Genome-Wide Association Mapping of Crown and Brown Rust Resistance in Perennial Ryegrass
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Phenotyping
2.2. Core Collection
2.3. DNA Isolation and Sequencing
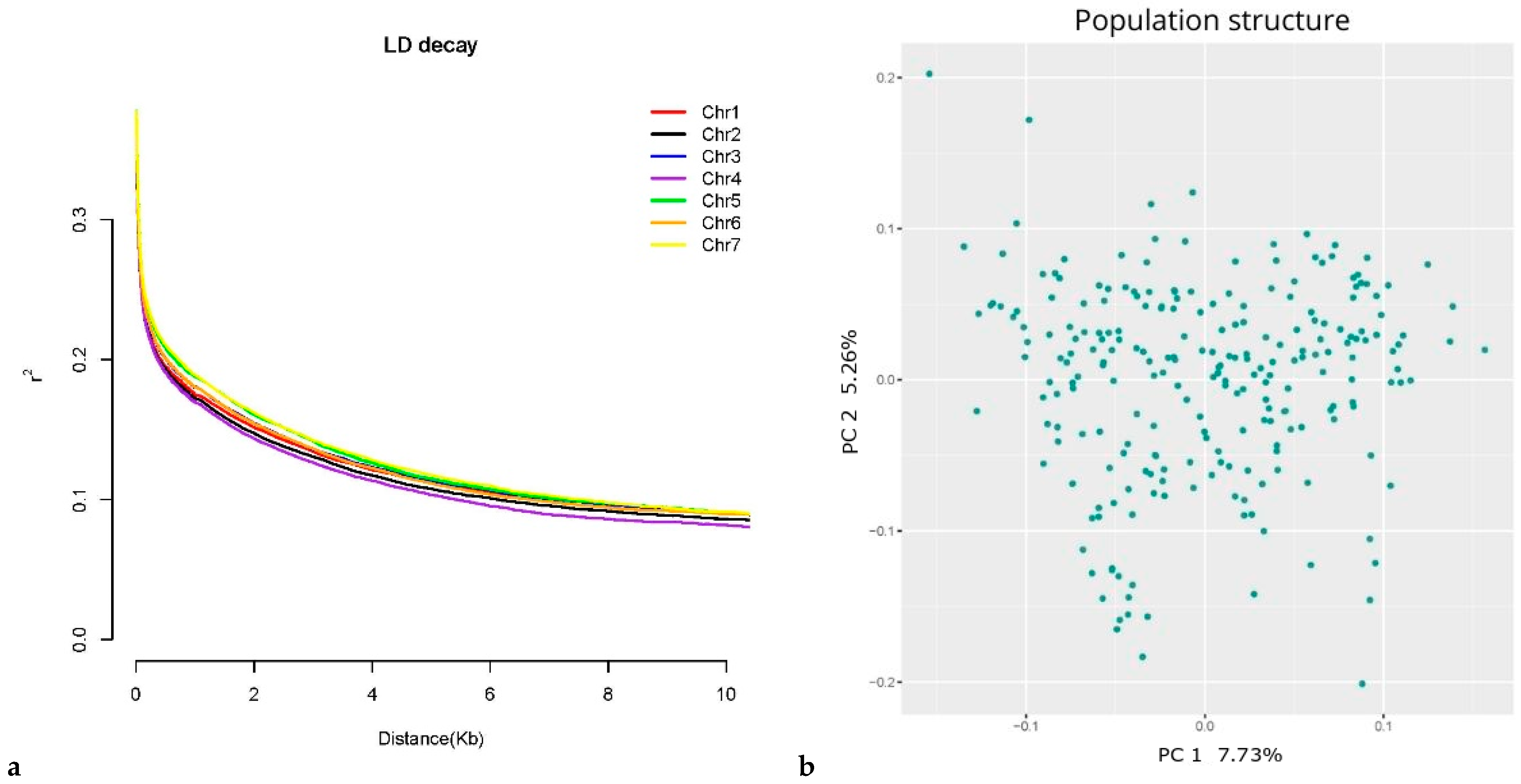
2.4. Genotyping, LD, and Population Structure
2.5. Genome-Wide Complex Trait Analysis (GCTA)
2.6. Multi-Marker Analysis of Genomic Annotation (MAGMA)
2.7. Annotation of QTL Regions
3. Results
3.1. Phenotypic Analysis
3.2. Molecular Markers, LD and Population Structure
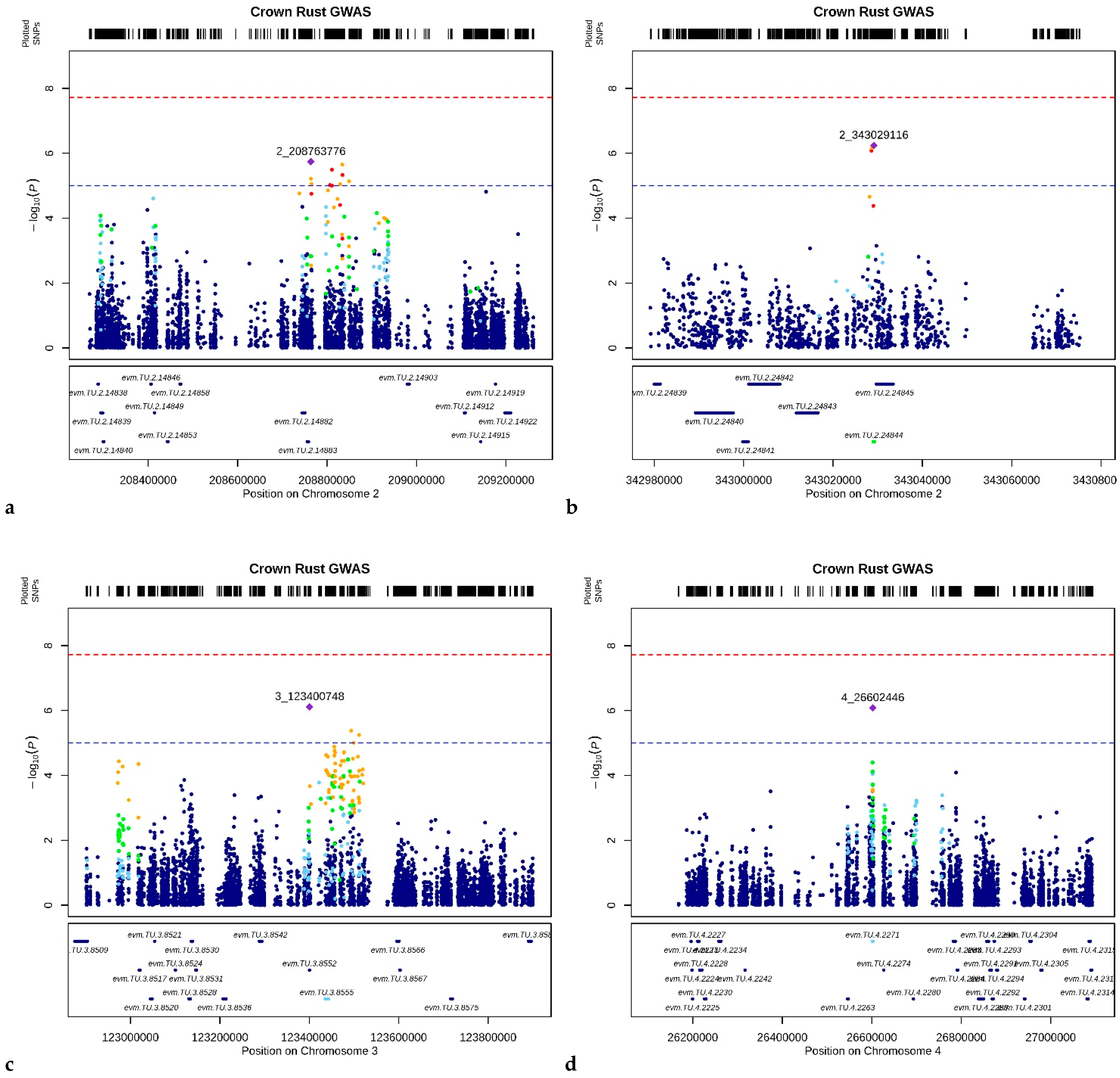
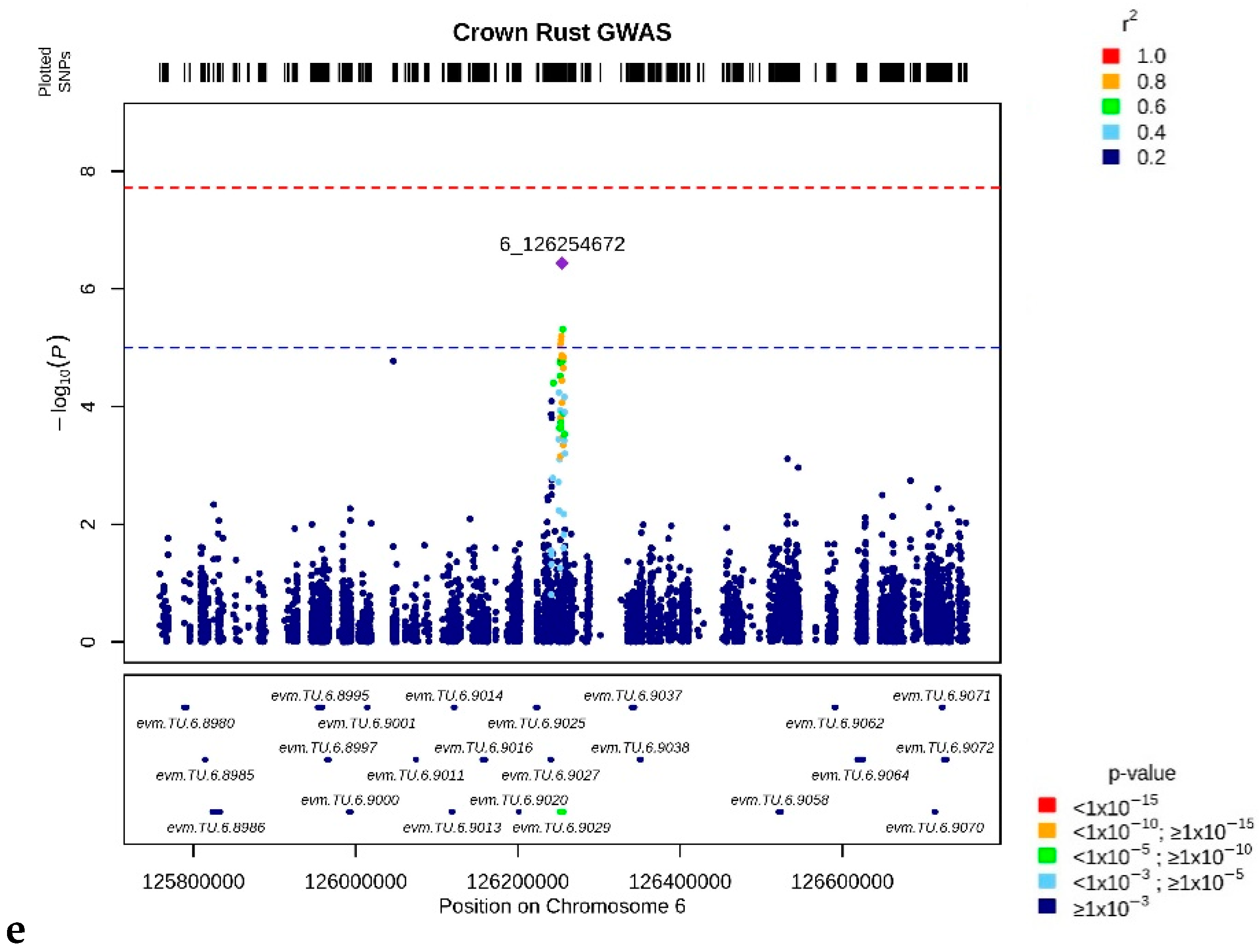
3.3. Crown Rust Resistance Loci
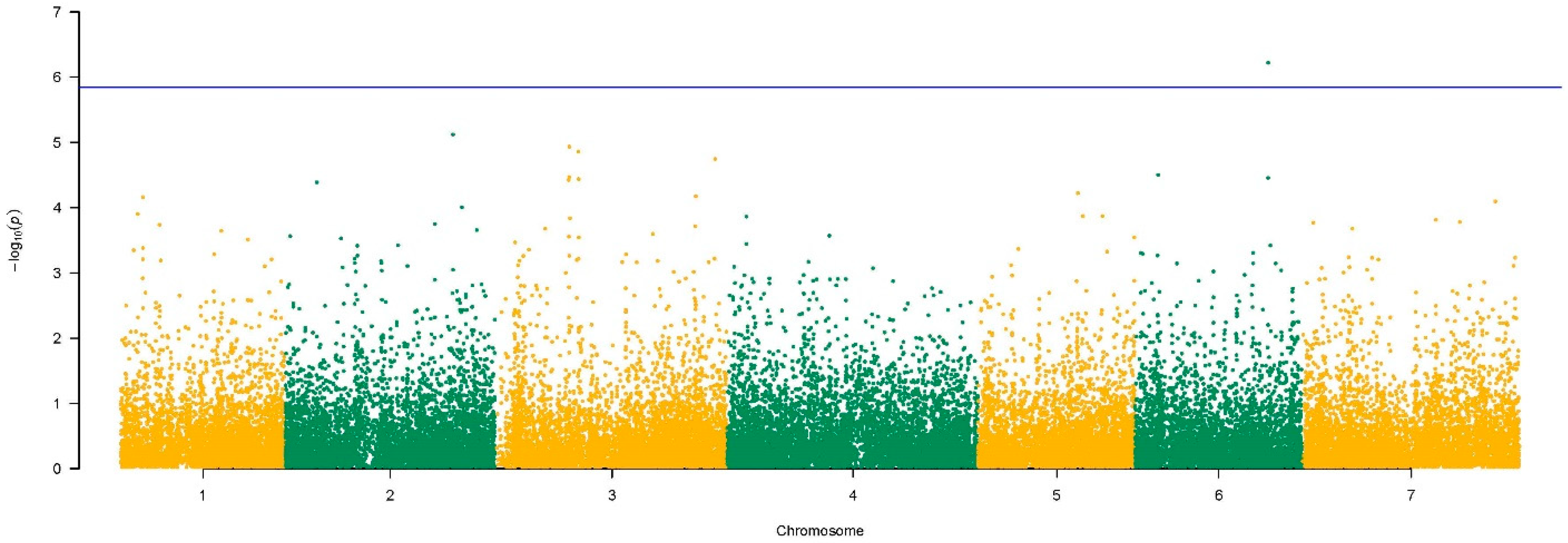
3.4. Brown Rust Resistance Loci
4. Discussion
4.1. Candidate Genes for Crown Rust Resistance
4.2. Candidate Genes for Brown Rust Resistance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Humphreys, M.O.; Feuerstein, U.; Vandewalle, M.; Baert, J. Ryegrasses. In Fodder Crops and Amenity Grasses. Handbook of Plant Breeding; Boller, B., Posselt, U.K., Veronesi, F., Eds.; Springer: New York, NY, USA, 2010; Volume 5, pp. 211–260. [Google Scholar]
- Duller, S.; Thorogood, D.; Bonos, S. (Eds.) Breeding Objectives in Amenity Grasses; Springer: New York, NY, USA, 2010. [Google Scholar]
