Osteocytic Pericellular Matrix (PCM): Accelerated Degradation under In Vivo Loading and Unloading Conditions Using a Novel Imaging Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Metabolic Labeling of the Osteocytic PCM In Vitro and In Vivo
2.2. Testing the Metabolic Labeling Method in Young vs. Old Bone
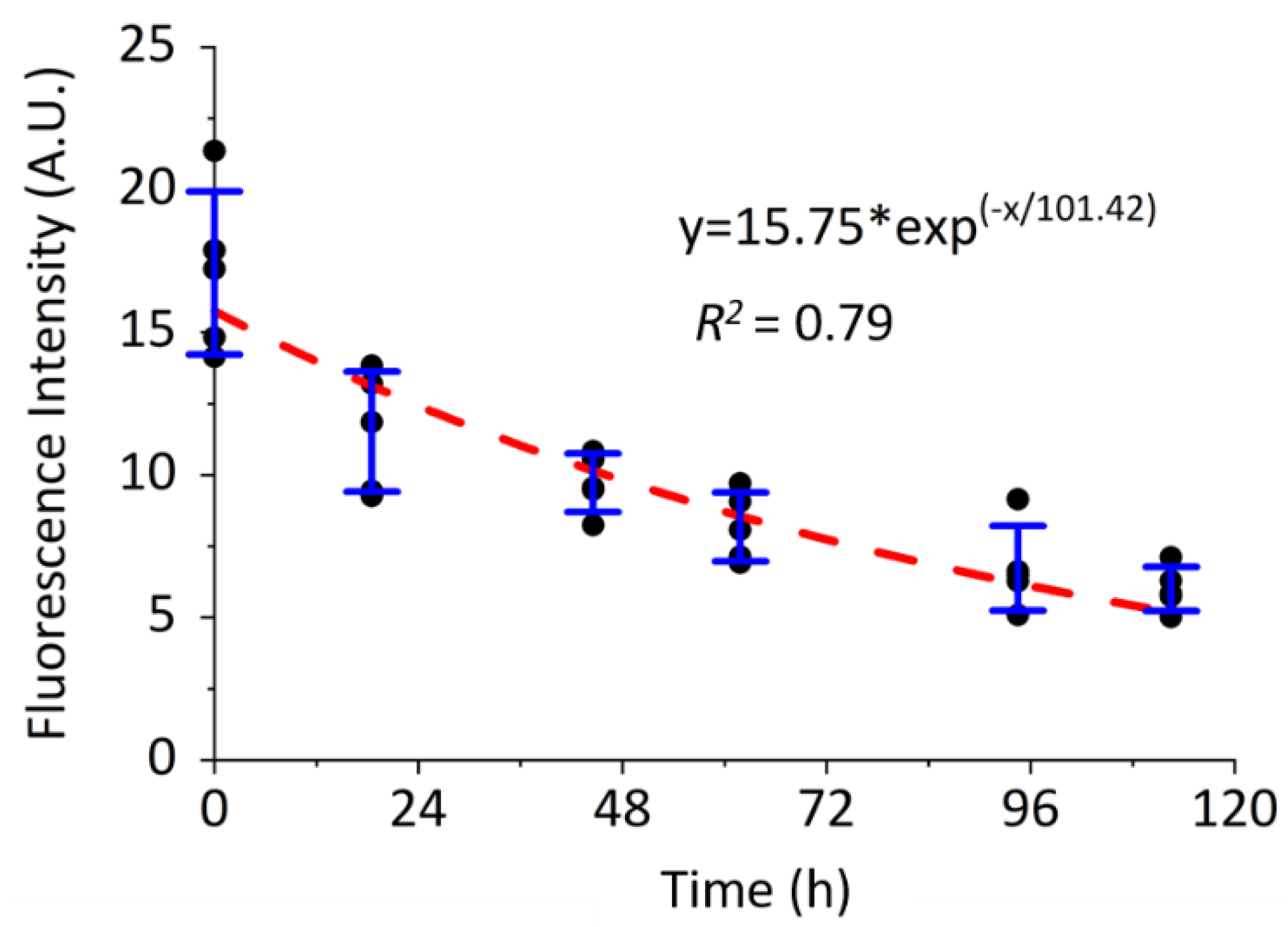
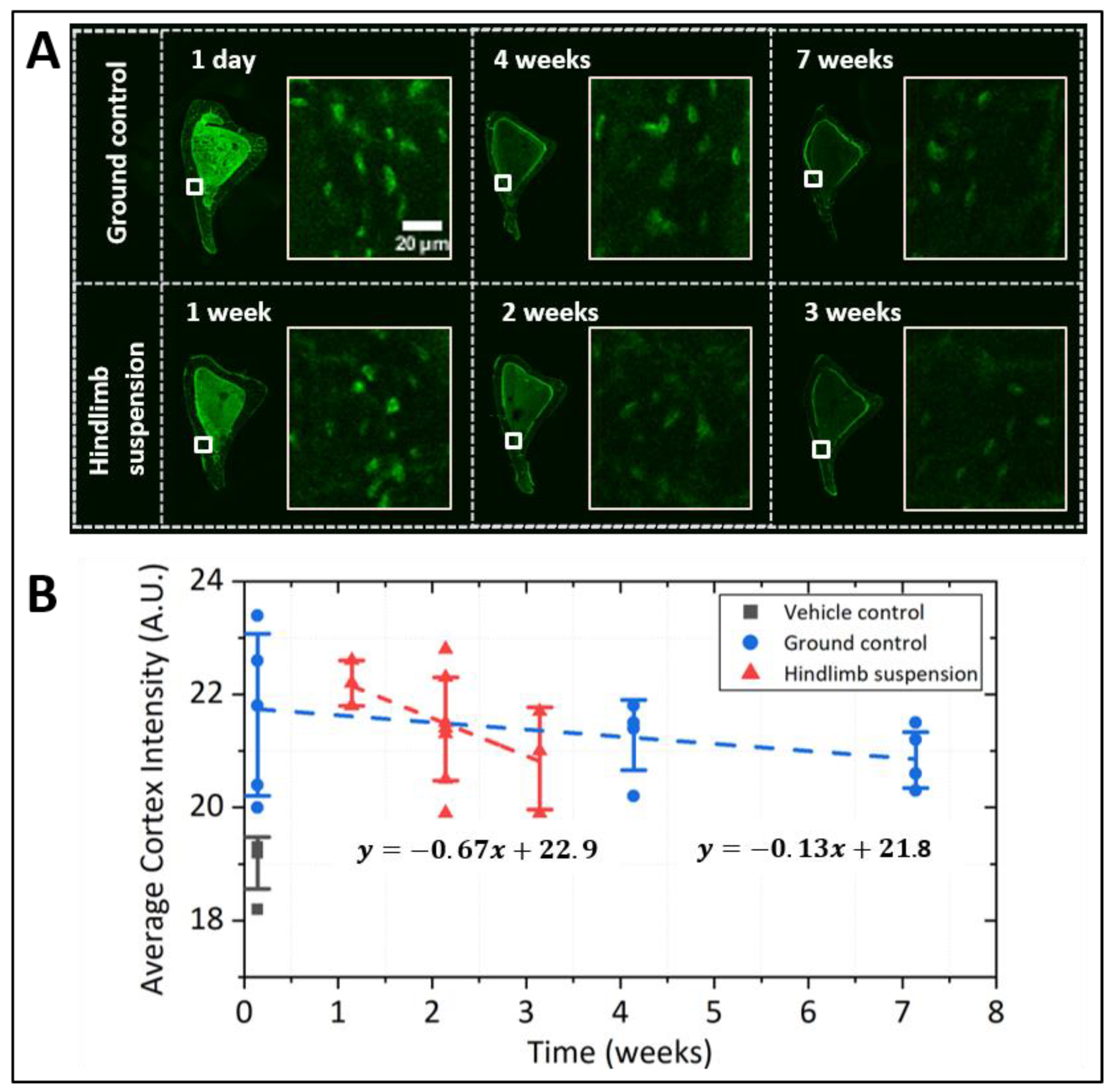
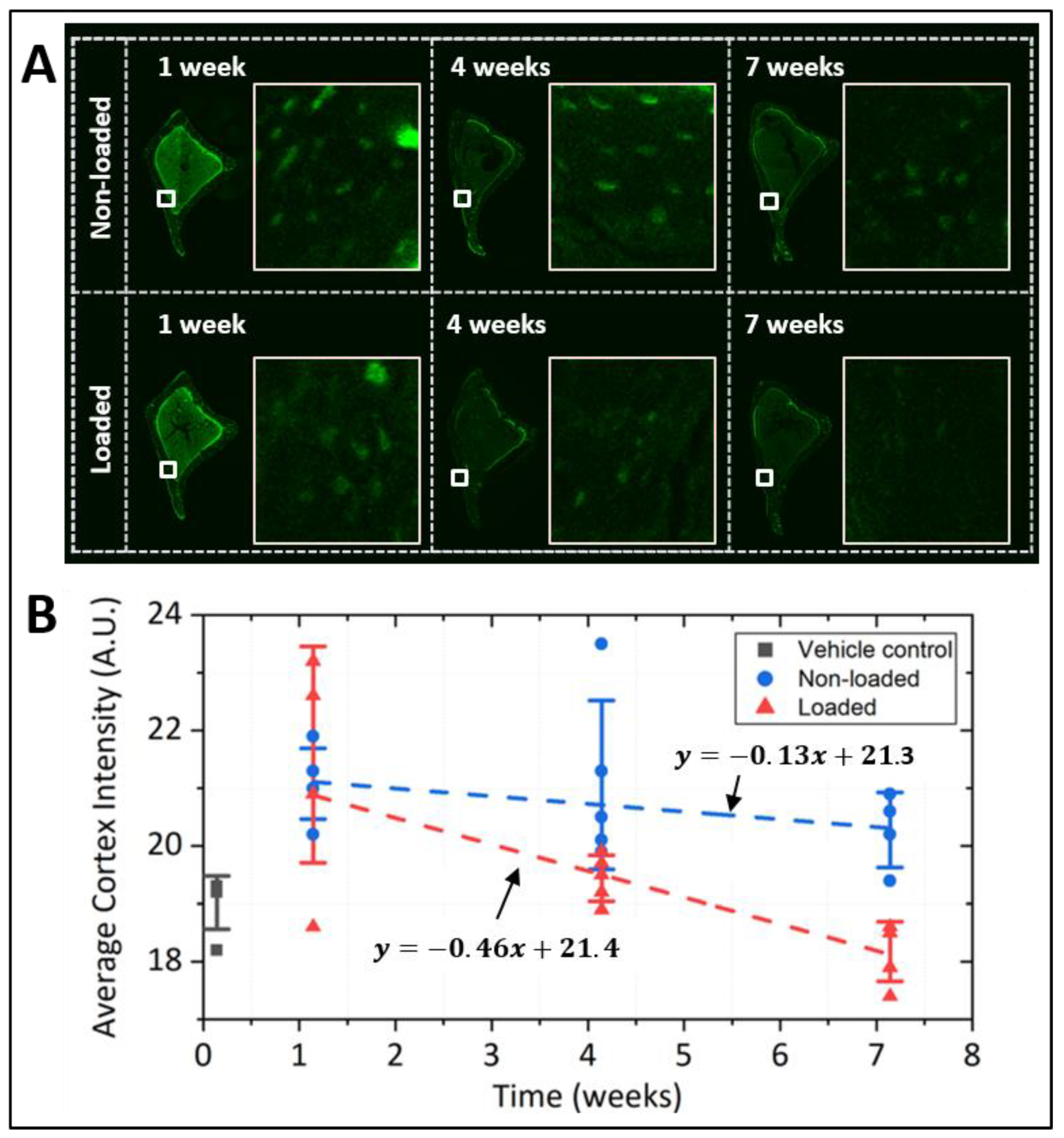
2.3. Pulse-Chase Experiments in Mice and Cultured Osteocytic Cells
2.4. RNA-Sequencing and Immunohistochemistry
3. Results
3.1. Metabolic Labeling of De Novo Osteocytic PCM in Cultured Cells and Living Bone
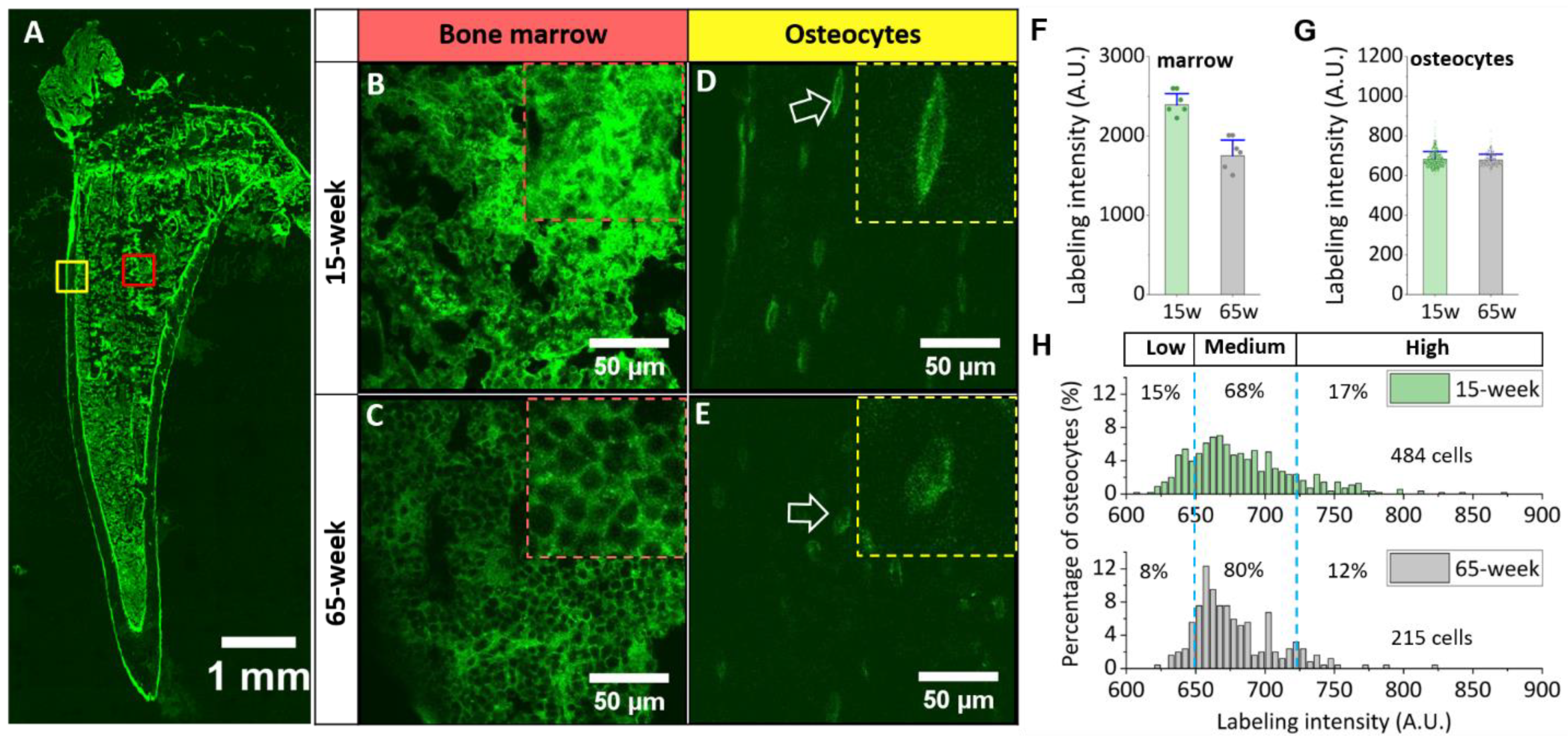
3.2. Different Labeling Patterns in Young vs. Old Bone
3.3. Different Labeling Patterns in Young vs. Old Bone
3.4. Mechanical Loading Increased Transcript and Protein Levels of MMP 14 in Bone
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vitkov, L.; Krautgartner, W.D.; Hannig, M.; Weitgasser, R.; Stoiber, W. Candida attachment to oral epithelium. Oral Microbiol. Immunol. 2002, 17, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Reitsma, S.; Slaaf, D.W.; Vink, H.; van Zandvoort, M.A.M.J.; oude Egbrink, M.G.A. The endothelial glycocalyx: Composition, functions, and visualization. Pflügers Arch. Eur. J. Physiol. 2007, 454, 345–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horiuchi, K.; Naito, I.; Nakano, K.; Nakatani, S.; Nishida, K.; Taguchi, T.; Ohtsuka, A. Three-dimensional ultrastructure of the brush border glycocalyx in the mouse small intestine: A high resolution scanning electron microscopic study. Arch. Histol. Cytol. 2005, 68, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Rilla, K.; Tiihonen, R.; Kultti, A.; Tammi, M.; Tammi, R. Pericellular Hyaluronan Coat Visualized in Live Cells with a Fluorescent Probe Is Scaffolded by Plasma Membrane Protrusions. J. Histochem. Cytochem. 2008, 56, 901–910. [Google Scholar] [CrossRef] [Green Version]
