Abstract
Staphylococcus aureus encodes 16 two-component systems (TCSs) that enable the bacteria to sense and respond to changing environmental conditions. Considering the function of these TCSs in bacterial survival and their potential role as drug targets, it is important to understand the exact mechanisms underlying signal perception. The differences between the sensing of appropriate signals and the transcriptional activation of the TCS system are often not well described, and the signaling mechanisms are only partially understood. Here, we review present insights into which signals are sensed by histidine kinases in S. aureus to promote appropriate gene expression in response to diverse environmental challenges.
1. Introduction
The human bacterial pathogen S. aureus asymptomatically colonizes approximately 20% of the general population [1]. However, S. aureus is also a major human pathogen that causes a variety of acute and chronic diseases [2,3]. The versatility of this organism arises due to its capacity to produce accessory molecules that mediate specific interactions with host cells and to quickly adapt to changing environments. Gene expression is tightly controlled by an interactive regulatory network. Two-component systems (TCSs) are central pillars of environmental sensing [4,5]. S. aureus encodes 16 TCSs that are evenly distributed on the bacterial chromosome, and some methicillin-resistant S. aureus (MRSA) encode one additional TCS on an accessory SCC-mec island [6]. Among the 16 systems, only WalRK is essential for bacterial growth, whereas the others can be deleted simultaneously in the same strain without affecting cell viability under laboratory conditions [6].
However, most TCSs contribute to better survival or modulation of virulence under certain infectious conditions [4,7]. Each TCS is mostly autonomous in sensing and responding to its cognate signals, with limited cross-regulation between the systems [6]. In many cases, the signals leading to TCS activation and the underlying molecular mechanisms are not well understood. In terms of function, a canonical TCS consists of a histidine kinase (HK) that is autophosphorylated at a conserved histidine residue and then transfers the phosphoryl group to an aspartate residue in the cognate response regulator (RR) [8]. Many HKs also elicit phosphatase activity, which is important for resetting the system [9]. HKs are usually membrane-bound homodimers that receive signals via defined transmembrane, extracellular, or intracellular sensing domains [10,11]. The cytoplasmic C-terminus includes a linker or HAMP domain (not always present), additional sensory domains, a conserved dimerization and histidine-containing domain (DHp), and a catalytic ATP binding (CA) domain. After sensing a signal, a conformational change generally occurs in the sensor portion of the protein that triggers the autophosphorylation of the conserved moieties of the protein [12].
In S. aureus, four HKs are classified as intramembrane-sensing histidine kinases (IM-HKs), namely, SaeS, VraS, GraS, and BraS [13]. One HK, AgrB, is a quorum sensor that enables the bacteria to monitor cell density [4,14]. PhoR and WalK are the only HKs in S. aureus that harbors a Per-ARNT-Sim (PAS) domain as a potential ligand interaction site [15]. Two S. aureus HKs, namely, AirS and NreB, are iron-sulfur (Fe-S) cluster-containing proteins (Figure 1).
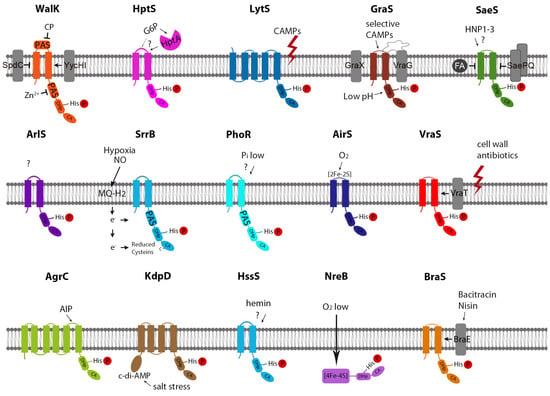
Figure 1.
Structure and Signaling-Mechanisms of S. aureus HKs. CP: Peptidoglycan cleavage products; CAMP: Cationic antimicrobial peptide; FA: fatty acid.
Many TCS gene clusters contain additional proteins, some of which are known to interact with the function of the TCS (Table 1). The TCS-encoding operons are often autoregulated, providing a positive feedback loop, or are under the control of other regulators. However, an increase in TCS expression does not necessarily result in an increase in target gene expression, e.g., under conditions when the native signal for HK activation is missing. Moreover, overexpression may also result in inhibition through the coactivation of accessory genes with inhibitory function (e.g., saePQ). Thus, the gene expression levels of TCSs are often not informative for identifying cognate signals, and analyses of defined reporter genes might be more suitable. In general, operon architecture and overall functions are well studied by phenotypic and genetic analyses of mutant strains. Target genes are often localized next to the TCSs but can also be scattered throughout the chromosome. The dedicated signals and the mechanism by which TCSs sense their signals are still largely unknown. Here, we will summarize recent knowledge on the signaling mechanism of individual S. aureus HKs.

Table 1.
Genetic organization, regulation, and function of S. aureus TCSs.
2. WalRK
The WalRK (also called YycGF) system was first described in Bacillus subtilis [16,17] and is highly conserved in low G+C Gram-positive bacteria [18,19]. In S. aureus, WalRK regulates the expression of genes involved in cell wall metabolism, thereby controlling autolysis, biofilm formation, and virulence [20,21,22]. WalRK function is strongly associated with the vancomycin-intermediate resistant S. aureus (VISA) phenotype. Clinical VISA strains, as well as resistant strains selected in vitro, often contain single nucleotide polymorphisms (SNPs) within the WalRK system [23,24,25]. The HK WalK is encoded within a 4-gene operon (walRKHI), which seems not to be autoregulated. WalK from S. aureus harbors an intracellular and an extracellular PAS domain presumably involved in sensing [26]. The cytoplasmic PAS domain in B. subtilis [27], and presumably also that in S. aureus [28], is essential to localize WalK to the division septum. This domain (amino acids: from 261 to 375) is conserved among members of Staphylococcus species and contains four critical and highly conserved residues (His 271, Asp 274, His 364, and Glu 368) that are involved in Zn2+ binding [29]. The WalKH271Y mutation abrogates metal binding, increasing WalK kinase activity and WalR phosphorylation. Thus, Zn2+ binding to WalK is important to restrict WalK activity. Recently, an altered WalK-H364 residue within the Zn2+ binding region, detected in VISA strains, was predicted to destabilize the protein and decrease WalK activity [30]. Interestingly, another VISA-related WalK mutation (Y306N) located in the cytoplasmic PAS domain increased WalK kinase activity in liposomes [31].
Deletion of the extracellular PAS domain of B. subtilis WalK results in constitutive signaling [27,32]. In this organism, peptidoglycan cleavage products generated by D,L-endopeptidase digestion specifically inhibit WalK activity, presumably by interaction with the extracellular PAS domain [32]. It is likely that the homologous PAS domain of S. aureus WalK also senses peptidoglycan degradation products. This would support a model in which WalK is inhibited when it binds to its signal and activated when unliganded.
WalK was previously proposed to be involved in sensing the D-Ala-D-Ala moiety of Lipid II as a signal for active cell wall synthesis [19]. This conclusion was mainly based on the observation that different cell wall-targeting antibiotics affect the WalRK regulon in B. subtilis. Recent studies suggest that this finding is likely mediated by serine/threonine kinase PknB activity. The interaction of Lipid II with PknB stimulates the phosphorylation of the RR WalR [33]. Amino acid T101, which represents the sole PknB phosphorylation site on WalR, is located in the vicinity of the dimerization interface of the regulator and might therefore influence dimerization and DNA binding behavior.
The genes that encode two additional membrane proteins, yycH and yycI, are co-transcribed with walRK, and the gene products modulate WalK activity. In contrast to B. subtilis, the disruption of the yycH and yycI genes in S. aureus led to a downregulation of the WalRK regulon [34]. Both proteins were predicted to interact with WalK in a bacterial two-hybrid system, most likely forming a ternary complex via their transmembrane domains [34]. Phosphorylation assays with full-length recombinant proteins in phospholipid liposomes confirmed that YycH and YycI stimulate WalK activity [35]. Recently, SpdC, an Abi (abortive infectivity)-domain membrane protein, was described to also interact with WalK at the division septum and to negatively control the expression of WalRK regulon genes [36].
Thus far, the main modulators of WalK activity (Zn2+, peptidoglycan cleavage products, SpdC) inhibit WalK. The question remains whether there is any true signal that activates WalK (possible acting via YycHI) or whether regulation primarily occurs via inhibition of an otherwise constitutively active (kinase-On) WalK system.
3. HptASR
The HptASR (hexose phosphate transport) system regulates glucose-6-phosphate (G6P) uptake by activating the hexose phosphate antiporter UhpT [37,38]. The system appears to be important for intracellular survival because the carbon source in the cytoplasm of host cells is limited to hexose phosphate. The hptASR mutant is impaired in survival/multiplication within various types of host cells and gives resistance to fosfomycin [37]. uhpT is also differentially expressed, e.g., in biofilm-forming bacteria, during acidification or oxidative stress, which likely involves control by the catabolite control protein A (CcpA) system. CcpA represses hptSR in a glucose-dependent manner, thereby coordinating G6P uptake and metabolism when glucose is limited [39].
G6P sensing of the HptASR system is mediated by HptA, a secreted protein that binds to G6P directly and then transfers the signal to the membrane-bound HK HptS. The solved structures of HptA in complex with the periplasmic domain of HptS showed that HptA forms a tetramer with HptS [40]. The G6P-free form of HptA binds to the distal membrane side of the HptS periplasmic domain. G6P binding to HptA switches the contact region to the membrane-proximal domain of HptS. This causes untwisting and tilting movement in the TM2 region of HptS. This conformational change is then transmitted to the HAMP domain to allow kinase activation. However, G6P might also directly interfere with HptS because a strain lacking HptA is completely abolished G6P sensing [38].