- Kopecký, D.; Havránková, M.; Loureiro, J.; Castro, S.; Lukaszewski, A.J.; Bartoš, J.; Kopecká, J.; Doležel, J. Physical Distribution of Homoeologous Recombination in Individual Chromosomes of Festuca pratensis in Lolium multiflorum. Cytogenet. Genome Res. 2010, 129, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Mattner, S.W.; Parbery, D.G. Rust-Enhanced Allelopathy of Perennial Ryegrass against White Clover. Agron. J. 2001, 93, 54–59. [Google Scholar] [CrossRef]
- Potter, L.R. Effect of crown rust on regrowth, competitive ability and nutritional quality of perennial and Italian ryegrasses. Plant Pathol. 1987, 36, 455–461. [Google Scholar] [CrossRef]
- Grimes, R.C.; Watkin, B.R.; Gallagher, J.R. The growth of lambs grazing on perennial ryegrass, tall fescue and cocksfoot, with and without white clover, as related to the botanical and chemical composition of the pasture and pattern of fermentation in the rumen. J. Agric. Sci. 1967, 68, 11–21. [Google Scholar] [CrossRef]
- Humphreys, M.O. Water-soluble carbohydrates in perennial ryegrass breeding. Grass Forage Sci. 1989, 44, 423–430. [Google Scholar] [CrossRef]
- McKenzie, E.H.C. Seasonal changes in fungal spore numbers in ryegrass white clover pasture, and the effects of benomyl on pasture fungi. N. Z. J. Agric. Res. 1971, 14, 379–392. [Google Scholar] [CrossRef][Green Version]