- Guilak, F.; Alexopoulos, L.G.; Upton, M.L.; Youn, I.; Choi, J.B.; Cao, L.; Setton, L.A.; Haider, M.A. The pericellular matrix as a transducer of biomechanical and biochemical signals in articular cartilage. Ann. N. Y. Acad. Sci. 2006, 1068, 498–512. [Google Scholar] [CrossRef]
- You, L.-D.; Weinbaum, S.; Cowin, S.C.; Schaffler, M.B. Ultrastructure of the osteocyte process and its pericellular matrix. Anat. Rec. 2004, 278A, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Thompson, W.R.; Modla, S.; Grindel, B.J.; Czymmek, K.J.; Kirn-Safran, C.B.; Wang, L.; Duncan, R.L.; Farach-Carson, M.C. Perlecan/Hspg2 deficiency alters the pericellular space of the lacunocanalicular system surrounding osteocytic processes in cortical bone. J. Bone Miner. Res. 2011, 26, 618–629. [Google Scholar] [CrossRef] [Green Version]
- Wang, L. Solute Transport in the Bone Lacunar-Canalicular System (LCS). Curr. Osteoporos. Rep. 2018, 16, 32–41. [Google Scholar] [CrossRef]
- Farach-Carson, M.C.; Warren, C.R.; Harrington, D.A.; Carson, D.D. Border patrol: Insights into the unique role of perlecan/heparan sulfate proteoglycan 2 at cell and tissue borders. Matrix Biol. 2014, 34, 64–79. [Google Scholar] [CrossRef]
- Weinbaum, S.; Cowin, S.C.; Zeng, Y. A model for the excitation of osteocytes by mechanical loading-induced bone fluid shear stresses. J. Biomech. 1994, 27, 339–360. [Google Scholar] [CrossRef]
- Aarden, E.M.; Burger, E.H.; Nijweide, P.J. Function of osteocytes in bone. J. Cell. Biochem. 1994, 55, 287–299. [Google Scholar] [CrossRef]
- Schaffler, M.B.; Cheung, W.-Y.; Majeska, R.; Kennedy, O. Osteocytes: Master Orchestrators of Bone. Calcif. Tissue Int. 2014, 94, 5–24. [Google Scholar] [CrossRef] [Green Version]
- Bonewald, L.F. The amazing osteocyte. J. Bone Miner. Res. 2011, 26, 229–238. [Google Scholar] [CrossRef]
- Lai, X.; Price, C.; Modla, S.; Thompson, W.R.; Caplan, J.; Kirn-Safran, C.B.; Wang, L. The dependences of osteocyte network on bone compartment, age, and disease. Bone Res. 2015, 3, 15009. [Google Scholar] [CrossRef] [Green Version]