4. LytSR
The LytSR system was identified as a TCS that affects autolysis in S. aureus through the activation of the lrgAB operon [41]. It was proposed that LrgA, together with the homologous CidA, functions as a holin/anti-holin system to regulate murein hydrolase activity and, ultimately, cell death. lytSR expression is positively modulated by the direct binding of SpoVG to a short consensus sequence within the 5’ noncoding region of lytSR [42]. The HK LytS harbors five transmembrane regions that are likely involved in sensing alterations in the membrane potential and thereby acting as a “voltmeter” [43,44]. His390 of LytS is the site of autophosphorylation, and Asn394 is a critical amino acid involved in phosphatase activity [45]. Cationic antimicrobial peptides (CAMPs) usually alter membrane potential; therefore, LytS might function as a universal CAMP sensor and responder. Consistent with this assumption, lytSR mutants were shown to be more sensitive to CAMP [43]. This makes the system important for in vivo survival because it enables a general response to CAMPs in contrast to the rather selective response mediated by the other CAMP-sensing TCSs, namely, GraRS and SaeRS.
Of note, under specific conditions, acetyl phosphate can act as phosphodonor for the RR LytR, thereby bypassing LytS [45,46].
5. GraRS
The GraRS system, also called ApsSR [47], mediates resistance to several CAMPs and vancomycin mainly via regulation of vraFG, mprF, and dltABCD [47,48,49,50]. The regulon was shown to be activated by indolicidin, mellitin, nisin, LL-37 [47], colistin [50], RP-1, and polymyxin B (PMB) but not by HNP-1, vancomycin, gentamycin, or daptomycin [51]. GraS belongs to the subset of IM-HK, in which two transmembrane helices frame a very short extracellular loop (EL) [13]. For the homologous GraS/ApsS HK of S. epidermidis, the predicted 9-amino-acid EL, which harbors a high density of negative charges, directly binds to CAMPs [52]. The ELs of GraS/ApsS of S. aureus and S. epidermidis show 33% similarity, resulting in different degrees of selectivity for CAMPs [47]. Direct interaction of the EL loop region (DYDFPIDSL) with CAMPs is also likely in S. aureus, since a soluble EL mimic of GraS protected S. aureus against CAMP-mediated death [53]. The proline at position 39 and the two aspartic acid residues at positions 37 and 41 located within the EL are important for full GraS activity [53,54,55].
GraRS is part of a three-gene operon (graXRS) localized immediately upstream of the ABC transporter genes vraF and vraG [56]. Whereas the graXRS operon does not seem to be autoregulated, vraFG expression is under the tight control of GraRS [56]. The auxiliary factor GraX is required for colistin-mediated activation of the system [56] and likely functions as a scaffold to promote protein interactions with GraS, GraR, and VraFG [57].
The GraR target gene vraG encodes a membrane permease, while vraF encodes an ATPase to provide energy for efflux. Interestingly, a vraG mutant phenocopied a graS mutant with respect to CAMP sensing [58]. Native VraG interacted with GraS to restrict its kinase activity [58]. VraG contains a single 200-residue EL located between the seventh and eighth transmembrane segments. Deletion of the VraG-EL domain reduced the interaction and resulted in GraS activation even without CAMPs. It is hypothesized that the tight interaction between VraG-EL and GraS-EL is weakened by the binding of CAMPs, resulting in the activation of GraS. Specific lysine residue(s) on the VraG-EL presumably reduces the sensing of cognate CAMPs by GraS-EL. Thus, VraG, more than an efflux pump, facilitates GraS-mediated sensing of HDPs [58].
Recent work has expanded the sensory capabilities of GraS to include the sensing of acidic pH [54,55,59], and GraS activation at low pH is mandatory for survival within macrophages [58]. Interestingly, pH-dependent modulation of GraS activity seems to be independent of the Gra-EL implicated in CAMP sensing [58]. Acidic pH reduces the charge on polar lipid head groups, which presumably impacts GraS activity. It was hypothesized that acidic pH should also reduce the charge on acidic amino acids in the EL of GraS, which would render them less effective in sensing CAMP at low pH [55]. An EL-independent mechanism of GraS activation under acidic conditions might be beneficial for the bacterium. Thus, low pH and CAMPs present during infection seems to independently signal through GraS to promote bacterial survival.
6. SaeRS
The SaeRS system first described by Giraudo et al. [60] regulates the expression of numerous virulence factors in S. aureus, including surface-bound and secreted proteins [60,61,62,63]; for a comprehensive review about the organization of the sae operon, consensus sequences, and regulated target genes, see [64]. The Sae system was shown to be induced by phagocytosis-related signals, of which human neutrophil peptides 1–3 (HNP1–3) showed the most pronounced effect [65]. SaeS activation by subinhibitory concentrations of HNP1–3 was confirmed in strains of the USA300 lineage [66,67]. However, the widely used strains 8325 and Col were found to be nonresponsive to HNP1–3 sensing [65].
SaeS is coded within a four-gene operon, saePQRS, transcribed from two promoters (P1 and P3). The mature full-length transcript is transcribed from the autoactivated P1 promoter, and a stable RNA is generated by RnaseY-dependent endoribonucleolytic cleavage between saeP and saeQ [68]. saePQRS expression is decreased in agr mutants [65,69]. However, Agr and Sae impact virulence gene expression primarily independent of each other [64]. The P1 promoter was also shown to be under the control of the metabolic regulator CodY, and CodY also indirectly regulates sae expression through Agr- and Rot-mediated repression of the sae P1 promoter [70]. One additional constitutive P3 promoter located within saeQ ensures a basal level of saeRS expression [71,72].
HK SaeS is a IM-HK [13] with a short 9-aa EL. A single amino acid substitution in the first transmembrane helix (L18P) of the EL in the Newman strain renders the system hyperactive [73]. The overall confirmation of the transmembrane domain, as well as the EL peptide composition, are involved in signal transduction [64,66].
It was proposed that in the signal transfer process, the entire N-terminal domain of SaeS (two transmembrane helices and the EL) works as a coherent unit in a tripwire manner. In this “tripwire” model, the overall conformation of the entire N-terminal domain is the key determinant in controlling the kinase activity of HK. Any stimulus that elicits conformational changes in the N-terminal domain is expected to either repress or activate the kinase activity of the HK, depending on the nature of the conformational change [64].
This model predicts that molecules that alter the conformation of the transmembrane domain can serve as signal transducers to regulate SaeS activity. However, despite the well-documented specific effect of HNPs on SaeS activation, it is still unclear whether these peptides directly bind to SaeS or whether they act via a hitherto-unidentified receptor molecule. The latter theory is supported by the observation that not all S. aureus strains are sensitive to HPN despite harboring identical SaeRS alleles [65].
In addition to HPNs, additional factors can modulate SaeS activity. The cotranscribed lipoprotein SaeP and the membrane protein SaeQ are dispensable for the activation of the Sae system [73,74]; however, they are required to induce SaeS phosphatase activity. Thus, SaePQ helps to set the system to a pre-activation state via a negative feedback mechanism. The membrane protein SpdC was proposed to interact with the transmembrane domains of WalK and SaeS [36]. The silkworm apolipophorin protein binds to lipoteichoic acid (LTA), and LTA complexed with the protein inhibits the kinase activity of SaeS by interacting with the transmembrane domain of SaeS [75]. In an in vitro enzyme assay, Cu, Zn and Fe ions were shown to inhibit the kinase activity of SaeS [76], possibly by competing with Mg. Furthermore, the fatty acid kinase VfrB was proposed to positively control SaeRS-mediated transcription of virulence factors [77].
Recently, excess levels of both external and cytoplasmic free fatty acids were proposed to modulate bacterial virulence factor production via SaeRS inhibition [77,78]. Unsaturated fatty acids are generally stronger inhibitors than saturated fatty acids [79]. Accordingly, deletion of respiratory dehydrogenase genes, which leads to altered NAD+/NADH ratios and free fatty acid accumulation, is accompanied by reduced SaeRS activity [80]. Fatty acid sensitivity occurs independently of SaePQ but requires the native SaeS transmembrane [79]. Thus, SaeS probably integrates membrane-active signals to either increase (HPN1) or decrease (fatty acids) SaeS kinase activity.
7. TCS-7, DesKR
One TCS has thus far not been characterized in S. aureus and is thus provisionally named TCS-7. A four-gene cluster that encodes an ABC transporter system and the TCS is conserved among Firmicutes. Recently, S. aureus HK (SA1313) was expressed in B. subtilis and shown to functionally complement the B. subtilis homologue DesK [81]. The TM domain of B. subtilis DesK was shown to be essential for signal detection and modulation of its catalytic activities and thought to be involved in temperature sensing [17]. Similar to B. subtilis DesK, the S. aureus homologue adopts a kinase-ON state at low temperature and phosphorylates DesR. However, both enzymes behave differently in the context of changes in the cytoplasmic membrane properties caused by isothermal conditions. Such differences likely occur due to major differences in the TM domain. The two co-transcribed genes (SauSa300_1217 and SauSa300_1218) encode putative ABC transporter systems with unknown functions. Further analyses in S. aureus are required to prove that DesKR is involved in temperature sensing and determine whether and how the putative ABC transporter is functionally linked to DesKR activity.