- Thomas, H.W.R.B.J. Infection of ryegrass by three rust fungi (Puccinia coronata, P. graminis and P. loliina) and some effects of temperature on the establishment of the disease and sporulation. Plant Pathol. 1997, 46, 751–761. [Google Scholar]
- Dracatos, P.M.; Cogan, N.O.I.; Keane, P.J.; Smith, K.F.; Forster, J.W. Biology and Genetics of Crown Rust Disease in Ryegrasses. Crop Sci. 2010, 50, 1605–1624. [Google Scholar] [CrossRef]
- Pasquali, E.; Barcaccia, G. Genomics Applied to the Analysis of Flowering Time, Abiotic Stress Tolerance and Disease Resistance: A Review of What We Have Learned in Lolium spp. Agriculture 2020, 10, 425. [Google Scholar] [CrossRef]
- Rietman, H.; Bijsterbosch, G.; Cano, L.M.; Lee, H.R.; Vossen, J.H.; Jacobsen, E.; Visser, R.G.; Kamoun, S.; Vleeshouwers, V.G. Qualitative and Quantitative Late Blight Resistance in the Potato Cultivar Sarpo Mira Is Determined by the Perception of Five Distinct RXLR Effectors. Mol. Plant-Microbe Interact. 2012, 25, 910–919. [Google Scholar] [CrossRef]
- Sanz, M.J.; Loarce, Y.; Fominaya, A.; Vossen, J.H.; Ferrer, E. Identification of RFLP and NBS/PK profiling markers for disease resistance loci in genetic maps of oats. Theor. Appl. Genet. 