- You, L.; Cowin, S.C.; Schaffler, M.B.; Weinbaum, S. A model for strain amplification in the actin cytoskeleton of osteocytes due to fluid drag on pericellular matrix. J. Biomech. 2001, 34, 1375–1386. [Google Scholar] [CrossRef]
- Gardinier, J.D.; Gangadharan, V.; Wang, L.; Duncan, R.L. Hydraulic pressure during fluid flow regulates purinergic signaling and cytoskeleton organization of osteoblasts. Cell. Mol. Bioeng. 2014, 7, 266–277. [Google Scholar] [CrossRef] [Green Version]
- Duncan, R.L.; Turner, C.H. Mechanotransduction and the functional response of bone to mechanical strain. Calcif. Tissue Int. 1995, 57, 344–358. [Google Scholar] [CrossRef]
- Robling, A.G.; Castillo, A.B.; Turner, C.H. Biomechanical and molecular regulation of bone remodeling. Annu. Rev. Biomed. Eng. 2006, 8, 455–498. [Google Scholar] [CrossRef]
- Jing, D.; Baik, A.D.; Lu, X.L.; Zhou, B.; Lai, X.; Wang, L.; Luo, E.; Guo, X.E. In situ intracellular calcium oscillations in osteocytes in intact mouse long bones under dynamic mechanical loading. FASEB J. 2014, 28, 1582–1592. [Google Scholar] [CrossRef]
- Pei, S.; Parthasarathy, S.; Parajuli, A.; Martinez, J.; Lv, M.; Jiang, S.; Wu, D.; Wei, S.; Lu, X.L.; Farach-Carson, M.C.; et al. Perlecan/Hspg2 deficiency impairs bone’s calcium signaling and associated transcriptome in response to mechanical loading. Bone 2020, 131, 115078. [Google Scholar] [CrossRef]
- Klein-Nulend, J.; Bacabac, R.G.; Bakker, A.D. Mechanical loading and how it affects bone cells: The role of the osteocyte cytoskeleton in maintaining our skeleton. Eur. Cells Mater. 2012, 24, 278–291. [Google Scholar] [CrossRef]
- Kelly, N.H.; Schimenti, J.C.; Ross, F.P.; van der Meulen, M.C.H. Transcriptional profiling of cortical versus cancellous bone from mechanically-loaded murine tibiae reveals differential gene expression. Bone 2016, 86, 22–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robling, A.G.; Niziolek, P.J.; Baldridge, L.A.; Condon, K.W.; Allen, M.R.; Alam, I.; Mantila, S.M.; Gluhak-Heinrich, J.; Bellido, T.M.; Harris, S.E.; et al. Mechanical Stimulation of Bone in Vivo Reduces Osteocyte Expression of Sost/Sclerostin. J. Biol. Chem. 2008, 283, 5866–5875. [Google Scholar] [CrossRef] [Green Version]