8. ArlRS
The ArlRS (autolysis-related locus) system [82] regulates the expression of several virulence factors [83]. After sensing a yet unknown signal, activated ArlR, in turn, drives the expression of the global regulators MgrA [84,85] and Spx [86]. The Spx regulator controls the S. aureus response to ß-lactam antibiotics and stress, while the global regulator MgrA directly impacts virulence by controlling the expression of over 100 effector genes. The expression of arlRS and some arl target genes is sensitive to DNA supercoiling [87,88]. However, little is known about a potential activating signal of the HK ArlS. There is no special motif predicted in the sensor domain, but the 118-amino acid extracellular region between the two membrane-spanning domains (InterPro prediction) could be a potential ligand interaction site. Recent studies revealed that ArlS is necessary for the activation of ArlR in response to manganese starvation caused by calprotectin and glucose limitation [89,90].
9. SrrAB
The SrrAB (staphylococcal respiratory response) system [91] regulates the expression of genes involved in anaerobic metabolism, nitrosative stress, and cytochrome biosynthesis [92,93]. The HK SrrB responds to hypoxia and nitric oxide (NO), possibly by sensing reduced menaquinone levels as a result of impaired electron flow in the electron transport chain [92,93,94]. SrrB contains an extracellular cache domain and a cytoplasmic HAMP-PAS region [95]. The structure of the catalytic domain revealed a unique intramolecular cysteine disulfide bond in the ATP-binding domain [95], rendering the SrrB kinase activity sensitive to the cysteine redox state. It was proposed that under aerobic conditions, cysteines are oxidized to keep kinase activity low. Under anaerobic conditions, the pool of reduced menaquinones increases, resulting in reduced SrrB cysteines and higher kinase activity. However, fully oxidized SrrB retains ~60% activity, and cysteines are not conserved in all staphylococcal species [95]. Thus, additional molecular mechanisms are likely involved in modulating SrrB activity. The PAS domain itself significantly influenced SrrB kinase and phosphatase activity in vitro [96]. Redox-active ligand binding to the PAS domain may further affect SrrB activity. Moreover, the role of the extracellular Cache domain in SrrB activation remains unclear.
10. PhoPR
Inorganic phosphate (Pi) is an essential nutrient required in multiple cellular processes, including energy metabolism and intracellular signaling. However, this nutrient is generally found at very low concentrations in its free form and is directly utilizable by bacterial cells. The PhoPR system is essential for phosphate homeostasis in S. aureus by sensing phosphate limitation and regulating the expression of three phosphate transporters, namely, PstSCAB, NptA, and PitA [97]. In addition to phosphate transporter expression, PhoPR controls factors contributing to virulence. In the homologous system in E. coli, the PhoR kinase interacts through its PAS domain with the negative regulator PhoU, which monitors the activity of the Pi-specific ATP-binding cassette transporter PstSCAB [98]. Low activity of the Pst transporter then results in a high activity of PhoR. Signal sensing is mediated through conformational changes of PstB in response to Pi availability [99]. In B. subtilis, no homologue to PhoU is present, and PhoR activity is responsive to biosynthetic intermediates of wall teichoic acid (WTA) metabolism [100]. Considering that the respective WTA intermediate is not present in S. aureus and that PhoU is not needed for the growth of S. aureus in Pi-depleted medium, Pi sensing in S. aureus is fundamentally different from that in E. coli and B. subtilis [97]. Nevertheless, the intracellular PAS domain can be assumed to be important for ligand interactions. The functionality of the phosphate homeostasis system appears important for bacterial pathogenesis, as cells expressing mutant phoPR showed an altered capacity to invade multiple organs in a mouse model of infection [97]. It should be noted that this phenomenon occurs independently of the presence of the phosphate transporter NptA. This observation might be related to the involvement of Pi homeostasis in the maintenance of cell wall structure in Gram-positive bacteria, as revealed by the impact of Pi on resistance to cell wall-active antibiotics, such as glycopeptides [101].
11. AirSR
The AirSR (anaerobic iron-sulfur cluster-containing redox sensor regulator) system [102], also called YhcSR [103], responds to oxidation signals. AirSR contributes to resistance to H2O2, autolysis, survival in blood, and vancomycin resistance [104,105]. The N-terminal sensor domain of HK AirS contains a Fe-S cluster essential for its kinase activity [103]. Under anaerobic conditions or in the absence of ROS, the Fe-S cluster is reduced, leading to low phosphorylation of AirS. When exposed to oxygen, [2Fe-2S]+ is oxidized to [2Fe-2S]2+, which leads to full kinase activity of AirS. Overoxidation and exposure to H2O2 or NO stress results in protein inactivation. Consistent with this model, only a few genes were affected by AirRS when bacteria were grown anaerobically [105] since the reduced AirS should have low kinase activity under this condition. The regulation occurred through direct binding to the promoter region of target genes. Surprisingly, in a previous study, it was found that AirR only affects gene expression under anaerobic conditions in strain Newman [102]. This discrepancy may suggest that the regulatory activity of AirR is strain-specific. Why AirSR acts so differently in different strains remains unclear. Regarding physiopathology, this TCS has been described to contribute to S. aureus survival in human blood by transcriptionally regulating the expression of sspABC, which encodes important proteases that mediate the hydrolysis of opsonins, thus inhibiting killing by professional phagocytic cells [104].
12. VraSR
The VraSR (vancomycin resistance associated) system facilitates resistance to cell wall antibiotics via the regulation of cell wall biosynthesis and antibiotic resistance genes [106]. Activation of VraSR target genes is triggered by cell wall synthesis inhibitors such as glycopeptides or ß-lactam antibiotics. A similar conserved VraSR-linked cell wall stress response can be evoked by inhibition of peptidoglycan ligase [107] or penicillin-binding protein pbpB [108]. The HK coding vraS is part of an autoregulated four-gene operon (vraUTSR) [109]. VraS also belong to the family of IM-HK [13]. The molecular mechanism involved in VraS sensing remains unclear; however, the interaction between VraT and VraS seems to be involved. Deletion of the putative membrane protein VraT leads to constitutive vra regulon expression [110]. It was suggested that cell wall damage modifies the VraT-VraS interaction to influence VraS kinase activity.
13. AgrCA
The AgrCA (accessory gene regulator) system [111] senses population density through the production of an autoinducing peptide (AIP) and serves as a regulator of global virulence mostly via the regulatory RNAIII and the transcription factor Rot (for reviews see [4,112,113,114,115]. The system is composed of a 4-gene operon (agrBDCA) and the divergently transcribed regulatory RNAIII, which encodes hld. AIP is a thiolactone peptide (8–9 aa) that is processed from the AgrD-encoded propeptide by AgrB [116]. There are several AIP derivatives that interact with cognate AgrC to activate kinase activity. For S. aureus, four Agr specificity groups were identified (Groups I–IV). Additionally, AIPs inhibit noncognate AgrC. The Agr circuit is conserved in many staphylococcal species, with each producing its own unique AIP [117]. Therefore, Agr polymorphisms contribute to strain interference between staphylococci of different Agr groups. The thiolactone ring of AIP is necessary for binding to AgrC, whereas the tail is critical for activation. Deletion of the tail converts AIP from an agonist into an antagonist molecule [118]. The structural features of both native AIPs and non-native analogues were revealed using NMR spectroscopy [119,120,121]. Two critical structural motifs within the AIP-III ligand were identified: (i) a hydrophobic patch (or “knob”) on the macrocycle essential for receptor binding and (ii) an additional hydrophobic contact or “anchor” on the N-terminal tail critical for receptor activation. In the absence of the anchor, peptides containing a hydrophobic knob were found to inhibit the AgrC-III receptor, presumably by outcompeting the native ligand.
The HK AgrC is classified as a member of the ‘‘HPK10’’ kinase subfamily [122] in which an Asn residue substitutes for the conserved ‘‘G1-box’’ Asp, which normally hydrogen-bonds to the N-6 amino group on the adenine base of the nucleotide. Therefore, the Asn residue is likely responsible for the exceptionally weak affinity between full-length AgrC and ATP, which renders the kinase activity of AgrC strongly dependent upon the cellular ATP level. Thus, when energy starvation decreases the cellular ATP level, the AgrC kinase activity will be diminished even in the presence of AIP activators.
The N-terminal sensor domain spans the membrane six times. A short peptide sequence with a high helical propensity, termed the S helix, links the sensor domain with the cytoplasmic DHp and CA domains. AIPs bind to AgrC-I at 2:2 stoichiometry. The binding of agonist or inverse-agonist peptides results in twisting of the linker in different directions, thus either promoting or inhibiting kinase activity. However, AIP signals do not regulate signaling at the level of RR dephosphorylation [122].
The crystal structure of the AgrC HK module trapped in the apo form showed that the CA domain docks against the DHp helices in a manner that prevents histidine autophosphorylation [123]. Noncovalent interaction between R238 and Q305 stabilizes AgrC in the “off” state. This “latch” is proposed to lift after the binding of AIP to the extracellular AgrC sensor domain. Constitutive mutations discovered in the HK module, e.g., R238, localize to the docking interface [124]. Naturally occurring mutations in AgrC are consistent with repositioning of key functional domains of AgrC [125].
The mechanism(s) by which ligand binding induces these structural changes has yet to be resolved. It was proposed that AIP binding stabilizes or induces discrete rotational conformations in the last helix of the sensor that is directly transduced into the DHp domain by the S helix [123].
AgrC activity is likely influenced by other poorly defined factors [113,120]. Recently, a putative metalloprotease, MroQ, was shown to be required for Agr activity [126,127]. MroQ is proposed to function at the level of the peptide-processing module (AgrBD) [127] or via interaction with AgrC [126].
