Dorsal Striatum Transcriptome Profile Profound Shift in Repeated Aggression Mouse Model Converged to Networks of 12 Transcription Factors after Fighting Deprivation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Experimental Procedures
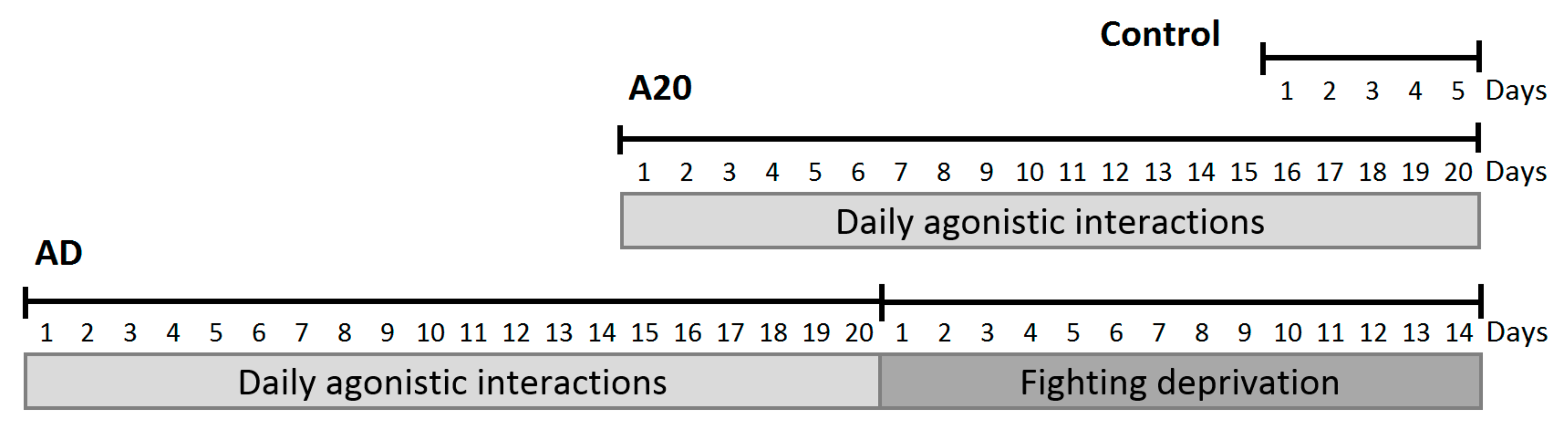
Modeling Repeated Aggression in Male Mice
2.3. RNA-Seq Data Collection and Processing
2.4. Statistical Analysis
3. Results
3.1. Detecting DEGs in Three Pairwise Comparisons
3.2. C_AD Comparison
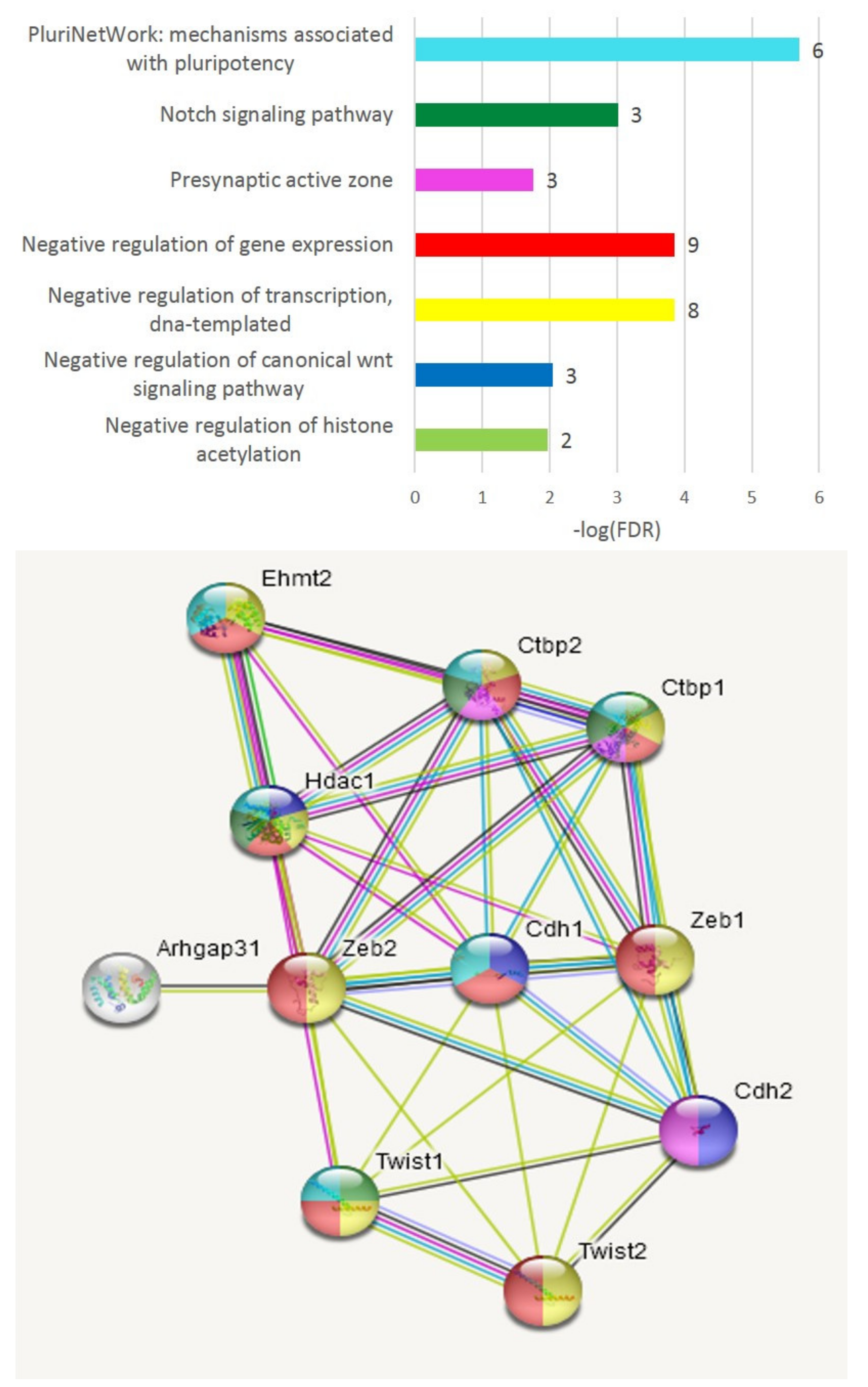
3.2.1. Expansion of TF Clusters by Co-Variation Analysis
3.2.2. Control Six-TF DEGs Assessment
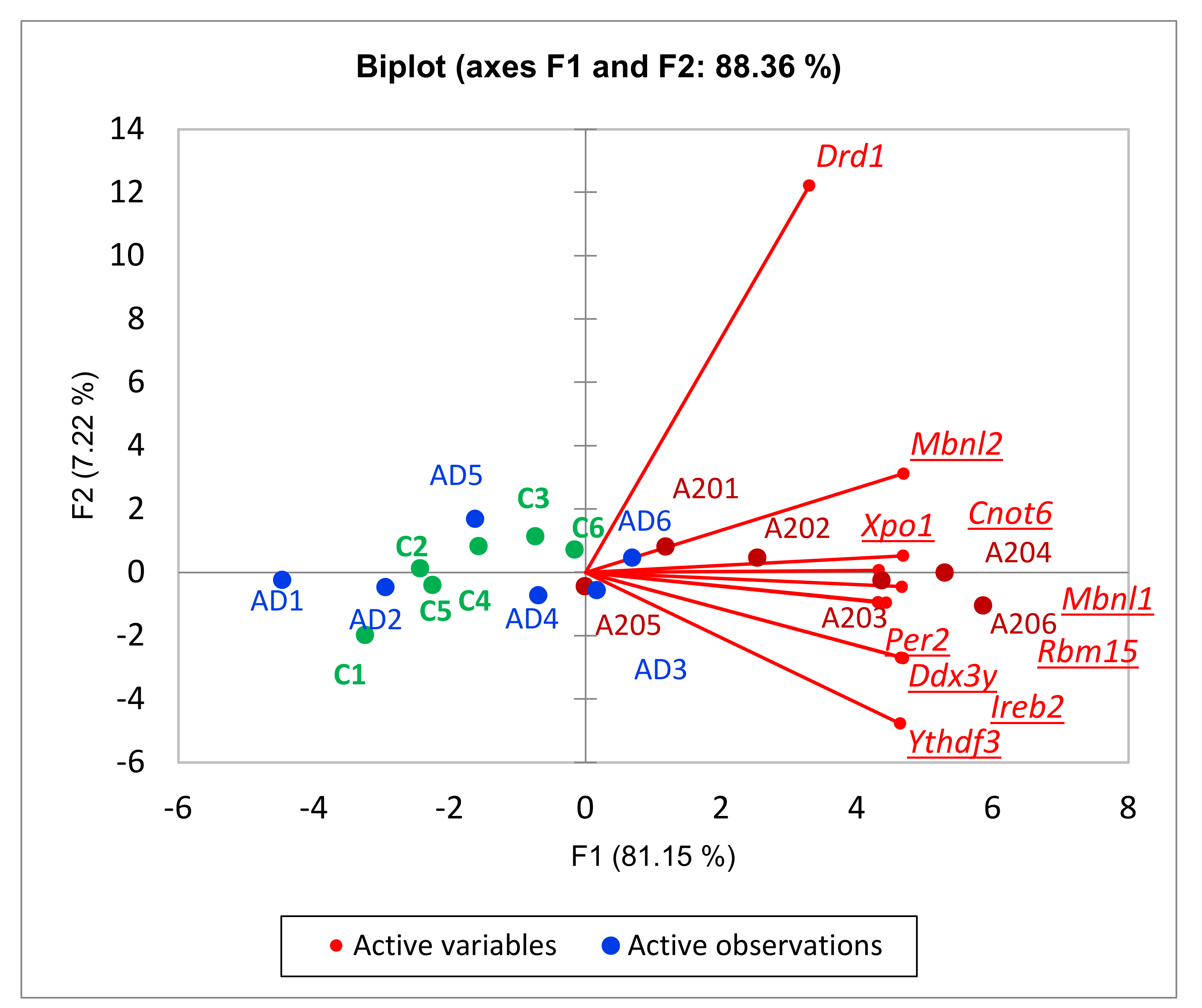
3.2.3. Circadian Rhythm DEG Analysis
3.2.4. Cluster Two Expansion
3.2.5. Third Cluster Featuring Joint A20/AD Species
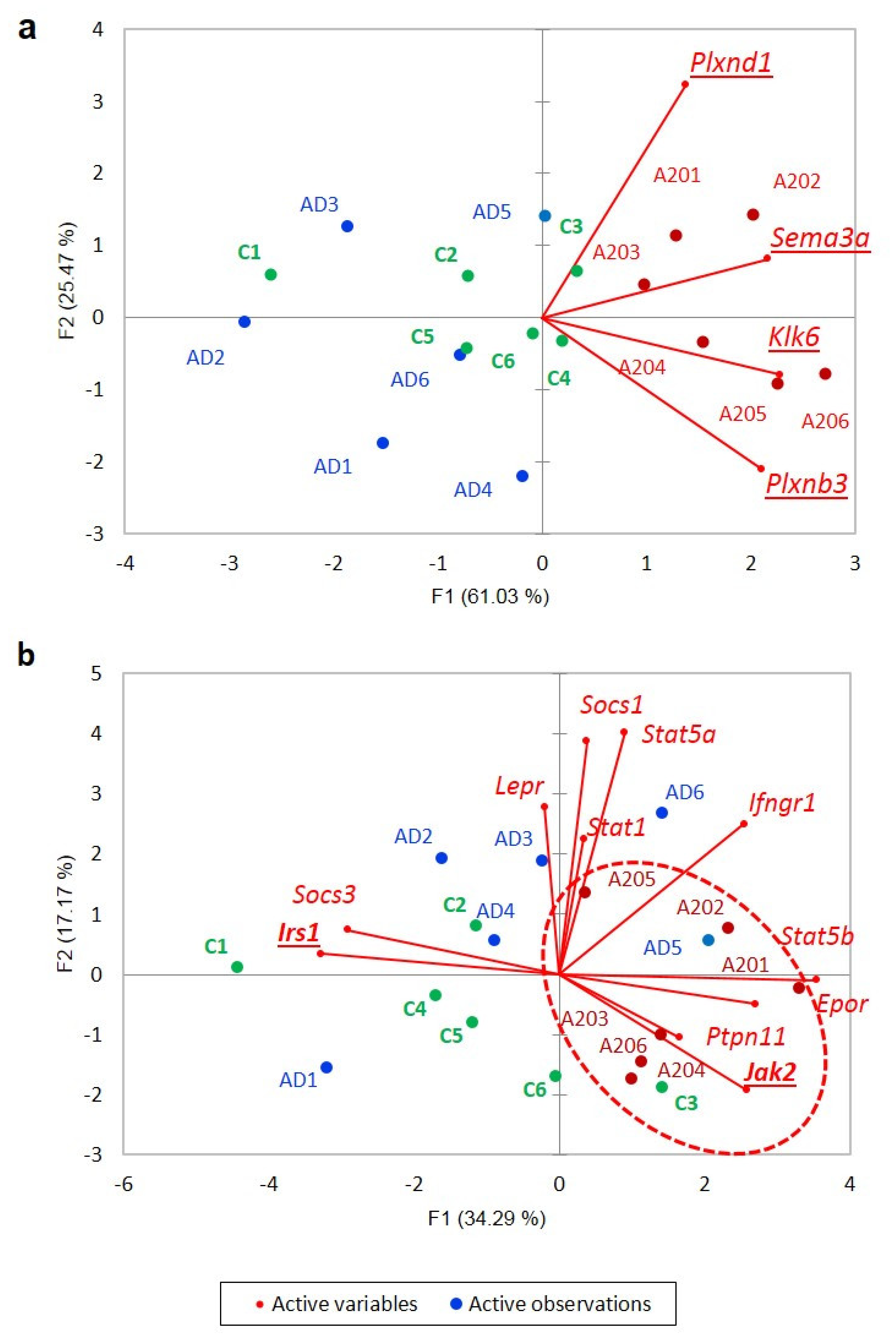
3.3. A20 vs. AD Comparison
3.3.1. Differentially Expressed Genes between Groups
3.3.2. Elaborating on A20-Associated Positive DEG Cluster
3.3.3. Dopamine-Mediated cAMP Signaling Cascade Balanced Gene Projection in A20 vs. AD Comparison
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DEGs | differentially expressed genes |
| FPKM | fragments per kilobase of transcript per million mapped reads |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes Pathway Database |
References
- Seto, M.C.; Healey, L.V.; Ahmed, A.G. Legally Mandated, Formally Pressured, or Voluntary Anger Treatment: Associations with Treatment Recommendations, Refusal, and Completion. J. Interpers. Violence 2021. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N. Straub tail, the deprivation effect and addiction to aggression. In Motivation of Health Behavior; O’Neal, P.W., Ed.; NOVA Science Publishers: New York, NY, USA, 2007; pp. 97–110. [Google Scholar]
- Kudryavtseva, N.N.; Smagin, D.A.; Bondar, N.P. Modeling fighting deprivation effect in mouse repeated aggression paradigm. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2011, 35, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Kudryavtseva, N.N. Positive fighting experience, addiction-like state, and relapse: Retrospective analysis of experimental studies. Aggress. Violent Behav. 2020, 52, 101403. [Google Scholar] [CrossRef]