2013, 126, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Mundt, C.C. Durable resistance: A key to sustainable management of pathogens and pests. Infect. Genet. Evol. 2014, 27, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Schielzeth, H.; Husby, A. Challenges and prospects in genome-wide quantitative trait loci mapping of standing genetic variation in natural populations. Ann. N. Y. Acad. Sci. 2014, 1320, 35–57. [Google Scholar] [CrossRef] [PubMed]
- Fè, D.; Cericola, F.; Byrne, S.; Lenk, I.; Ashraf, B.H.; Pedersen, M.G.; Roulund, N.; Asp, T.; Janss, L.; Jensen, C.S.; et al. Genomic dissection and prediction of heading date in perennial ryegrass. BMC Genom. 2015, 16, 921. [Google Scholar] [CrossRef] [PubMed]
- Kovi, M.R.; Fjellheim, S.; Sandve, S.R.; Larsen, A.; Rudi, H.; Asp, T.; Kent, M.P.; Rognli, O.A. Population Structure, Genetic Variation, and Linkage Disequilibrium in Perennial Ryegrass Populations Divergently Selected for Freezing Tolerance. Front. Plant Sci. 2015, 6, 929. [Google Scholar] [CrossRef]
- Xing, Y.; Frei, U.; Schejbel, B.; Asp, T.; Lubberstedt, T. Nucleotide diversity and linkage disequilibrium in 11 expressed resistance candidate genes in Lolium perenne. BMC Plant Biol. 2007, 7, 43. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef]
- The Nordic Public Private Partnership. 2018. Available online: https://www.nordgen.org/en/our-work/nordic-public-private-partnership-ppp/ppp-projects/ (accessed on 1 February 2020).