14. KdpDE
KdpDE TCSs are conserved in many pathogens and widely studied for their regulatory role in potassium (K+) transport and virulence gene expression [128]. In S. aureus, KdpDE activates the expression of the high-affinity potassium uptake system KdpABC and several virulence factors, such as the capsular biosynthesis gene cluster [129,130,131,132]. kdpDE is localized next to kdpABC but transcribed from a different promoter. KdpDE contributes to survival in human blood or in the presence of professional phagocytes [129]. In addition, KdpDE activates the expression of sigS, which encodes an alternative sigma factor [133]. KdpDE expression is increased by osmotic stress caused by NaCl or sucrose but decreased by K+. Transcriptional regulation of kdpDE expression seems not to be autoregulated since it occurs independently of the response regulator KdpE [129]. KdpDE expression is also positively regulated by Agr through the transcription factor Rot [129].
KdpD HK directly binds the second messenger molecule c-di-AMP through its intracellular universal stress protein domain [134]. A conserved SXS-X20-FTAXY motif is important for this binding, and c-di-AMP binding inhibits KdpDE signaling. It is proposed that under salt stress conditions, c-di-AMP inhibition is relieved, leading to KdpD activation. Of note, some MRSA strains harboring staphylococcal cassette chromosome mec type II (SCCmecII) contain a second kdp operon [135].
15. HssRS
The HssRS (heme sensor) system senses the level of extracellular hemin and activates the expression of hrtAB, which encodes a heme-regulated efflux pump, thereby sustaining heme homeostasis in S. aureus [136,137]. Genomics-based approaches have not revealed a potential heme-binding domain in HssS [136]. It was hypothesized that HssS might sense any toxic effect caused by heme [137]. The system is important to efficiently protect S. aureus from the toxic effects of heme.
16. NreBC
NreBC (nitrogen regulation) is important for nitrate and nitrite reduction [138]. The system is part of the narGYJI-nreABC operon that encodes nitrate reductase and the GAF (cGMP-specific phosphodiesterases, adenylyl cyclases, and FhlA)-domain-containing NO3− sensor NreA and is best studied in the nonpathogenic Staphylococcus carnosus [139,140]. NreB is a bifunctional HK with integral cryptic phosphatase activity. Activation of phosphatase activity and dephosphorylation of RR, NreC-P requires NreA as a cofactor [140]. In the presence of NO3−, the NreA-NO3− complex shows decreased interaction with NreB, allowing NreB phosphorylation. NreB is the only S. aureus HK with a cytoplasmic sensor kinase. NreB is an oxygen-sensing protein containing a Fe-S cluster at the N-terminus. Under anoxic conditions, NreB contains an oxygen-sensitive [4Fe-4S]2+ cluster and is phosphorylated at the conserved histidine residue. Disassembly of the [4Fe-4S]2+ cluster to a [2Fe-2S]2+ cluster upon exposure to oxygen leads to decreased autophosphorylation of NreB [139]. Thus, under anoxic conditions, NreB with the [4Fe-4S]2+ cluster exists in the active state, but when no NO3− is available, it remains inhibited by NreA.
17. BraRS
The BraRS (bacitracin resistance-associated) system [141], also called NsaRS (nisin susceptibility associated) [142], is essential for bacitracin and nisin resistance in S. aureus. The sensor kinase BraS is classified as an IM-HK and harbors an extremely short extracellular loop of only three amino acids [141]. The ABC transporters BraDE and VraDE are involved in bacitracin resistance, and their operon expression is in turn induced by bacitracin and nisin through braRS. BraDE is important for the detection of antibiotics, whereas VraDE confers resistance by functioning as an efflux pump. Bacitracin is sensed by BraDE, and the signal is transduced to BraRS through a mechanism that remains to be elucidated, but that likely involves interaction between BraE and BraS [141]. Nisin resistance can be selected by braRS mutations, including SNPs in the promoter region of braRS. This results in constitutive induction of VraDE expression even in the presence of the unphosphorylated form of the BraR mutant protein [143]. Although the system is only involved in bacitracin and nisin resistance, other antimicrobials targeting the cell wall also activate its expression [144]. Thus, BraRS is involved in sensing disruptions of the cell envelope of S. aureus. Significant homologies have been found between the braRS and graSR systems in various staphylococcal species. Interestingly, bsaRS homologues are mainly present in skin-colonizing staphylococci [145].
18. Conclusions and Outlook
The synthesis of many factors involved in virulence and stress resistance is controlled by TCSs. TCSs allow the bacteria to properly adapt to the diverse stresses encountered during infection. The mechanisms by which HKs sense are only partially understood. Structural and biochemical analyses of purified S. aureus HKs are still rare, largely because most of the HKs are membrane proteins that are difficult to purify. HK phosphorylation activity in living bacteria cannot be measured directly and is often implied from gene expression analysis of selected target genes. However, the expression of many target genes is controlled by various regulators [7], which often impedes the interpretation of the data. Thus, the development of well-validated sensors based on confirmed or modified target genes would significantly improve the available experimental tools. Such genes should primarily be responsive to a single RR with an approved RR binding motif. To allow better readout, additional motifs in the selected promoter regions can be modified to improve the specificity of synthetic biosensors. In vitro assays to analyze signaling mechanisms in reconstituted liposomes and their use in structural studies are needed to gain further insights into the mechanisms underlying signal transduction.
An improved understanding of HK function can guide screens to search for new TCS inhibitors. TCSs are attractive candidate targets in the development of new antimicrobials. Specific inhibitors are assumed to have few undesirable side effects in mammals and might be used to specifically block virulence and/or resistance. Such blockers are thought to be less prone to the development of resistance because bacterial growth is usually not inhibited. Non-native small molecules and peptides capable of interfering with TCSs have been analyzed and have shown significant value as research tools and as potential antivirulence agents [146]. “Quorum quenchers” (inhibitors of quorum sensing) are of special interest and have been shown to block S. aureus virulence in animal models [115,147,148,149]. However, they may also promote biofilm formation, which has to be considered before clinical application. Inhibitors of other S. aureus TCSs, such as VraS [150], SaeS [149], or WalK [151], are also under investigation.
Author Contributions
All authors drafted, commented, and edited the manuscript, which was in addition coordinated by C.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation; TR156 grant number 246807620 to CW).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The work was supported by infrastructural funding from the Deutsche Forschungsgemeinschaft (DFG), Cluster of Excellence EXC 2124 “Controlling Microbes to Fight Infections.”
Conflicts of Interest
The authors declare no conflict of interest.
References
- Weidenmaier, C.; Goerke, C.; Wolz, C. Staphylococcus aureus determinants for nasal colonization. Trends Microbiol. 2012, 20, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G., Jr. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Raineri, E.J.M.; Altulea, D.; van Dijl, J.M. Staphylococcal trafficking and infection—from ‘nose to gut’ and back. FEMS Microbiol. Rev. 2021. online ahead of print. [Google Scholar] [CrossRef]
- Wang, B.; Muir, T.W. Regulation of virulence in Staphylococcus aureus: Molecular mechanisms and remaining puzzles. Cell Chem. Biol. 2016, 23, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Rapun-Araiz, B.; Haag, A.F.; Solano, C.; Lasa, I. The impact of two-component sensorial network in staphylococcal speciation. Curr. Opin. Microbiol. 2020, 55, 40–47. [Google Scholar] [CrossRef]
- Villanueva, M.; Garcia, B.; Valle, J.; Rapun, B.; Ruiz de Los Mozos, I.; Solano, C.; Marti, M.; Penades, J.R.; Toledo-Arana, A.; Lasa, I. Sensory deprivation in Staphylococcus aureus. Nat. Commun. 2018, 9, 523. [Google Scholar] [CrossRef]