- Basile, G.A.; Bertino, S.; Bramanti, A.; Ciurleo, R.; Anastasi, G.P.; Milardi, D.; Cacciola, A. Striatal topographical organization: Bridging the gap between molecules, connectivity and behavior. Eur. J. Histochem. 2021, 65, 3284. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.W.; Jonkman, S.; Difeliceantonio, A.G.; O’Connor, R.M.; Ghoshal, S.; Romano, M.F.; Everitt, B.J.; Kenny, P.J. Opposing roles for striatonigral and striatopallidal neurons in dorsolateral striatum in consolidating new instrumental actions. Nat. Commun. 2021, 12, 5121. [Google Scholar] [CrossRef]
- Plattner, F.; Hayashi, K.; Hernandez-Cortes, A.; Benavides, D.R.; Tassin, T.C.; Tan, C.; Day, J.P.; Fina, M.W.; Yuen, E.Y.; Yan, Z.; et al. The role of ventral striatal cAMP signaling in stress-induced behaviors. Nat. Neurosci. 2015, 18, 1094–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishi, A.; Matamales, M.; Musante, V.; Valjent, E.; Kuroiwa, M.; Kitahara, Y.; Rebholz, H.; Greengard, P.; Girault, J.-A.; Nairn, A.C.; et al. Glutamate Counteracts Dopamine/PKA Signaling via Dephosphorylation of DARPP-32 Ser-97 and Alteration of Its Cytonuclear Distribution. J. Biol. Chem. 2017, 292, 1462–1476. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, A.; Chung, J.-R.; Zhang, S.; Zhang, H.; Grossman, Y.; Aleyasin, H.; Flanigan, M.E.; Pfau, M.L.; Menard, C.; Dumitriu, D.; et al. Establishment of a repeated social defeat stress model in female mice. Sci. Rep. 2017, 7, 12838. [Google Scholar] [CrossRef] [Green Version]
- Yapo, C.; Nair, A.G.; Clement, L.; Castro, L.R.; Kotaleski, J.H.; Vincent, P. Detection of phasic dopamine by D1 and D2 striatal medium spiny neurons. J. Physiol. 2017, 595, 7451–7475. [Google Scholar] [CrossRef] [Green Version]