- Xiong, J.; Onal, M.; Jilka, R.L.; Weinstein, R.S.; Manolagas, S.C.; O’Brien, C.A. Matrix-embedded cells control osteoclast formation. Nat. Med. 2011, 17, 1235–1241. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Lai, X.; Price, C.; Thompson, W.R.; Li, W.; Quabili, T.R.; Tseng, W.-J.; Liu, X.S.; Zhang, H.; Pan, J.; et al. Perlecan-Containing Pericellular Matrix Regulates Solute Transport and Mechanosensing Within the Osteocyte Lacunar-Canalicular System. J. Bone Miner. Res. 2014, 29, 878–891. [Google Scholar] [CrossRef] [Green Version]
- Guilak, F.; Nims, R.J.; Dicks, A.; Wu, C.-L.; Meulenbelt, I. Osteoarthritis as a disease of the cartilage pericellular matrix. Matrix Biol. 2018, 71–72, 40–50. [Google Scholar] [CrossRef]
- Sieve, I.; Münster-Kühnel, A.K.; Hilfiker-Kleiner, D. Regulation and function of endothelial glycocalyx layer in vascular diseases. Vascul. Pharmacol. 2018, 100, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Nieuwdorp, M.; van Haeften, T.W.; Gouverneur, M.C.L.G.; Mooij, H.L.; van Lieshout, M.H.P.; Levi, M.; Meijers, J.C.M.; Holleman, F.; Hoekstra, J.B.L.; Vink, H.; et al. Loss of Endothelial Glycocalyx During Acute Hyperglycemia Coincides with Endothelial Dysfunction and Coagulation Activation In Vivo. Diabetes 2006, 55, 480–486. [Google Scholar] [CrossRef] [Green Version]
- Oberleithner, H.; Peters, W.; Kusche-Vihrog, K.; Korte, S.; Schillers, H.; Kliche, K.; Oberleithner, K. Salt overload damages the glycocalyx sodium barrier of vascular endothelium. Pflügers Arch. Eur. J. Physiol. 2011, 462, 519–528. [Google Scholar] [CrossRef] [Green Version]
- Ewald, C.Y. The Matrisome during Aging and Longevity: A Systems-Level Approach toward Defining Matreotypes Promoting Healthy Aging. Gerontology 2020, 66, 266–274. [Google Scholar] [CrossRef]