- Rapun-Araiz, B.; Haag, A.F.; De Cesare, V.; Gil, C.; Dorado-Morales, P.; Penades, J.R.; Lasa, I. Systematic Reconstruction of the Complete Two-Component Sensorial Network in Staphylococcus aureus. mSystems 2020, 5, e00511-20. [Google Scholar] [CrossRef]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef]
- Huynh, T.N.; Stewart, V. Negative control in two-component signal transduction by transmitter phosphatase activity. Mol. Microbiol. 2011, 82, 275–286. [Google Scholar] [CrossRef]
- Mascher, T.; Helmann, J.D.; Unden, G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 2006, 70, 910–938. [Google Scholar] [CrossRef]
- Abriata, L.A.; Albanesi, D.; Dal Peraro, M.; de Mendoza, D. Signal sensing and transduction by histidine kinases as unveiled through studies on a temperature sensor. Acc. Chem. Res. 2017, 50, 1359–1366. [Google Scholar] [CrossRef]
- Mitrophanov, A.Y.; Groisman, E.A. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008, 22, 2601–2611. [Google Scholar] [CrossRef] [PubMed]
- Mascher, T. Intramembrane-sensing histidine kinases: A new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 2006, 264, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Ross, H.F.; Projan, S.J.; Kornblum, J.; Kreiswirth, B.; Moghazeh, S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993, 12, 3967–3975. [Google Scholar] [CrossRef]
- Ulrich, L.E.; Zhulin, I.B. MiST: A microbial signal transduction database. Nucl. Acids Res. 2007, 35, D386–D390. [Google Scholar] [CrossRef] [PubMed]
- Fabret, C.; Feher, V.A.; Hoch, J.A. Two-component signal transduction in Bacillus subtilis: How one organism sees its world. J. Bacteriol. 1999, 181, 1975–1983. [Google Scholar] [CrossRef]
- Fernandez, P.; Porrini, L.; Albanesi, D.; Abriata, L.A.; Dal Peraro, M.; de Mendoza, D.; Mansilla, M.C. Transmembrane prolines mediate signal sensing and decoding in Bacillus subtilis DesK histidine kinase. mBio 2019, 10, e02564-19. [Google Scholar] [CrossRef] [PubMed]
- Dubrac, S.; Msadek, T. Tearing down the wall: Peptidoglycan metabolism and the WalK/WalR (YycG/YycF) essential two-component system. Adv. Exp. Med. Biol. 2008, 631, 214–228. [Google Scholar] [CrossRef]
- Dubrac, S.; Bisicchia, P.; Devine, K.M.; Msadek, T. A matter of life and death: Cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 2008, 70, 1307–1322. [Google Scholar] [CrossRef]
- Dubrac, S.; Msadek, T. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 2004, 186, 1175–1181. [Google Scholar] [CrossRef]
- Dubrac, S.; Boneca, I.G.; Poupel, O.; Msadek, T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 2007, 189, 8257–8269. [Google Scholar] [CrossRef] [PubMed]
- Delaune, A.; Dubrac, S.; Blanchet, C.; Poupel, O.; Mader, U.; Hiron, A.; Leduc, A.; Fitting, C.; Nicolas, P.; Cavaillon, J.M.; et al. The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect. Immun. 2012, 80, 3438–3453. [Google Scholar] [CrossRef] [PubMed]
- Machado, H.; Seif, Y.; Sakoulas, G.; Olson, C.A.; Hefner, Y.; Anand, A.; Jones, Y.Z.; Szubin, R.; Palsson, B.O.; Nizet, V.; et al. Environmental conditions dictate differential evolution of vancomycin resistance in Staphylococcus aureus. Commun. Biol. 2021, 4, 793. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; Davies, J.K.; Johnson, P.D.; Stinear, T.P.; Grayson, M.L. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: Resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 2010, 23, 99–139. [Google Scholar] [CrossRef]
- Castro, B.E.; Berrio, M.; Vargas, M.L.; Carvajal, L.P.; Millan, L.V.; Rios, R.; Hernandez, A.K.; Rincon, S.; Cubides, P.; Forero, E.; et al. Detection of heterogeneous vancomycin intermediate resistance in MRSA isolates from Latin America. J. Antimicrob. Chemother. 2020, 75, 2424–2431. [Google Scholar] [CrossRef]
- Kim, T.; Choi, J.; Lee, S.; Yeo, K.J.; Cheong, H.K.; Kim, K.K. Structural studies on the extracellular domain of sensor histidine kinase YycG from Staphylococcus aureus and its functional implications. J. Mol. Biol. 2016, 428, 3074–3089. [Google Scholar] [CrossRef]
- Fukushima, T.; Szurmant, H.; Kim, E.J.; Perego, M.; Hoch, J.A. A sensor histidine kinase co-ordinates cell wall architecture with cell division in Bacillus subtilis. Mol. Microbiol. 2008, 69, 621–632. [Google Scholar] [CrossRef]
- Poupel, O.; Moyat, M.; Groizeleau, J.; Antunes, L.C.; Gribaldo, S.; Msadek, T.; Dubrac, S. Transcriptional analysis and subcellular protein localization reveal specific features of the essential WalKR system in Staphylococcus aureus. PLoS ONE 2016, 11, e0151449. [Google Scholar] [CrossRef]
- Monk, I.R.; Shaikh, N.; Begg, S.L.; Gajdiss, M.; Sharkey, L.K.R.; Lee, J.Y.H.; Pidot, S.J.; Seemann, T.; Kuiper, M.; Winnen, B.; et al. Zinc-binding to the cytoplasmic PAS domain regulates the essential WalK histidine kinase of Staphylococcus aureus. Nat. Commun. 2019, 10, 3067. [Google Scholar] [CrossRef]
- Baseri, N.; Najar-Peerayeh, S.; Bakhshi, B. Investigating the effect of an identified mutation within a critical site of PAS domain of WalK protein in a vancomycin-intermediate resistant Staphylococcus aureus by computational approaches. BMC Microbiol. 2021, 21, 240. [Google Scholar] [CrossRef]
- Turck, M.; Bierbaum, G. Purification and activity testing of the full-length YycFGHI proteins of Staphylococcus aureus. PLoS ONE 2012, 7, e30403. [Google Scholar] [CrossRef]
- Dobihal, G.S.; Brunet, Y.R.; Flores-Kim, J.; Rudner, D.Z. Homeostatic control of cell wall hydrolysis by the WalRK two-component signaling pathway in Bacillus subtilis. Elife 2019, 8, e52088. [Google Scholar] [CrossRef]
- Hardt, P.; Engels, I.; Rausch, M.; Gajdiss, M.; Ulm, H.; Sass, P.; Ohlsen, K.; Sahl, H.G.; Bierbaum, G.; Schneider, T.; et al. The cell wall precursor lipid II acts as a molecular signal for the Ser/Thr kinase PknB of Staphylococcus aureus. Int. J. Med. Microbiol. 2017, 307, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cameron, D.R.; Jiang, J.H.; Kostoulias, X.; Foxwell, D.J.; Peleg, A.Y. Vancomycin susceptibility in methicillin-resistant Staphylococcus aureus is mediated by YycHI activation of the WalRK essential two-component regulatory system. Sci. Rep. 2016, 6, 30823. [Google Scholar] [CrossRef]
- Gajdiss, M.; Monk, I.R.; Bertsche, U.; Kienemund, J.; Funk, T.; Dietrich, A.; Hort, M.; Sib, E.; Stinear, T.P.; Bierbaum, G. YycH and YycI Regulate Expression of Staphylococcus aureus Autolysins by Activation of WalRK Phosphorylation. Microorganisms 2020, 8, 870. [Google Scholar] [CrossRef] [PubMed]
- Poupel, O.; Proux, C.; Jagla, B.; Msadek, T.; Dubrac, S. SpdC, a novel virulence factor, controls histidine kinase activity in Staphylococcus aureus. PLoS Pathog. 2018, 14, e1006917. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Kim, J.W.; Moon, B.Y.; Lee, J.; Fortin, Y.J.; Austin, F.W.; Yang, S.J.; Seo, K.S. Characterization of a novel two-component regulatory system, HptRS, the regulator for the hexose phosphate transport system in Staphylococcus aureus. Infect. Immun. 2015, 83, 1620–1628. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, H.; Liu, X.; Wang, M.; Xue, T.; Sun, B. Regulatory mechanism of the three-component system HptRSA in glucose-6-phosphate uptake in Staphylococcus aureus. Med. Microbiol. Immunol. 2016, 205, 241–253. [Google Scholar] [CrossRef]
- Reed, J.M.; Olson, S.; Brees, D.F.; Griffin, C.E.; Grove, R.A.; Davis, P.J.; Kachman, S.D.; Adamec, J.; Somerville, G.A. Coordinated regulation of transcription by CcpA and the Staphylococcus aureus two-component system HptRS. PLoS ONE 2018, 13, e0207161. [Google Scholar] [CrossRef]
- Wang, M.; Guo, Q.; Zhu, K.; Fang, B.; Yang, Y.; Teng, M.; Li, X.; Tao, Y. Interface switch mediates signal transmission in a two-component system. Proc. Natl. Acad. Sci. USA 2020, 117, 30433–30440. [Google Scholar] [CrossRef] [PubMed]
- Brunskill, E.W.; Bayles, K.W. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 1996, 178, 611–618. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, S.; Sun, B. SpoVG regulates cell wall metabolism and oxacillin resistance in methicillin-resistant Staphylococcus aureus strain N315. Antimicrob. Agents Chemother. 2016, 60, 3455–3461. [Google Scholar] [CrossRef]
- Yang, S.J.; Xiong, Y.Q.; Yeaman, M.R.; Bayles, K.W.; Abdelhady, W.; Bayer, A.S. Role of the LytSR two-component regulatory system in adaptation to cationic antimicrobial peptides in Staphylococcus aureus. Antimicrob. Agents Chemother. 2013, 57, 3875–3882. [Google Scholar] [CrossRef] [PubMed]
- Patton, T.G.; Yang, S.J.; Bayles, K.W. The role of proton motive force in expression of the Staphylococcus aureus cid and lrg operons. Mol. Microbiol. 2006, 59, 1395–1404. [Google Scholar] [CrossRef]