- Babenko, V.N.; Galyamina, A.G.; Rogozin, I.B.; Smagin, D.A.; Kudryavtseva, N.N. Dopamine response gene pathways in dorsal striatum MSNs from a gene expression viewpoint: cAMP-mediated gene networks. BMC Neurosci. 2020, 21, 12–14. [Google Scholar] [CrossRef] [Green Version]
- Hikida, T.; Morita, M.; Kuroiwa, M.; Macpherson, T.; Shuto, T.; Sotogaku, N.; Niwa, M.; Sawa, A.; Nishi, A. Adolescent psychosocial stress enhances sensitization to cocaine exposure in genetically vulnerable mice. Neurosci. Res. 2020, 151, 38–45. [Google Scholar] [CrossRef]
- Ophuis, R.J.A.O.; Boender, A.J.; Van Rozen, A.J.; Adan, R.A.H. Cannabinoid, melanocortin and opioid receptor expression on DRD1 and DRD2 subpopulations in rat striatum. Front. Neuroanat. 2014, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- Bazov, I.; Sarkisyan, D.; Kononenko, O.; Watanabe, H.; Yakovleva, T.; Hansson, A.C.; Sommer, W.H.; Spanagel, R.; Bakalkin, G. Dynorphin and κ-Opioid Receptor Dysregulation in the Dopaminergic Reward System of Human Alcoholics. Mol. Neurobiol. 2018, 55, 7049–7061. [Google Scholar] [CrossRef] [Green Version]
- Gipson, C.D.; Kupchik, Y.M.; Kalivas, P.W. Rapid, transient synaptic plasticity in addiction. Neuropharmacology 2014, 76, 276–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiga, S.; Mulas, G.; Piras, F.; Diana, M. The “addicted” spine. Front. Neuroanat. 2014, 8, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hotulainen, P.; Hoogenraad, C.C. Actin in dendritic spines: Connecting dynamics to function. J. Cell Biol. 2010, 189, 619–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spijker, S.; Houtzager, S.W.J.; de Gunst, M.C.M.; de Boer, W.P.H.; Schoffelmeer, A.N.M.; Smit, A.B. Morphine exposure and abstinence define specific stages of gene expression in the rat nucleus accumbens. FASEB J. 2004, 18, 848–850. [Google Scholar] [CrossRef]