- Sletten, E.M.; Bertozzi, C.R. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chemie Int. Ed. 2009, 48, 6974–6998. [Google Scholar] [CrossRef] [Green Version]
- Baskin, J.M.; Prescher, J.A.; Laughlin, S.T.; Agard, N.J.; Chang, P.V.; Miller, I.A.; Lo, A.; Codelli, J.A.; Bertozzi, C.R. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. USA 2007, 104, 16793–16797. [Google Scholar] [CrossRef] [Green Version]
- Rong, J.; Han, J.; Dong, L.; Tan, Y.; Yang, H.; Feng, L.; Wang, Q.-W.; Meng, R.; Zhao, J.; Wang, S.; et al. Glycan Imaging in Intact Rat Hearts and Glycoproteomic Analysis Reveal the Upregulation of Sialylation during Cardiac Hypertrophy. J. Am. Chem. Soc. 2014, 136, 17468–17476. [Google Scholar] [CrossRef]
- Kostrominova, T.Y. Application of WGA lectin staining for visualization of the connective tissue in skeletal muscle, bone, and ligament/tendon studies. Microsc. Res. Tech. 2011, 74, 18–22. [Google Scholar] [CrossRef] [Green Version]
- Fedarko, N.S.; Termine, J.D.; Young, M.F.; Robey, P.G. Temporal regulation of hyaluronan and proteoglycan metabolism by human bone cells in vitro. J. Biol. Chem. 1990, 265, 12200–12209. [Google Scholar] [CrossRef]
- Simon, E.; Kornitzer, D. Pulse-Chase Analysis to Measure Protein Degradation. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 536, pp. 65–75. ISBN 9780124200708. [Google Scholar]
- Varki, A.; Freeze, H.H.; Manzi, A.E. Overview of Glycoconjugate Analysis. Curr. Protoc. Protein Sci. 2009, 57, 12.1.1–12.1.10. [Google Scholar] [CrossRef]
- Dyment, N.A.; Jiang, X.; Chen, L.; Hong, S.-H.; Adams, D.J.; Ackert-Bicknell, C.; Shin, D.-G.; Rowe, D.W. High-Throughput, Multi-Image Cryohistology of Mineralized Tissues. J. Vis. Exp. 2016, 115, 54468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parajuli, A.; Liu, C.; Li, W.; Gu, X.; Lai, X.; Pei, S.; Price, C.; You, L.; Lu, X.L.; Wang, L. Bone’s responses to mechanical loading are impaired in type 1 diabetes. Bone 2015, 81, 152–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijeratne, S.S.; Martinez, J.R.; Grindel, B.J.; Frey, E.W.; Li, J.; Wang, L.; Farach-Carson, M.C.; Kiang, C.