- Lehman, M.K.; Bose, J.L.; Sharma-Kuinkel, B.K.; Moormeier, D.E.; Endres, J.L.; Sadykov, M.R.; Biswas, I.; Bayles, K.W. Identification of the amino acids essential for LytSR-mediated signal transduction in Staphylococcus aureus and their roles in biofilm-specific gene expression. Mol. Microbiol. 2015, 95, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.; Golemi-Kotra, D. Signaling mechanism by the Staphylococcus aureus two-component system LytSR: Role of acetyl phosphate in bypassing the cell membrane electrical potential sensor LytS. F1000Research 2015, 4, 79. [Google Scholar] [CrossRef]
- Li, M.; Cha, D.J.; Lai, Y.; Villaruz, A.E.; Sturdevant, D.E.; Otto, M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 2007, 66, 1136–1147. [Google Scholar] [CrossRef]
- Meehl, M.; Herbert, S.; Gotz, F.; Cheung, A. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 2679–2689. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Z.; Xu, T.; Ge, H.; Zhou, F.; Zhu, X.; Li, X.; Qu, D.; Zheng, C.; Wu, Y.; et al. The Role of graRS in regulating virulence and antimicrobial resistance in methicillin-resistant Staphylococcus aureus. Front. Microbiol. 2021, 12, 727104. [Google Scholar] [CrossRef] [PubMed]
- Falord, M.; Mader, U.; Hiron, A.; Debarbouille, M.; Msadek, T. Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS ONE 2011, 6, e21323. [Google Scholar] [CrossRef]
- Yang, S.J.; Bayer, A.S.; Mishra, N.N.; Meehl, M.; Ledala, N.; Yeaman, M.R.; Xiong, Y.Q.; Cheung, A.L. The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect. Immun. 2012, 80, 74–81. [Google Scholar] [CrossRef]
- Li, M.; Lai, Y.; Villaruz, A.E.; Cha, D.J.; Sturdevant, D.E.; Otto, M. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. USA 2007, 104, 9469–9474. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.L.; Bayer, A.S.; Yeaman, M.R.; Xiong, Y.Q.; Waring, A.J.; Memmi, G.; Donegan, N.; Chaili, S.; Yang, S.J. Site-specific mutation of the sensor kinase GraS in Staphylococcus aureus alters the adaptive response to distinct cationic antimicrobial peptides. Infect. Immun. 2014, 82, 5336–5345. [Google Scholar] [CrossRef]
- Cheung, A.L.; Cho, J.; Bayer, A.S.; Yeaman, M.R.; Xiong, Y.Q.; Donegan, N.P.; Mikheyeva, I.V.; Lee, G.Y.; Yang, S.J. Role of the Staphylococcus aureus extracellular loop of GraS in resistance to distinct human defense peptides in PMN and invasive cardiovascular infections. Infect. Immun. 2021, 89, e0034721. [Google Scholar] [CrossRef]
- Kuiack, R.C.; Veldhuizen, R.A.W.; McGavin, M.J. Novel functions and signaling specificity for the GraS sensor kinase of Staphylococcus aureus in response to acidic pH. J. Bacteriol. 2020, 202, e00219-20. [Google Scholar] [CrossRef]
- Falord, M.; Karimova, G.; Hiron, A.; Msadek, T. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 1047–1058. [Google Scholar] [CrossRef]
- Muzamal, U.; Gomez, D.; Kapadia, F.; Golemi-Kotra, D. Diversity of two-component systems: Insights into the signal transduction mechanism by the Staphylococcus aureus two-component system GraSR. F1000Research 2014, 3, 252. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Costa, S.K.; Wierzbicki, R.M.; Rigby, W.F.C.; Cheung, A.L. The extracellular loop of the membrane permease VraG interacts with GraS to sense cationic antimicrobial peptides in Staphylococcus aureus. PLoS Pathog. 2021, 17, e1009338. [Google Scholar] [CrossRef] [PubMed]
- Flannagan, R.S.; Kuiack, R.C.; McGavin, M.J.; Heinrichs, D.E. Staphylococcus aureus uses the GraXRS regulatory system to sense and sdapt to the acidified phagolysosome in macrophages. mBio 2018, 9, e01143-18. [Google Scholar] [CrossRef]
- Giraudo, A.T.; Raspanti, C.G.; Calzolari, A.; Nagel, R. Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can. J. Microbiol. 1994, 40, 677–681. [Google Scholar] [CrossRef]
- Rogasch, K.; Ruhmling, V.; Pane-Farre, J.; Hoper, D.; Weinberg, C.; Fuchs, S.; Schmudde, M.; Broker, B.M.; Wolz, C.; Hecker, M.; et al. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 2006, 188, 7742–7758. [Google Scholar] [CrossRef]
- Steinhuber, A.; Goerke, C.; Bayer, M.G.; Doring, G.; Wolz, C. Molecular architecture of the regulatory Locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 2003, 185, 6278–6286. [Google Scholar] [CrossRef]
- Nygaard, T.K.; Borgogna, T.R.; Sward, E.W.; Guerra, F.E.; Dankoff, J.G.; Collins, M.M.; Pallister, K.B.; Chen, L.; Kreiswirth, B.N.; Voyich, J.M. Aspartic Acid Residue 51 of SaeR Is Essential for Staphylococcus aureus Virulence. Front. Microbiol. 2018, 9, 3085. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yeo, W.S.; Bae, T. The SaeRS Two-Component System of Staphylococcus aureus. Genes 2016, 7, 81. [Google Scholar] [CrossRef]
- Geiger, T.; Goerke, C.; Mainiero, M.; Kraus, D.; Wolz, C. The virulence regulator Sae of Staphylococcus aureus: Promoter activities and response to phagocytosis-related signals. J. Bacteriol. 2008, 190, 3419–3428. [Google Scholar] [CrossRef] [PubMed]
- Flack, C.E.; Zurek, O.W.; Meishery, D.D.; Pallister, K.B.; Malone, C.L.; Horswill, A.R.; Voyich, J.M. Differential regulation of staphylococcal virulence by the sensor kinase SaeS in response to neutrophil-derived stimuli. Proc. Natl. Acad. Sci. USA 2014, 111, E2037–E2045. [Google Scholar] [CrossRef]
- Liu, Q.; Cho, H.; Yeo, W.S.; Bae, T. The extracytoplasmic linker peptide of the sensor protein SaeS tunes the kinase activity required for staphylococcal virulence in response to host signals. PLoS Pathog. 2015, 11, e1004799. [Google Scholar] [CrossRef]
- Marincola, G.; Schafer, T.; Behler, J.; Bernhardt, J.; Ohlsen, K.; Goerke, C.; Wolz, C. RNase Y of Staphylococcus aureus and its role in the activation of virulence genes. Mol. Microbiol. 2012, 85, 817–832. [Google Scholar] [CrossRef]
- Novick, R.P.; Jiang, D. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 2003, 149, 2709–2717. [Google Scholar] [CrossRef]
- Mlynek, K.D.; Sause, W.E.; Moormeier, D.E.; Sadykov, M.R.; Hill, K.R.; Torres, V.J.; Bayles, K.W.; Brinsmade, S.R. Nutritional regulation of the Sae two-component system by CodY in Staphylococcus aureus. J. Bacteriol. 2018, 200, e00012-18. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Cheung, A. Repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect. Immun. 2008, 76, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.W.; Cho, H.; Lee, H.; Li, C.; Garza, J.; Fried, M.; Bae, T. Identification of the P3 promoter and distinct roles of the two promoters of the SaeRS two-component system in Staphylococcus aureus. J. Bacteriol. 2011, 193, 4672–4684. [Google Scholar] [CrossRef]
- Mainiero, M.; Goerke, C.; Geiger, T.; Gonser, C.; Herbert, S.; Wolz, C. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J. Bacteriol. 2010, 192, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.W.; Cho, H.; Jones, M.B.; Shatzkes, K.; Sun, F.; Ji, Q.; Liu, Q.; Peterson, S.N.; He, C.; Bae, T. The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol. Microbiol. 2012, 86, 331–348. [Google Scholar] [CrossRef] [PubMed]
- Omae, Y.; Hanada, Y.; Sekimizu, K.; Kaito, C. Silkworm apolipophorin protein inhibits hemolysin gene expression of Staphylococcus aureus via binding to cell surface lipoteichoic acids. J. Biol. Chem. 2013, 288, 25542–25550. [Google Scholar] [CrossRef]
- Cho, H.; Jeong, D.W.; Liu, Q.; Yeo, W.S.; Vogl, T.; Skaar, E.P.; Chazin, W.J.; Bae, T. Calprotectin increases the activity of the SaeRS two component system and murine mortality during Staphylococcus aureus Infections. PloS Pathog. 2015, 11, e1005026. [Google Scholar] [CrossRef]
- Krute, C.N.; Rice, K.C.; Bose, J.L. VfrB is a key activator of the Staphylococcus aureus SaeRS two-component system. J. Bacteriol. 2017, 199, E00828-16. [Google Scholar] [CrossRef]
- Ericson, M.E.; Subramanian, C.; Frank, M.W.; Rock, C.O. Role of fatty acid kinase in cellular lipid homeostasis and SaeRS-dependent virulence factor expression in Staphylococcus aureus. mBio 2017, 8, e00988-17. [Google Scholar] [CrossRef]
- DeMars, Z.R.; Krute, C.N.; Ridder, M.J.; Gilchrist, A.K.; Menjivar, C.; Bose, J.L. Fatty acids can inhibit Staphylococcus aureus SaeS activity at the membrane independent of alterations in respiration. Mol. Microbiol. 2021, 116, 1378–1391. [Google Scholar] [CrossRef]
- Schurig-Briccio, L.A.; Parraga Solorzano, P.K.; Lencina, A.M.; Radin, J.N.; Chen, G.Y.; Sauer, J.D.; Kehl-Fie, T.E.; Gennis, R.B. Role of respiratory NADH oxidation in the regulation of Staphylococcus aureus virulence. EMBO Rep. 2020, 21, e45832. [Google Scholar] [CrossRef]