- Russo, S.J.; Dietz, D.; Dumitriu, D.; Morrison, J.H.; Malenka, R.C.; Nestler, E.J. The addicted synapse: Mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 2010, 33, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Chung, W.-S.; Clarke, L.E.; Wang, G.X.; Stafford, B.K.; Sher, A.; Chakraborty, C.; Joung, J.; Foo, L.C.; Thompson, A.; Chen, C.; et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 2013, 504, 394–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, B.L.; Oscar-Berman, M.; Kaplan, G.B. Opioid-induced structural and functional plasticity of medium-spiny neurons in the nucleus accumbens. Neurosci. Biobehav. Rev. 2021, 120, 417–430. [Google Scholar] [CrossRef]
- Robinson, T.E.; Berridge, K.C. Addiction. Annu. Rev. Psychol. 2003, 54, 25–53. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N.; Smagin, D.; Kovalenko, I.L.; Vishnivetskaya, G.B. Repeated positive fighting experience in male inbred mice. Nat. Protoc. 2014, 9, 2705–2717. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N. The sensory contact model for the study of aggressive and submissive behaviors in male mice. Aggress. Behav. 1991, 17, 285–291. [Google Scholar] [CrossRef]
- The Allen Mouse Brain Atlas. Available online: http://mouse.brain-map.org/static/atlas (accessed on 24 April 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11.14.1–11.14.19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Castañeda, T.R.; de Prado, B.M.; Prieto, D.; Mora, F. Circadian rhythms of dopamine, glutamate and GABA in the striatum and nucleus accumbens of the awake rat: Modulation by light. J. Pineal Res. 2004, 36, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Ogura, H.; Aruga, J.; Mikoshiba, K. Behavioral Abnormalities of Zic1 and Zic2 Mutant Mice: Implications as Models for Human Neurological Disorders. Behav. Genet. 2001, 31, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Maruotti, N.; Corrado, A.; Neve, A.; Cantatore, F.P. Systemic effects of Wnt signaling. J. Cell. Physiol. 2013, 228, 1428–1432. [Google Scholar] [CrossRef]