-H. Single molecule force measurements of perlecan/HSPG2: A key component of the osteocyte pericellular matrix. Matrix Biol. 2016, 50, 27–38. [Google Scholar] [CrossRef] [Green Version]
- Tiede-Lewis, L.A.M.; Dallas, S.L. Changes in the osteocyte lacunocanalicular network with aging. Bone 2019, 122, 101–113. [Google Scholar] [CrossRef]
- Fedarko, N.S.; Vetter, U.K.; Weinstein, S.; Robey, P.G. Age-related changes in hyaluronan, proteoglycan, collagen, and osteonectin synthesis by human bone cells. J. Cell. Physiol. 1992, 151, 215–227. [Google Scholar] [CrossRef]
- Boskey, A.L.; Coleman, R. Aging and Bone. J. Dent. Res. 2010, 89, 1333–1348. [Google Scholar] [CrossRef] [PubMed]
- Grynpas, M.D.; Hunter, G.K. Bone mineral and glycosaminoglycans in newborn and mature rabbits. J. Bone Miner. Res. 1988, 3, 159–164. [Google Scholar] [CrossRef]
- Hagan, M.L.; Yu, K.; Zhu, J.; Vinson, B.N.; Roberts, R.L.; Montesinos Cartagena, M.; Johnson, M.H.; Wang, L.; Isales, C.M.; Hamrick, M.W.; et al. Decreased pericellular matrix production and selection for enhanced cell membrane repair may impair osteocyte responses to mechanical loading in the aging skeleton. Aging Cell 2020, 19, e13056. [Google Scholar] [CrossRef] [Green Version]
- Santer, V.; White, R.J.; Roughley, P.J. O-Linked oligosaccharides of human articular cartilage proteoglycan. Biochim. Biophys. Acta 1982, 716, 277–282. [Google Scholar] [CrossRef]
- Farr, J.N.; Fraser, D.G.; Wang, H.; Jaehn, K.; Ogrodnik, M.B.; Weivoda, M.M.; Drake, M.T.; Tchkonia, T.; LeBrasseur, N.K.; Kirkland, J.L.; et al. Identification of Senescent Cells in the Bone Microenvironment. J. Bone Miner. Res. 2016, 31, 1920–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherk, V.D.; Rosen, C.J. Senescent and apoptotic osteocytes and aging: Exercise to the rescue? Bone 2019, 121, 255–258. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Han, Y.; Henderson, S.C.; Majeska, R.J.; Weinbaum, S.; Schaffler, M.B. In situ measurement of solute transport in the bone lacunar-canalicular system. Proc. Natl. Acad. Sci. USA 2005, 102, 11911–11916. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; You, L.; Schaffler, M.B.; Wang, L. The dependency of solute diffusion on molecular weight and shape in intact bone. Bone 2009, 45, 1017–1023. [Google Scholar] [CrossRef] [Green Version]
- Price, C.; Zhou, X.; Li, W.; Wang, L. Real-time measurement of solute transport within the lacunar-canalicular system of mechanically loaded bone: Direct evidence for load-induced fluid flow. J. Bone Miner. Res. 2011, 26, 277–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, C.; Sun, Y.-Q.; Hadjiargyrou, M.; McLeod, K. Increased expression of matrix metalloproteinase-1 in osteocytes precedes bone resorption as stimulated by disuse: Evidence for autoregulation of the cell’s mechanical environment? J. Orthop. Res. 1999, 17, 354–361. [Google Scholar] [CrossRef]
- Visigalli, D.; Strangio, A.; Palmieri, D.; Manduca, P. Hind limb unloading of mice modulates gene expression at the protein and mRNA level in mesenchymal bone cells. BMC Musculoskelet. Disord. 2010, 11, 147. [Google Scholar] [CrossRef] [Green Version]
- Klein-Nulend, J.; Roelofsen, J.; Semeins, C.M.; Bronckers, A.L.J.J.; Burger, E.H. Mechanical stimulation of osteopontin mRNA expression and synthesis in bone cell cultures. J. Cell. Physiol. 1997, 170, 174–181. [Google Scholar] [CrossRef]
- Mantila Roosa, S.M.; Liu, Y.; Turner, C.H. Gene expression patterns in bone following mechanical loading. J. Bone Miner. Res. 2011, 26, 100–112. [Google Scholar] [CrossRef] [Green Version]
- Sternlicht, M.D.; Werb, Z. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 2001, 17, 463–516. [Google Scholar] [CrossRef] [Green Version]
- Itoh, Y. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 2015, 44–46, 207–223. [Google Scholar] [CrossRef]