- Fernandez, P.; Diaz, A.R.; Re, M.F.; Porrini, L.; de Mendoza, D.; Albanesi, D.; Mansilla, M.C. Identification of Novel thermosensors in Gram-Positive pathogens. Front. Mol. Biosci. 2020, 7, 592747. [Google Scholar] [CrossRef]
- Fournier, B.; Hooper, D.C. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 2000, 182, 3955–3964. [Google Scholar] [CrossRef] [PubMed]
- Fournier, B.; Klier, A.; Rapoport, G. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 2001, 41, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Crosby, H.A.; Tiwari, N.; Kwiecinski, J.M.; Xu, Z.; Dykstra, A.; Jenul, C.; Fuentes, E.J.; Horswill, A.R. The Staphylococcus aureus ArlRS two-component system regulates virulence factor expression through MgrA. Mol. Microbiol. 2020, 113, 103–122. [Google Scholar] [CrossRef]
- Luong, T.T.; Lee, C.Y. The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology 2006, 152, 3123–3131. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Zhu, X.; Zhao, K.; Yan, Y.; Xu, T.; Wang, J.; Zheng, J.; Huang, W.; Shi, L.; Shang, Y.; et al. The role of ArlRS in regulating oxacillin susceptibility in methicillin-resistant Staphylococcus aureus indicates it is a potential target for antimicrobial resistance breakers. Emerg. Microbes Infect. 2019, 8, 503–515. [Google Scholar] [CrossRef]
- Fournier, B.; Klier, A. Protein A gene expression is regulated by DNA supercoiling which is modified by the ArlS-ArlR two-component system of Staphylococcus aureus. Microbiology 2004, 150, 3807–3819. [Google Scholar] [CrossRef]
- Schroder, W.; Bernhardt, J.; Marincola, G.; Klein-Hitpass, L.; Herbig, A.; Krupp, G.; Nieselt, K.; Wolz, C. Altering gene expression by aminocoumarins: The role of DNA supercoiling in Staphylococcus aureus. BMC Genomics 2014, 15, 291. [Google Scholar] [CrossRef] [PubMed]
- Parraga Solorzano, P.K.; Shupe, A.C.; Kehl-Fie, T.E. The sensor histidine kinase ArlS Is necessary for Staphylococcus aureus to activate ArlR in response to nutrient availability. J. Bacteriol. 2021, 203, e0042221. [Google Scholar] [CrossRef]
- Parraga Solorzano, P.K.; Yao, J.; Rock, C.O.; Kehl-Fie, T.E. Disruption of glycolysis by nutritional immunity activates a two-component system that coordinates a metabolic and antihost response by Staphylococcus aureus. mBio 2019, 10, e01321-19. [Google Scholar] [CrossRef]
- Yarwood, J.M.; McCormick, J.K.; Schlievert, P.M. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 2001, 183, 1113–1123. [Google Scholar] [CrossRef] [PubMed]
- Kinkel, T.L.; Roux, C.M.; Dunman, P.M.; Fang, F.C. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio 2013, 4, e00696-13. [Google Scholar] [CrossRef]
- Wilde, A.D.; Snyder, D.J.; Putnam, N.E.; Valentino, M.D.; Hammer, N.D.; Lonergan, Z.R.; Hinger, S.A.; Aysanoa, E.E.; Blanchard, C.; Dunman, P.M.; et al. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog. 2015, 11, e1005341. [Google Scholar] [CrossRef] [PubMed]
- Windham, I.H.; Chaudhari, S.S.; Bose, J.L.; Thomas, V.C.; Bayles, K.W. SrrAB modulates Staphylococcus aureus cell death through regulation of cidABC transcription. J. Bacteriol. 2016, 198, 1114–1122. [Google Scholar] [CrossRef]
- Tiwari, N.; Lopez-Redondo, M.; Miguel-Romero, L.; Kulhankova, K.; Cahill, M.P.; Tran, P.M.; Kinney, K.J.; Kilgore, S.H.; Al-Tameemi, H.; Herfst, C.A.; et al. The SrrAB two-component system regulates Staphylococcus aureus pathogenicity through redox sensitive cysteines. Proc. Natl. Acad. Sci. USA 2020, 117, 10989–10999. [Google Scholar] [CrossRef]
- Tiwari, K.B.; Sen, S.; Gatto, C.; Wilkinson, B.J. Fluorescence Polarization (FP) Assay for measuring Staphylococcus aureus membrane fluidity. Methods Mol. Biol. 2021, 2341, 55–68. [Google Scholar] [CrossRef]
- Kelliher, J.L.; Radin, J.N.; Kehl-Fie, T.E. PhoPR contributes to Staphylococcus aureus growth during phosphate starvation and pathogenesis in an environment-specific manner. Infect. Immun. 2018, 86, e00371-18. [Google Scholar] [CrossRef] [PubMed]
- Gardner, S.G.; Johns, K.D.; Tanner, R.; McCleary, W.R. The PhoU protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. J. Bacteriol. 2014, 196, 1741–1752. [Google Scholar] [CrossRef] [PubMed]
- Vuppada, R.K.; Hansen, C.R.; Strickland, K.A.P.; Kelly, K.M.; McCleary, W.R. Phosphate signaling through alternate conformations of the PstSCAB phosphate transporter. BMC Microbiol. 2018, 18, 8. [Google Scholar] [CrossRef]
- Devine, K.M. Activation of the PhoPR-mediated response to phosphate limitation is regulated by wall teichoic acid metabolism in Bacillus subtilis. Front. Microbiol. 2018, 9, 2678. [Google Scholar] [CrossRef]
- Santos-Beneit, F.; Ordonez-Robles, M.; Martin, J.F. Glycopeptide resistance: Links with inorganic phosphate metabolism and cell envelope stress. Biochem. Pharmacol. 2017, 133, 74–85. [Google Scholar] [CrossRef][Green Version]
- Sun, F.; Ji, Q.; Jones, M.B.; Deng, X.; Liang, H.; Frank, B.; Telser, J.; Peterson, S.N.; Bae, T.; He, C. AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J. Am. Chem. Soc. 2012, 134, 305–314. [Google Scholar] [CrossRef]
- Sun, J.; Zheng, L.; Landwehr, C.; Yang, J.; Ji, Y. Identification of a novel essential two-component signal transduction system, YhcSR, in Staphylococcus aureus. J. Bacteriol. 2005, 187, 7876–7880. [Google Scholar] [CrossRef]
- Hall, J.W.; Yang, J.; Guo, H.; Ji, Y. The AirSR two-component system contributes to Staphylococcus aureus survival in human blood and transcriptionally regulates sspABC operon. Front. Microbiol. 2015, 6, 682. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Yang, Y.; Xue, T.; Sun, B. Modulation of cell wall synthesis and susceptibility to vancomycin by the two-component system AirSR in Staphylococcus aureus NCTC8325. BMC Microbiol. 2013, 13, 286. [Google Scholar] [CrossRef]
- Kuroda, M.; Kuroda, H.; Oshima, T.; Takeuchi, F.; Mori, H.; Hiramatsu, K. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 2003, 49, 807–821. [Google Scholar] [CrossRef]
- Sobral, R.G.; Jones, A.E.; Des Etages, S.G.; Dougherty, T.J.; Peitzsch, R.M.; Gaasterland, T.; Ludovice, A.M.; de Lencastre, H.; Tomasz, A. Extensive and genome-wide changes in the transcription profile of Staphylococcus aureus induced by modulating the transcription of the cell wall synthesis gene murF. J. Bacteriol. 2007, 189, 2376–2391. [Google Scholar] [CrossRef]
- Gardete, S.; Wu, S.W.; Gill, S.; Tomasz, A. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 3424–3434. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Daum, R.S.; Boyle-Vavra, S. VraSR two-component regulatory system and its role in induction of pbp2 and vraSR expression by cell wall antimicrobials in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Boyle-Vavra, S.; Yin, S.; Daum, R.S. The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 2006, 262, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Kornblum, J.; Kreiswirth, B.; Projan, S.J.; Ross, H.F.; Novick, R.P. Agr: A polycistronic locus regulating exoprotein synthesis in S. aureus. In Molecular Biology of the Staphylococci; Novick, R.P., Ed.; VCH Publishers: New York, NY, USA, 1990; Volume 535, pp. 373–402. [Google Scholar] [CrossRef]
- Novick, R.P.; Geisinger, E. Quorum sensing in staphylococci. Annu. Rev. Genet. 2008, 42, 541–564. [Google Scholar] [CrossRef]
- Kavanaugh, J.S.; Horswill, A.R. Impact of environmental cues on Staphylococcal quorum sensing and biofilm development. J. Biol. Chem. 2016, 291, 12556–12564. [Google Scholar] [CrossRef] [PubMed]
- Bronesky, D.; Wu, Z.; Marzi, S.; Walter, P.; Geissmann, T.; Moreau, K.; Vandenesch, F.; Caldelari, I.; Romby, P. Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu. Rev. Microbiol. 2016, 70, 299–316. [Google Scholar] [CrossRef]
- Horswill, A.R.; Gordon, C.P. Structure-activity relationship studies of small molecule modulators of the Staphylococcal accessory gene regulator. J. Med. Chem. 2020, 63, 2705–2730. [Google Scholar] [CrossRef]
- Ji, G.; Beavis, R.C.; Novick, R.P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 1995, 92, 12055–12059. [Google Scholar] [CrossRef] [PubMed]
- Gless, B.H.; Bejder, B.S.; Monda, F.; Bojer, M.S.; Ingmer, H.; Olsen, C.A. Rearrangement of Thiodepsipeptides by S --> N Acyl shift delivers homodetic autoinducing peptides. J. Am. Chem. Soc. 2021, 143, 10514–10518. [Google Scholar] [CrossRef]
- Lyon, G.J.; Mayville, P.; Muir, T.W.; Novick, R.P. Rational design of a global inhibitor of the virulence response in Staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. Proc. Natl. Acad. Sci. USA 2000, 97, 13330–13335. [Google Scholar] [CrossRef]