- Lashgari, N.-A.; Roudsari, N.M.; Momtaz, S.; Sathyapalan, T.; Abdolghaffari, A.H.; Sahebkar, A. The involvement of JAK/STAT signaling pathway in the treatment of Parkinson’s disease. J. Neuroimmunol. 2021, 361, 577758. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.-D.; Gainetdinov, R.; Arbuckle, M.I.; Sotnikova, T.D.; Cyr, M.; Beaulieu, J.-M.; Torres, G.E.; Grant, S.; Caron, M.G. Identification of PSD-95 as a Regulator of Dopamine-Mediated Synaptic and Behavioral Plasticity. Neuron 2004, 41, 625–638. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xu, T.-X.; Hallett, P.J.; Watanabe, M.; Grant, S.G.N.; Isacson, O.; Yao, W.-D. PSD-95 Uncouples Dopamine-Glutamate Interaction in the D1/PSD-95/NMDA Receptor Complex. J. Neurosci. 2009, 29, 2948–2960. [Google Scholar] [CrossRef]
- Zhang, J.; Saur, T.; Duke, A.N.; Grant, S.G.N.; Platt, N.M.; Rowlett, J.K.; Isacson, O.; Yao, W.-D. Motor impairments, striatal degeneration, and altered dopamine-glutamate interplay in mice lacking PSD-95. J. Neurogenet. 2014, 28, 98–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ökvist, A.; Fagergren, P.; Whittard, J.; Garcia-Osta, A.; Drakenberg, K.; Horvath, M.C.; Schmidt, C.J.; Keller, E.; Bannon, M.J.; Hurd, Y.L. Dysregulated postsynaptic density and endocytic zone in the amygdala of human heroin and cocaine abusers. Biol. Psychiatry 2011, 69, 245–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Guan, Y.; Ma, Y.; Xue, L.; Zhang, P.; Yang, X.; Chong, T. Bioinformatic analyses and experimental validation of the role of m6A RNA methylation regulators in progression and prognosis of adrenocortical carcinoma. Aging 2021, 13, 11919–11941. [Google Scholar] [CrossRef] [PubMed]
- Just, L.; Olenik, C.; Meyer, D.K. Glial Expression of the Proenkephalin Gene in Slice Cultures of the Subventricular Zone. J. Mol. Neurosci. 1998, 11, 57–66. [Google Scholar] [CrossRef]
- Cingolani, L.A.; Goda, Y. Actin in action: The interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 2008, 9, 344–356. [Google Scholar] [CrossRef]