- Douglas, B.; Martin, M.c.h.l.e.r.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using {lme4}. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar]
- De Beukelaer, H.; Davenport, G.F.; Fack, V. Core Hunter 3: Flexible core subset selection. BMC Bioinform. 2018, 19, 203. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 1985, 5, 69–76. [Google Scholar] [CrossRef]
- Andrews, S. FASTQC (Version 0.11.9). Babraham Bioinforma. 2018. Available online: https://www.bioinformatics.babraham.ac.uk/projects (accessed on 5 February 2020).
- Krueger, F.; James, F.; Ewels, P.; Ebrahim Afyounian, B.S.-B. TrimGalore: v0.6.7. 2021. Available online: https://doi.org/10.5281/zenodo.5127899 (accessed on 20 August 2021). [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Picard. Available online: http://broadinstitute.github.io/picard (accessed on 1 September 2019).
- De Pristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.D. qqman; an R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Jing, Y.; Lin, R. The VQ Motif-Containing Protein Family of Plant-Specific Transcriptional Regulators. Plant Physiol. 2015, 169, 371–378. [Google Scholar] [CrossRef]
- Leveau, A.; Reed, J.; Qiao, X.; Stephenson, M.J.; Mugford, S.T.; Melton, R.E.; Rant, J.C.; Vickerstaff, R.; Langdon, T.; Osbourn, A. Towards take-all control: A C-21β oxidase required for acylation of triterpene defence compounds in oat. New Phytol. 2019, 221, 1544–1555. [Google Scholar] [CrossRef]
- Desrousseaux, D.; Sandron, F.; Siberchicot, A.; Cierco-Ayrolles, C.; Mangin, B. LDcorSV: Linkage Disequilibrium Corrected by the Structure and the Relatedness. R Package Version 1.3.3. 2020. Available online: https://cran.r-project.org/web/packages/LDcorSV/LDcorSV.pdf (accessed on 20 September 2020).
- Liu, W.; Ghouri, F.; Yu, H.; Li, X.; Yu, S.; Shahid, M.Q.; Liu, X. Genome wide re-sequencing of newly developed Rice Lines from common wild rice (Oryza rufipogon Griff.) for the identification of NBS-LRR genes. PLoS ONE 2017, 12, e0180662. [Google Scholar] [CrossRef]
- Hong, E.P.; Park, J.W. Sample size and statistical power calculation in genetic association studies. Genom. Inf. 2012, 10, 117–122. [Google Scholar] [CrossRef]
- Wang, M.; Xu, S. Statistical power in genome-wide association studies and quantitative trait locus mapping. Heredity 2019, 123, 287–306. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, L.; Luo, W.; Jin, Y.; Gong, F.; He, J.; Liu, D.; Zheng, Y.; Wu, B. Transcriptome analysis provides insights into the mechanisms underlying wheat cultivar Shumai126 responding to stripe rust. Gene 2021, 768, 145290. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Bhaganagare, G.; Bandopadhyay, R.; Prabhu, K.V.; Gupta, P.K.; Mukhopadhyay, K. Targeted spatio-temporal expression based characterization of state of infection and time-point of maximum defense in wheat NILs during leaf rust infection. Mol. Biol. Rep. 2012, 39, 9373–9382. [Google Scholar] [CrossRef] [PubMed]
- Zeisler-Diehl, V.V.; Barthlott, W.; Schreiber, L. Plant Cuticular Waxes: Composition, Function, and Interactions with Microorganisms. In Hydrocarbons, Oils and Lipids: Diversity, Origin, Chemistry and Fate; Springer: Cham, Switzerland, 2018; pp. 123–138. [Google Scholar] [CrossRef]
- Agrios, G.N. chapter six—How Plants Defend Themselves against Pathogens. In Plant Pathology, 5th ed.; Agrios, G.N., Ed.; Academic Press: Cambridge, MA, USA, 2005; pp. 207–248. [Google Scholar]
- Dumsday, J.L.; Smith, K.F.; Forster, J.W.; Jones, E.S. SSR-based genetic linkage analysis of resistance to crown rust (Puccinia coronata f. sp. lolii) in perennial ryegrass (Lolium perenne). Plant Pathol. 2003, 52, 628–637. [Google Scholar] [CrossRef]