- Tal-Gan, Y.; Ivancic, M.; Cornilescu, G.; Blackwell, H.E. Characterization of structural elements in native autoinducing peptides and non-native analogues that permit the differential modulation of AgrC-type quorum sensing receptors in Staphylococcus aureus. Org. Biomol. Chem. 2016, 14, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Tal-Gan, Y.; Ivancic, M.; Cornilescu, G.; Cornilescu, C.C.; Blackwell, H.E. Structural characterization of native autoinducing peptides and abiotic analogues reveals key features essential for activation and inhibition of an AgrC quorum sensing receptor in Staphylococcus aureus. J. Am. Chem. Soc. 2013, 135, 18436–18444. [Google Scholar] [CrossRef]
- West, K.H.J.; Shen, W.; Eisenbraun, E.L.; Yang, T.; Vasquez, J.K.; Horswill, A.R.; Blackwell, H.E. Non-native peptides capable of pan-activating the agr quorum sensing system across multiple specificity groups of Staphylococcus epidermidis. ACS Chem. Biol. 2021, 16, 1070–1078. [Google Scholar] [CrossRef]
- Wang, B.; Zhao, A.; Novick, R.P.; Muir, T.W. Activation and inhibition of the receptor histidine kinase AgrC occurs through opposite helical transduction motions. Mol. Cell 2014, 53, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Zhao, A.; Jeffrey, P.D.; Kim, M.K.; Bassler, B.L.; Stone, H.A.; Novick, R.P.; Muir, T.W. Identification of a molecular latch that regulates staphylococcal virulence. Cell. Chem. Biol. 2019, 26, 548–558.e4. [Google Scholar] [CrossRef]
- Geisinger, E.; George, E.A.; Chen, J.; Muir, T.W.; Novick, R.P. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J. Biol. Chem. 2008, 283, 8930–8938. [Google Scholar] [CrossRef] [PubMed]
- Sloan, T.J.; Murray, E.; Yokoyama, M.; Massey, R.C.; Chan, W.C.; Bonev, B.B.; Williams, P. Timing is everything: Impact of naturally occurring Staphylococcus aureus AgrC cytoplasmic domain adaptive mutations on autoinduction. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed]
- Marroquin, S.; Gimza, B.; Tomlinson, B.; Stein, M.; Frey, A.; Keogh, R.A.; Zapf, R.; Todd, D.A.; Cech, N.B.; Carroll, R.K.; et al. MroQ Is a novel Abi-Domain Protein that influences virulence gene expression in Staphylococcus aureus via modulation of Agr activity. Infect. Immun. 2019, 87, e00002-19. [Google Scholar] [CrossRef]
- Cosgriff, C.J.; White, C.R.; Teoh, W.P.; Grayczyk, J.P.; Alonzo, F., 3rd. Control of Staphylococcus aureus quorum sensing by a membrane-embedded peptidase. Infect. Immun. 2019, 87, e00019-19. [Google Scholar] [CrossRef]
- Freeman, Z.N.; Dorus, S.; Waterfield, N.R. The KdpD/KdpE two-component system: Integrating K(+) homeostasis and virulence. PLoS Pathog. 2013, 9, e1003201. [Google Scholar] [CrossRef]
- Xue, T.; You, Y.; Hong, D.; Sun, H.; Sun, B. The Staphylococcus aureus KdpDE two-component system couples extracellular K+ sensing and Agr signaling to infection programming. Infect. Immun. 2011, 79, 2154–2167. [Google Scholar] [CrossRef]
- Price-Whelan, A.; Poon, C.K.; Benson, M.A.; Eidem, T.T.; Roux, C.M.; Boyd, J.M.; Dunman, P.M.; Torres, V.J.; Krulwich, T.A. Transcriptional profiling of Staphylococcus aureus during growth in 2 M NaCl leads to clarification of physiological roles for Kdp and Ktr K+ uptake systems. mBio 2013, 4, e00407-13. [Google Scholar] [CrossRef] [PubMed]
- Gries, C.M.; Bose, J.L.; Nuxoll, A.S.; Fey, P.D.; Bayles, K.W. The Ktr potassium transport system in Staphylococcus aureus and its role in cell physiology, antimicrobial resistance and pathogenesis. Mol. Microbiol. 2013, 89, 760–773. [Google Scholar] [CrossRef]
- Zhao, L.; Xue, T.; Shang, F.; Sun, H.; Sun, B. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect. Immun. 2010, 78, 3506–3515. [Google Scholar] [CrossRef] [PubMed]
- Burda, W.N.; Miller, H.K.; Krute, C.N.; Leighton, S.L.; Carroll, R.K.; Shaw, L.N. Investigating the genetic regulation of the ECF sigma factor sigmaS in Staphylococcus aureus. BMC Microbiol. 2014, 14, 280. [Google Scholar] [CrossRef] [PubMed]
- Moscoso, J.A.; Schramke, H.; Zhang, Y.; Tosi, T.; Dehbi, A.; Jung, K.; Grundling, A. Binding of Cyclic Di-AMP to the Staphylococcus aureus sensor kinase KdpD occurs via the universal stress protein domain and downregulates the expression of the Kdp potassium transporter. J. Bacteriol. 2016, 198, 98–110. [Google Scholar] [CrossRef]
- Hanssen, A.M.; Ericson Sollid, J.U. SCCmec in staphylococci: Genes on the move. FEMS Immunol. Med. Microbiol. 2006, 46, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Torres, V.J.; Stauff, D.L.; Pishchany, G.; Bezbradica, J.S.; Gordy, L.E.; Iturregui, J.; Anderson, K.L.; Dunman, P.M.; Joyce, S.; Skaar, E.P. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 2007, 1, 109–119. [Google Scholar] [CrossRef]
- Stauff, D.L.; Skaar, E.P. The heme sensor system of Staphylococcus aureus. Contrib. Microbiol. 2009, 16, 120–135. [Google Scholar] [CrossRef]
- Schlag, S.; Fuchs, S.; Nerz, C.; Gaupp, R.; Engelmann, S.; Liebeke, M.; Lalk, M.; Hecker, M.; Gotz, F. Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J. Bacteriol. 2008, 190, 7847–7858. [Google Scholar] [CrossRef]
- Reinhart, F.; Huber, A.; Thiele, R.; Unden, G. Response of the oxygen sensor NreB to air in vivo: Fe-S-containing NreB and apo-NreB in aerobically and anaerobically growing Staphylococcus carnosus. J. Bacteriol. 2010, 192, 86–93. [Google Scholar] [CrossRef]
- Klein, R.; Kretzschmar, A.K.; Unden, G. Control of the bifunctional O2 -sensor kinase NreB of Staphylococcus carnosus by the nitrate sensor NreA: Switching from kinase to phosphatase state. Mol. Microbiol. 2020, 113, 369–380. [Google Scholar] [CrossRef]
- Hiron, A.; Falord, M.; Valle, J.; Debarbouille, M.; Msadek, T. Bacitracin and nisin resistance in Staphylococcus aureus: A novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol. Microbiol. 2011, 81, 602–622. [Google Scholar] [CrossRef]
- Blake, K.L.; Randall, C.P.; O’Neill, A.J. In vitro studies indicate a high resistance potential for the lantibiotic nisin in Staphylococcus aureus and define a genetic basis for nisin resistance. Antimicrob. Agents Chemother. 2011, 55, 2362–2368. [Google Scholar] [CrossRef]
- Arii, K.; Kawada-Matsuo, M.; Oogai, Y.; Noguchi, K.; Komatsuzawa, H. Single mutations in BraRS confer high resistance against nisin A in Staphylococcus aureus. Microbiologyopen 2019, 8, e791. [Google Scholar] [CrossRef]
- Kolar, S.L.; Nagarajan, V.; Oszmiana, A.; Rivera, F.E.; Miller, H.K.; Davenport, J.E.; Riordan, J.T.; Potempa, J.; Barber, D.S.; Koziel, J.; et al. NsaRS is a cell-envelope-stress-sensing two-component system of Staphylococcus aureus. Microbiology 2011, 157, 2206–2219. [Google Scholar] [CrossRef] [PubMed]
- Coates-Brown, R.; Moran, J.C.; Pongchaikul, P.; Darby, A.C.; Horsburgh, M.J. Comparative genomics of Staphylococcus reveals determinants of speciation and diversification of antimicrobial defense. Front. Microbiol. 2018, 9, 2753. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Phillips-Jones, M.K. Membrane sensor histidine kinases: Insights from structural, ligand and inhibitor studies of full-length proteins and signalling domains for antibiotic discovery. Molecules 2021, 26, 5110. [Google Scholar] [CrossRef]
- Xie, Q.; Wiedmann, M.M.; Zhao, A.; Pagan, I.R.; Novick, R.P.; Suga, H.; Muir, T.W. Discovery of quorum quenchers targeting the membrane-embedded sensor domain of the Staphylococcus aureus receptor histidine kinase, AgrC. Chem. Commun. 2020, 56, 11223–11226. [Google Scholar] [CrossRef] [PubMed]
- Khan, B.A.; Yeh, A.J.; Cheung, G.Y.; Otto, M. Investigational therapies targeting quorum-sensing for the treatment of Staphylococcus aureus infections. Expert Opin. Investig. Drugs 2015, 24, 689–704. [Google Scholar] [CrossRef]
- Yeo, W.S.; Arya, R.; Kim, K.K.; Jeong, H.; Cho, K.H.; Bae, T. The FDA-approved anti-cancer drugs, streptozotocin and floxuridine, reduce the virulence of Staphylococcus aureus. Sci. Rep. 2018, 8, 2521. [Google Scholar] [CrossRef]
- Lee, H.; Boyle-Vavra, S.; Ren, J.; Jarusiewicz, J.A.; Sharma, L.K.; Hoagland, D.T.; Yin, S.; Zhu, T.; Hevener, K.E.; Ojeda, I.; et al. Identification of small molecules exhibiting oxacillin synergy through a novel assay for inhibition of vraTSR expression in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2019, 63, E02593-18. [Google Scholar] [CrossRef]
- Velikova, N.; Bem, A.E.; van Baarlen, P.; Wells, J.M.; Marina, A. WalK, the path towards new antibacterials with low potential for resistance development. ACS Med. Chem. Lett. 2013, 4, 891–894. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).