- Roy, K.; Bhattacharyya, P.; Deb, I. Naloxone precipitated morphine withdrawal and clock genes expression in striatum: A comparative study in three different protocols for the development of morphine dependence. Neurosci. Lett. 2018, 685, 24–29. [Google Scholar] [CrossRef]
- Kim, M.; Custodio, R.J.; Botanas, C.J.; De La Peña, J.B.; Sayson, L.V.; Abiero, A.; Ryoo, Z.Y.; Cheong, J.H.; Kim, H.J. The circadian gene, Per2, influences methamphetamine sensitization and reward through the dopaminergic system in the striatum of mice. Addict. Biol. 2019, 24, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Perreau-Lenz, S.; Sanchis-Segura, C.; Leonardi-Essmann, F.; Schneider, M.; Spanagel, R. Development of morphine-induced tolerance and withdrawal: Involvement of the clock gene mPer2. Eur. Neuropsychopharmacol. 2010, 20, 509–517. [Google Scholar] [CrossRef]
- Piechota, M.; Korostynski, M.; Sikora, M.; Golda, S.; Dzbek, J.; Przewlocki, R. Common transcriptional effects in the mouse striatum following chronic treatment with heroin and methamphetamine. Genes Brain Behav. 2012, 11, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; Maji, D.; Deb, I. Increase of Cry 1 expression is a common phenomenon of the disturbed circadian clock in ischemic stroke and opioid addiction. Biochem. Biophys. Res. Commun. 2021, 558, 8–13. [Google Scholar] [CrossRef]
- Tamura, E.K.; Oliveira-Silva, K.S.; Ferreira-Moraes, F.A.; Marinho, E.A.; Guerrero-Vargas, N.N. Circadian rhythms and substance use disorders: A bidirectional relationship. Pharmacol. Biochem. Behav. 2021, 201, 173105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Moye, L.S.; Southey, B.R.; Dripps, I.; Sweedler, J.V.; Pradhan, A.; Rodriguez-Zas, S.L. Opioid-Induced Hyperalgesia Is Associated with Dysregulation of Circadian Rhythm and Adaptive Immune Pathways in the Mouse Trigeminal Ganglia and Nucleus Accumbens. Mol. Neurobiol. 2019, 56, 7929–7949. [Google Scholar] [CrossRef] [PubMed]
- Tordjman, S.; Carlier, M.; Cohen, D.; Cesselin, F.; Bourgoin, S.; Colas-Linhart, N.; Petiet, A.; Perez-Diaz, F.; Hamon, M.; Roubertoux, P.L. Aggression and the three opioid families (endorphins, enkephalins, and dynorphins) in mice. Behav. Genet. 2003, 33, 529–536. [Google Scholar] [CrossRef]