- Muylle, H.; Baert, J.; Van Bockstaele, E.; Pertijs, J.; Roldán-Ruiz, I. Four QTLs determine crown rust (Puccinia coronata f. sp. lolii) resistance in a perennial ryegrass (Lolium perenne) population. Heredity 2005, 95, 348–357. [Google Scholar] [CrossRef][Green Version]
- Rödiger, A.; Galonska, J.; Bergner, E.; Agne, B.; Helm, S.; Alseekh, S.; Fernie, A.R.; Thieme, D.; Hoehenwarter, W.; Hause, G.; et al. Working day and night: Plastid casein kinase 2 catalyses phosphorylation of proteins with diverse functions in light- and dark-adapted plastids. Plant J. 2020, 104, 546–558. [Google Scholar] [CrossRef]
- Carretero, R.; Bancal, M.O.; Miralles, D.J. Effect of leaf rust (Puccinia triticina) on photosynthesis and related processes of leaves in wheat crops grown at two contrasting sites and with different nitrogen levels. Eur. J. Agron. 2011, 35, 237–246. [Google Scholar] [CrossRef]
- Zhang, H.; Fu, Y.; Guo, H.; Zhang, L.; Wang, C.; Song, W.; Yan, Z.; Wang, Y.; Ji, W. Transcriptome and Proteome-Based Network Analysis Reveals a Model of Gene Activation in Wheat Resistance to Stripe Rust. Int. J. Mol. Sci. 2019, 20, 1106. [Google Scholar] [CrossRef]
- Zhao, D.; Glynn, N.C.; Glaz, B.; Comstock, J.C.; Sood, S. Orange Rust Effects on Leaf Photosynthesis and Related Characters of Sugarcane. Plant Dis. 2011, 95, 640–647. [Google Scholar] [CrossRef]
- Wang, Y.; Chang, H.; Hu, S.; Lu, X.; Yuan, C.; Zhang, C.; Wang, P.; Xiao, W.; Xiao, L.; Xue, G.P.; et al. Plastid casein kinase 2 knockout reduces abscisic acid (ABA) sensitivity, thermotolerance, and expression of ABA- and heat-stress-responsive nuclear genes. J. Exp. Bot. 2014, 65, 4159–4175. [Google Scholar] [CrossRef]
- Kanneganti, V.; Gupta, A.K. Wall associated kinases from plants—An overview. Physiol. Mol. Biol. Plants 2008, 14, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Ashikawa, I.; Hayashi, N.; Abe, F.; Wu, J.; Matsumoto, T. Characterization of the rice blast resistance gene Pik cloned from Kanto51. Mol. Breed. 2012, 30, 485–494. [Google Scholar] [CrossRef]
- Chen, S.; Guo, Y.; Briggs, J.; Dubach, F.; Chao, S.; Zhang, W.; Rouse, M.N.; Dubcovsky, J. Mapping and characterization of wheat stem rust resistance genes SrTm5 and Sr60 from Triticum monococcum. Theor. Appl. Genet. 2018, 131, 625–635. [Google Scholar] [CrossRef]
- Cloutier, S.; McCallum, B.D.; Loutre, C.; Banks, T.W.; Wicker, T.; Feuillet, C.; Keller, B.; Jordan, M.C. Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol. Biol. 2007, 65, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting Mechanisms of Defense Against Biotrophic and Necrotrophic Pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Chiba, Y.; Shimizu, T.; Miyakawa, S.; Kanno, Y.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J. Plant Res. 2015, 128, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Fu, X.; Yang, C.; Tang, X.; Guo, L.; Li, C.; Xu, C.; Luo, K. Genome-wide investigation of pentatricopeptide repeat gene family in poplar and their expression analysis in response to biotic and abiotic stresses. Sci. Rep. 2018, 8, 2817. [Google Scholar] [CrossRef]
- Schejbel, B.; Jensen, L.B.; Xing, Y.; Lübberstedt, T. QTL analysis of crown rust resistance in perennial ryegrass under conditions of natural and artificial infection. Plant Breed. 2007, 126, 347–352. [Google Scholar] [CrossRef]
- Montilla-Bascón, G.; Rispail, N.; Sánchez-Martín, J.; Rubiales, D.; Mur, L.A.; Langdon, T.; Howarth, C.J.; Prats, E. Genome-wide association study for crown rust (Puccinia coronata f. sp. avenae) and powdery mildew (Blumeria graminis f. sp. avenae) resistance in an oat (Avena sativa) collection of commercial varieties and landraces. Front. Plant Sci. 2015, 6, 103. [Google Scholar] [CrossRef]