- Thathiah, A.; Carson, D.D. MT1-MMP mediates MUC1 shedding independent of TACE/ADAM17. Biochem. J. 2004, 382, 363–373. [Google Scholar] [CrossRef] [Green Version]
- Gibor, G.; Ilan, N.; Journo, S.; Sharabi, A.; Dreyer, J.; Gertel, S.; Singh, P.; Menachem, A.; Snir, N.; Elkayam, O.; et al. Heparanase is expressed in adult human osteoarthritic cartilage and drives catabolic responses in primary chondrocytes. Osteoarthr. Cartil. 2018, 26, 1110–1117. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Zheng, T.; Lopez-Aguilar, A.; Feng, L.; Kopp, F.; Marlow, F.L.; Wu, P. Monitoring Dynamic Glycosylation in Vivo Using Supersensitive Click Chemistry. Bioconjug. Chem. 2014, 25, 698–706. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, S.; Wang, S.; Martinez, J.R.; Parajuli, A.; Kirn-Safran, C.B.; Farach-Carson, M.C.; Lu, X.L.; Wang, L. Osteocytic Pericellular Matrix (PCM): Accelerated Degradation under In Vivo Loading and Unloading Conditions Using a Novel Imaging Approach. Genes 2022, 13, 72. https://doi.org/10.3390/genes13010072
Pei S, Wang S, Martinez JR, Parajuli A, Kirn-Safran CB, Farach-Carson MC, Lu XL, Wang L. Osteocytic Pericellular Matrix (PCM): Accelerated Degradation under In Vivo Loading and Unloading Conditions Using a Novel Imaging Approach. Genes. 2022; 13(1):72. https://doi.org/10.3390/genes13010072
Chicago/Turabian StylePei, Shaopeng, Shubo Wang, Jerahme R. Martinez, Ashutosh Parajuli, Catherine B. Kirn-Safran, Mary C. Farach-Carson, X. Lucas Lu, and Liyun Wang. 2022. "Osteocytic Pericellular Matrix (PCM): Accelerated Degradation under In Vivo Loading and Unloading Conditions Using a Novel Imaging Approach" Genes 13, no. 1: 72. https://doi.org/10.3390/genes13010072
APA StylePei, S., Wang, S., Martinez, J. R., Parajuli, A., Kirn-Safran, C. B., Farach-Carson, M. C., Lu, X. L., & Wang, L. (2022). Osteocytic Pericellular Matrix (PCM): Accelerated Degradation under In Vivo Loading and Unloading Conditions Using a Novel Imaging Approach. Genes, 13(1), 72. https://doi.org/10.3390/genes13010072