| Number of DEGs (FDR < 0.05) | Number of Non-Zero Expressed Gene Pairs | |
|---|---|---|
| Control vs. A20 | 1030 | 24,321 |
| Control vs. AD | 62 | 23,924 |
| A20 vs. AD | 1009 | 23,763 |
| Variables | Nr1d1 | Cry2 | Per1 | Npas2 | Arntl | Cry1 | Clock | Per2 |
|---|---|---|---|---|---|---|---|---|
| Nr1d1 | 1 | 0.603 | 0.312 | −0.247 | −0.620 | −0.288 | −0.305 | 0.025 |
| Cry2 | 0.603 | 1 | 0.656 | −0.087 | −0.392 | 0.179 | −0.138 | 0.447 |
| Per1 | 0.312 | 0.656 | 1 | −0.226 | −0.474 | 0.297 | 0.126 | 0.669 |
| Npas2 | −0.247 | −0.087 | −0.226 | 1 | 0.552 | 0.510 | 0.223 | 0.065 |
| Arntl | −0.620 | −0.392 | −0.474 | 0.552 | 1 | 0.325 | 0.369 | −0.067 |
| Cry1 | −0.288 | 0.179 | 0.297 | 0.510 | 0.325 | 1 | 0.323 | 0.786 |
| Clock | −0.305 | −0.138 | 0.126 | 0.223 | 0.369 | 0.323 | 1 | 0.274 |
| Per2 | 0.025 | 0.447 | 0.669 | 0.065 | –0.067 | 0.786 | 0.274 | 1 |
| Family | Min (q_Value) | Max (q_Value) | Stddev (q_Value) | # DEGs per Family |
|---|---|---|---|---|
| Slc | 1.35 × 10−3 | 3.63 × 10−2 | 1.05 × 10−2 | 29 |
| Kcn | 1.35 × 10−3 | 4.92 × 10−2 | 1.31 × 10−2 | 22 |
| Tme | 1.35 × 10−3 | 4.88 × 10−2 | 1.50 × 10−2 | 17 |
| Zfp | 1.35 × 10−3 | 4.97 × 10−2 | 1.38 × 10−2 | 15 |
| Col | 1.35 × 10−3 | 4.39 × 10−2 | 1.28 × 10−2 | 10 |
| Arh | 1.35 × 10−3 | 4.88 × 10−2 | 1.66 × 10−2 | 8 |
| Ptp | 1.35 × 10−3 | 2.59 × 10−2 | 1.04 × 10−2 | 7 |
| Map | 1.35 × 10−3 | 2.31 × 10−2 | 7.40 × 10−3 | 7 |
| Gpr | 1.35 × 10−3 | 2.10 × 10−2 | 7.21 × 10−3 | 7 |
| Kif | 1.35 × 10−3 | 2.24 × 10−2 | 7.86 × 10−3 | 6 |
| Cac | 1.35 × 10−3 | 4.56 × 10−2 | 1.89 × 10−2 | 6 |
| Tri | 1.35 × 10−3 | 3.10 × 10−2 | 1.30 × 10−2 | 6 |
| Ank | 1.35 × 10−3 | 4.56 × 10−2 | 1.51 × 10−2 | 6 |
| Fam | 1.35 × 10−3 | 1.37 × 10−2 | 4.58 × 10−3 | 6 |
| Cdk | 1.35 × 10−3 | 4.45 × 10−2 | 1.72 × 10−2 | 5 |
| Rgs | 1.35 × 10−3 | 4.67 × 10−2 | 1.57 × 10−2 | 5 |