- Wang, X.; Mace, E.; Hunt, C.; Cruickshank, A.; Henzell, R.; Parkes, H.; Jordan, D. Two distinct classes of QTL determine rust resistance in sorghum. BMC Plant Biol. 2014, 14, 366. [Google Scholar] [CrossRef]
- Asnaghi, C.; Roques, D.; Ruffel, S.; Kaye, C.; Hoarau, J.Y.; Telismart, H.; Girard, J.C.; Raboin, L.M.; Risterucci, A.M.; Grivet, L.; et al. Targeted mapping of a sugarcane rust resistance gene (Bru1) using bulked segregant analysis and AFLP markers. Theor. Appl. Genet. 2004, 108, 759–764. [Google Scholar] [PubMed]
- Dixon, M.S.; Jones, D.A.; Keddie, J.S.; Thomas, C.M.; Harrison, K.; Jones, J.D.G. The Tomato Cf-2 Disease Resistance Locus Comprises Two Functional Genes Encoding Leucine-Rich Repeat Proteins. Cell 1996, 84, 451–459. [Google Scholar] [PubMed]
- Wang, W.; Zhang, Y.; Wen, Y.; Berkey, R.; Ma, X.; Pan, Z.; Bendigeri, D.; King, H.; Zhang, Q.; Xiao, S. A comprehensive mutational analysis of the Arabidopsis resistance protein RPW8.2 reveals key amino acids for defense activation and protein targeting. Plant Cell 2013, 25, 4242–4261. [Google Scholar] [PubMed]
- Berdy, S.E.; Kudla, J.; Gruissem, W.; Gillaspy, G.E. Molecular Characterization of At5PTase1, an Inositol Phosphatase Capable of Terminating Inositol Trisphosphate Signaling. Plant Physiol. 2001, 126, 801. [Google Scholar]
- Golani, Y.; Kaye, Y.; Gilhar, O.; Ercetin, M.; Gillaspy, G.; Levine, A. Inositol polyphosphate phosphatidylinositol 5-phosphatase9 (At5ptase9) controls plant salt tolerance by regulating endocytosis. Mol. Plant 2013, 6, 1781–1794. [Google Scholar]
- Diener, A.C.; Ausubel, F.M. Resistance to Fusarium Oxysporum 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 2005, 171, 305–321. [Google Scholar]
- Li, H.; Zhou, S.-Y.; Zhao, W.-S.; Su, S.-C.; Peng, Y.-L. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 2009, 69, 337–346. [Google Scholar]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.V.V.; Jugulam, M. Role of Cytochrome P450 Enzymes in Plant Stress Response. Antioxidants 2020, 9, 454. [Google Scholar]
- Lee, O.R.; Pulla, R.K.; Kim, Y.-J.; Balusamy, S.R.D.; Yang, D.-C. Expression and stress tolerance of PR10 genes from Panax ginseng CA Meyer. Mol. Biol. Rep. 2012, 39, 2365–2374. [Google Scholar]
- Chen, Z.Y.; Brown, R.L.; Damann, K.E.; Cleveland, T.E. PR10 expression in maize and its effect on host resistance against Aspergillus flavus infection and aflatoxin production. Mol. Plant Pathol. 2010, 11, 69–81. [Google Scholar]
- Pulla, R.K.; Lee, O.R.; In, J.-G.; Kim, Y.-J.; Senthil, K.; Yang, D.-C. Expression and functional characterization of pathogenesis-related protein family 10 gene, PgPR10-2, from Panax ginseng C.A. Meyer. Physiol. Mol. Plant Pathol. 2010, 74, 323–329. [Google Scholar] [CrossRef]
- Kidwai, M.; Ahmad, I.; Chakrabarty, D. Class III peroxidase: An indispensable enzyme for biotic/abiotic stress tolerance and a potent candidate for crop improvement. Plant Cell Rep. 2020, 39, 1381–1393. [Google Scholar] [CrossRef]
- Czolpinska, M.; Rurek, M. Plant Glycine-Rich Proteins in Stress Response: An Emerging, Still Prospective Story. Front. Plant Sci. 2018, 9, 302. [Google Scholar] [CrossRef]
- Zhang, L.; You, J.; Chan, Z. Identification and characterization of TIFY family genes in Brachypodium distachyon. J. Plant Res. 2015, 128, 995–1005. [Google Scholar] [CrossRef]
- Zhang, X.; González-Carranza, Z.; Zhang, S.; Miao, Y.; Liu, C.-J.; Roberts, J. F-Box Proteins in Plants. Annu. Plant Rev. 2019, 2, 307–328. [Google Scholar] [CrossRef]
- Änkö, M.-L. Regulation of gene expression programmes by serine–arginine rich splicing factors. Semin. Cell Dev. Biol. 2014, 32, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Palusa, S.G.; Ali, G.S.; Reddy, A.S. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 2007, 49, 1091–1107. [Google Scholar] [CrossRef]
