrence of atrial fibrillation after coronary artery bypass graft surgery. Cardiovasc. Pathol. 2016, 25, 232–236. [Google Scholar] [CrossRef]
- He, X.; Zhang, K.; Gao, X.; Li, L.; Tan, H.; Chen, J.; Zhou, Y. Rapid atrial pacing induces myocardial fibrosis by down-regulating smad7 via microrna-21 in rabbit. Heart Vessel. 2016, 31, 1696–1708. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Chen, X.J.; Qian, C.; Dong, Q.; Ding, D.; Wu, Q.F.; Li, J.; Wang, H.F.; Li, W.H.; Xie, Q.; et al. Signal transducer and activator of transcription 3/microrna-21 feedback loop contributes to atrial fibrillation by promoting atrial fibrosis in a rat sterile pericarditis model. Circ. Arrhythm. Electrophysiol. 2016, 9. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The genecards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
- Chaldoupi, S.M.; Loh, P.; Hauer, R.N.; de Bakker, J.M.; van Rijen, H.V. The role of connexin40 in atrial fibrillation. Cardiovasc. Res. 2009, 84, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Choi, E.K.; Zhao, Y.; Everett, T.H.T.; Chen, P.S. Ganglionated plexi as neuromodulation targets for atrial fibrillation. J. Cardiovasc. Electrophysiol. 2017, 28, 1485–1491. [Google Scholar] [CrossRef] [Green Version]
- Antzelevitch, C.; Pollevick, G.D.; Cordeiro, J.M.; Casis, O.; Sanguinetti, M.C.; Aizawa, Y.; Guerchicoff, A.; Pfeiffer, R.; Oliva, A.; Wollnik, B.; et al. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by st-segment elevation, short qt intervals, and sudden cardiac death. Circulation 2007, 115, 442–449. [Google Scholar] [CrossRef]
- Mandyam, M.C.; Soliman, E.Z.; Alonso, A.; Dewland, T.A.; Heckbert, S.R.; Vittinghoff, E.; Cummings, S.R.; Ellinor, P.T.; Chaitman, B.R.; Stocke, K.; et al. The qt interval and risk of incident atrial fibrillation. Heart Rhythm 2013, 10, 1562–1568. [Google Scholar] [CrossRef] [Green Version]
- Mostafa, A.; El-Haddad, M.A.; Shenoy, M.; Tuliani, T. Atrial fibrillation post cardiac bypass surgery. Avicenna J. Med. 2012, 2, 65–70. [Google Scholar] [CrossRef]
- Saxena, A.; Dinh, D.T.; Smith, J.A.; Shardey, G.C.; Reid, C.M.; Newcomb, A.E. Usefulness of postoperative atrial fibrillation as an independent predictor for worse early and late outcomes after isolated coronary artery bypass grafting (multicenter australian study of 19,497 patients). Am. J. Cardiol. 2012, 109, 219–225. [Google Scholar] [CrossRef]
- Shah, R.; Tanriverdi, K.; Levy, D.; Larson, M.; Gerstein, M.; Mick, E.; Rozowsky, J.; Kitchen, R.; Murthy, V.; Mikalev, E.; et al. Discordant expression of circulating microrna from cellular and extracellular sources. PLoS ONE 2016, 11, e0153691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godoy, P.M.; Barczak, A.J.; DeHoff, P.; Srinivasan, S.; Etheridge, A.; Galas, D.; Das, S.; Erle, D.J.; Laurent, L.C. Comparison of reproducibility, accuracy, sensitivity, and specificity of mirna quantification platforms. Cell Rep. 2019, 29, 4212–4222.e4215. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Baseline Characteristics * | Total Study Population n = 1999 | Men n = 858 | Women n = 1141 | p-Value † |
|---|---|---|---|---|
| Age, years | 71.7 ± 7.6 | 71.4 ± 7.3 | 71.9 ± 7.8 | 0.116 |
| Women, n (%) | 1141 (57.1) | NA | 1141 (100) | |
| Body mass index, kg/m2 | 27.7 ± 4.1 | 27.6 ± 3.4 | 27.7 ± 4.6 | 0.382 |
| Total cholesterol, mmol/L ‡ | 5.6 ± 1.0 | 5.3 ± 1.0 | 5.9 ± 1.0 | <0.001 |
| High-density lipoprotein cholesterol, mmol/L ‡ | 1.4 ± 0.4 | 1.0 ± 0.3 | 1.6 ± 0.4 | <0.001 |
| Hypertension, n (%) | 1558 (77.9) | 654 (76.2) | 904 (79.2) | 0.109 |
| Smoking status, n (%) | <0.001 | |||
| Never | 599 (30.0) | 117 (13.6) | 482 (42.2) | |
| Former | 1094 (54.7) | 592 (69.0) | 502 (44.0) | |
| Current | 306 (15.3) | 149 (17.4) | 157 (13.8) | |
| History of diabetes mellitus, n (%) | 268 (13.4) | 145 (16.9) | 123 (10.8) | <0.001 |
| History of coronary heart disease, n (%) | 213 (10.7) | 145 (16.9) | 68 (6.0) | <0.001 |
| History of heart failure, n (%) | 101 (5.1) | 50 (5.8) | 51 (4.5) | 0.170 |
| Left ventricular hypertrophy, n (%) | 108 (5.4) | 62 (7.2) | 46 (4.0) | 0.002 |
| Cardiac medication, n (%) | 210 (10.5) | 102 (11.9) | 108 (9.5) | 0.108 |
| Lipid lowering medication, n (%) | 450 (22.5) | 208 (24.2) | 242 (21.2) | 0.080 |
| OR (95% CI) | ||||
|---|---|---|---|---|
| Model 1 * | p-Value | Model 2 † | p-Value | |
| Total Study Population | ||||
| miR-4798-3p | 0.64 (0.44–0.97) | 0.028033 | 0.63 (0.42–0.99) | 0.034433 |
| Men | ||||
| miR-4798-3p | 0.42 (0.27–0.69) | 0.000254 | 0.39 (0.24–0.66) | 0.000248 |
| Women | ||||
| miR-4798-3p | 1.53 (0.71–3.70) | 0.311964 | 1.84 (0.76–4.97) | 0.203587 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geurts, S.; Mens, M.M.J.; Bos, M.M.; Ikram, M.A.; Ghanbari, M.; Kavousi, M. Circulatory MicroRNAs in Plasma and Atrial Fibrillation in the General Population: The Rotterdam Study. Genes 2022, 13, 11. https://doi.org/10.3390/genes13010011
Geurts S, Mens MMJ, Bos MM, Ikram MA, Ghanbari M, Kavousi M. Circulatory MicroRNAs in Plasma and Atrial Fibrillation in the General Population: The Rotterdam Study. Genes. 2022; 13(1):11. https://doi.org/10.3390/genes13010011
Chicago/Turabian StyleGeurts, Sven, Michelle M. J. Mens, Maxime M. Bos, M. Arfan Ikram, Mohsen Ghanbari, and Maryam Kavousi. 2022. "Circulatory MicroRNAs in Plasma and Atrial Fibrillation in the General Population: The Rotterdam Study" Genes 13, no. 1: 11. https://doi.org/10.3390/genes13010011
APA StyleGeurts, S., Mens, M. M. J., Bos, M. M., Ikram, M. A., Ghanbari, M., & Kavousi, M. (2022). Circulatory MicroRNAs in Plasma and Atrial Fibrillation in the General Population: The Rotterdam Study. Genes, 13(1), 11. https://doi.org/10.3390/genes13010011