| Prr | 1.35 × 10−3 | 3.76 × 10−2 | 1.38 × 10−2 | 5 |
| Aka | 1.35 × 10−3 | 2.51 × 10−2 | 9.33 × 10−3 | 5 |
| Ple | 1.35 × 10−3 | 4.78 × 10−2 | 1.94 × 10−2 | 5 |
| Cep | 1.35 × 10−3 | 4.63 × 10−3 | 1.28 × 10−3 | 5 |
| Adc | 1.35 × 10−3 | 4.63 × 10−3 | 1.39 × 10−3 | 5 |
| Doc | 1.35 × 10−3 | 9.39 × 10−3 | 3.13 × 10−3 | 5 |
| Variables | Darpp-32 | Penk | Drd2 | Drd1 | Pdyn |
|---|---|---|---|---|---|
| Darpp-32 | 1 | 0.858 | 0.914 | 0.863 | 0.723 |
| Penk | 0.858 | 1 | 0.925 | 0.838 | 0.867 |
| Drd2 | 0.914 | 0.925 | 1 | 0.886 | 0.777 |
| Drd1 | 0.863 | 0.838 | 0.886 | 1 | 0.717 |
| Pdyn | 0.723 | 0.867 | 0.777 | 0.717 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babenko, V.; Redina, O.; Smagin, D.; Kovalenko, I.; Galyamina, A.; Babenko, R.; Kudryavtseva, N. Dorsal Striatum Transcriptome Profile Profound Shift in Repeated Aggression Mouse Model Converged to Networks of 12 Transcription Factors after Fighting Deprivation. Genes 2022, 13, 21. https://doi.org/10.3390/genes13010021
Babenko V, Redina O, Smagin D, Kovalenko I, Galyamina A, Babenko R, Kudryavtseva N. Dorsal Striatum Transcriptome Profile Profound Shift in Repeated Aggression Mouse Model Converged to Networks of 12 Transcription Factors after Fighting Deprivation. Genes. 2022; 13(1):21. https://doi.org/10.3390/genes13010021
Chicago/Turabian StyleBabenko, Vladimir, Olga Redina, Dmitry Smagin, Irina Kovalenko, Anna Galyamina, Roman Babenko, and Natalia Kudryavtseva. 2022. "Dorsal Striatum Transcriptome Profile Profound Shift in Repeated Aggression Mouse Model Converged to Networks of 12 Transcription Factors after Fighting Deprivation" Genes 13, no. 1: 21. https://doi.org/10.3390/genes13010021
APA StyleBabenko, V., Redina, O., Smagin, D., Kovalenko, I., Galyamina, A., Babenko, R., & Kudryavtseva, N. (2022). Dorsal Striatum Transcriptome Profile Profound Shift in Repeated Aggression Mouse Model Converged to Networks of 12 Transcription Factors after Fighting Deprivation. Genes, 13(1), 21. https://doi.org/10.3390/genes13010021