- Abiri, R.; Shaharuddin, N.A.; Maziah, M.; Yusof, Z.N.; Atabaki, N.; Sahebi, M.; Valdiani, A.; Kalhori, N.; Azizi, P.; Hanafi, M.M. Role of ethylene and the APETALA 2/ethylene response factor superfamily in rice under various abiotic and biotic stress conditions. Environ. Exp. Bot. 2017, 134, 33–44. [Google Scholar] [CrossRef]
- Grennan, A.K. Ethylene Response Factors in Jasmonate Signaling and Defense Response. Plant Physiol. 2008, 146, 1457. [Google Scholar] [CrossRef] [PubMed]
- van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of Inducible Defense-related Proteins in Infected Plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed]



| Traits | BR | CR |
|---|---|---|
| Min | 2 | 0 |
| Max | 9 | 9 |
| Mean | 6 | 4 |
| Variance | 3.09 | 4.21 |
| SD | 1.76 | 2.05 |
| Cvar (%) | 29 | 48 |
| Repeatability | 0.78 | 0.70 |
| Chromosome | Chr Size (bp) | SNPs/Chr | SNP/bp |
|---|---|---|---|
| Chr1 | 271,335,344 | 1,781,903 | 152 |
| Chr2 | 346,255,425 | 2,254,571 | 154 |
| Chr3 | 383,839,144 | 2,224,970 | 173 |
| Chr4 | 414,259,934 | 2,753,346 | 150 |
| Chr5 | 259,831,545 | 1,545,724 | 168 |
| Chr6 | 276,808,772 | 1,774,834 | 156 |
| Chr7 | 359,413,018 | 1,944,409 | 185 |
| Variables | V(A) | V(P) | V(e) | h2SNP |
|---|---|---|---|---|
| CR | 2.08 (1.82) | 4.97 (0.46) | 2.82 (1.75) | 0.42 (0.35) |
| BR | 2.88 (1.42) | 3.56 (0.33) | 0.68 (1.31) | 0.81 (0.37) |
| Gene ID | Chr | Start | Stop | N SNPs | −log10(p-Value) |
|---|---|---|---|---|---|
| evm.TU.2.24844 | 2 | 343,028,981 | 343,029,284 | 3 | 5.72 |
| evm.TU.3.8555 | 3 | 123,435,108 | 123,443,087 | 31 | 4.63 |
| evm.TU.3.20827 | 3 | 295,634,059 | 295,635,654 | 25 | 5.22 |
| evm.TU.4.2271 | 4 | 26,600,949 | 26,602,802 | 58 | 4.65 |
| evm.TU.6.9029 | 6 | 126,251,598 | 126,256,062 | 48 | 6.39 |
| evm.TU.7.24966 | 7 | 350,687,605 | 350,692,741 | 143 | 4.58 |
| Gene ID | Chr | Start | Stop | N SNPs | −log10(p-Value) |
|---|---|---|---|---|---|
| evm.TU.2.3894 | 2 | 52,178,587 | 52,182,215 | 131 | 4.39 |
| evm.TU.2.19756 | 2 | 277,159,670 | 277,161,603 | 34 | 5.12 |
| evm.TU.3.8432 | 3 | 121,925,172 | 121,925,559 | 1 | 4.43 |
| evm.TU.3.8507 | 3 | 122,870,603 | 122,872,466 | - | 4.47 |
| evm.TU.3.8509 | 3 | 122,875,455 | 122,903,941 | 34 | 4.93 |
| evm.TU.3.9565 | 3 | 138,177,359 | 138,179,818 | 14 | 4.86 |
| evm.TU.3.23458 | 3 | 332,049,981 | 332,054,690 | - | 4.17 |
| evm.TU.3.25826 | 3 | 363,848,840 | 363,852,037 | 60 | 4.74 |
| evm.TU.5.11920 | 5 | 164,892,059 | 164,894,440 | - | 4.22 |
| evm.TU.6.3022 | 6 | 37,956,882 | 37,960,523 | 58 | 4.50 |
| evm.TU.6.15799 | 6 | 219,584,326 | 219,586,833 | 56 | 6.22 |
| evm.TU.6.15800 | 6 | 219,587,444 | 219,589,409 | 55 | 4.46 |
| evm.TU.7.22707 | 7 | 319,181,319 | 319,183,513 | 59 | 4.10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fois, M.; Bellucci, A.; Malinowska, M.; Greve, M.; Ruud, A.K.; Asp, T. Genome-Wide Association Mapping of Crown and Brown Rust Resistance in Perennial Ryegrass. Genes 2022, 13, 20. https://doi.org/10.3390/genes13010020
Fois M, Bellucci A, Malinowska M, Greve M, Ruud AK, Asp T. Genome-Wide Association Mapping of Crown and Brown Rust Resistance in Perennial Ryegrass. Genes. 2022; 13(1):20. https://doi.org/10.3390/genes13010020
Chicago/Turabian StyleFois, Mattia, Andrea Bellucci, Marta Malinowska, Morten Greve, Anja Karine Ruud, and Torben Asp. 2022. "Genome-Wide Association Mapping of Crown and Brown Rust Resistance in Perennial Ryegrass" Genes 13, no. 1: 20. https://doi.org/10.3390/genes13010020
APA StyleFois, M., Bellucci, A., Malinowska, M., Greve, M., Ruud, A. K., & Asp, T. (2022). Genome-Wide Association Mapping of Crown and Brown Rust Resistance in Perennial Ryegrass. Genes, 13(1), 20. https://doi.org/10.3390/genes13010020







