Comprehensive Analysis of Innate Immunophenotyping Based on Immune Score Predicting Immune Alterations and Prognosis in Breast Cancer Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Download and Preprocessing
2.2. Identification of the Innate Immune-Related Signatures
2.3. Consensus Clustering Analysis
2.4. Functional Enrichment Analysis and Gene Set Enrichment Analysis
2.5. Constructed and Calculated the Innate-Cluster-Immune (ICI) Score
2.6. Establishment and Validation of the Risk Prognosis Model
2.7. Statistical Analysis
3. Results
3.1. Identification of the Innate Immune-Related Prognostic Signatures in Breast Cancer Patients
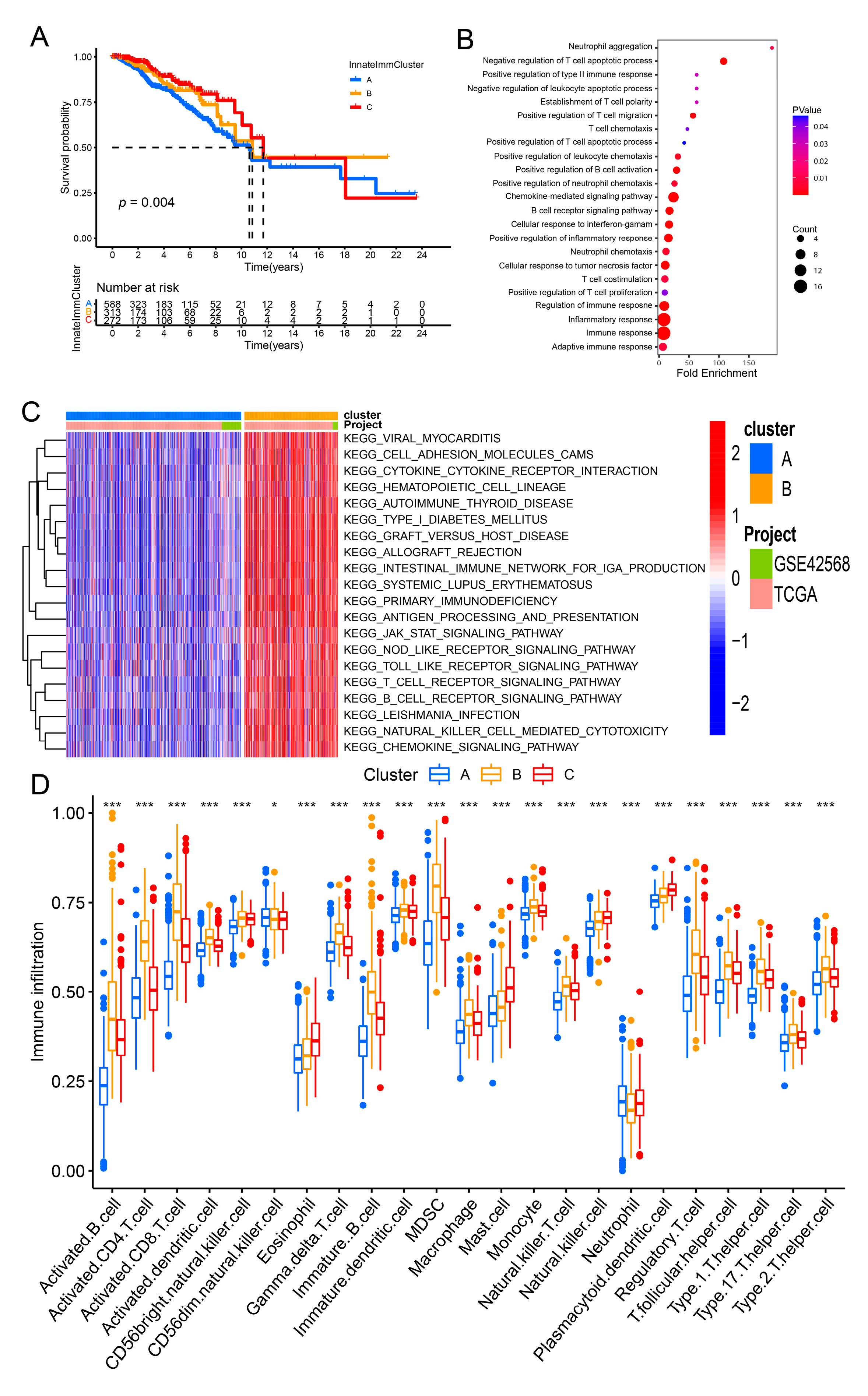
3.2. Construction of the InnateImmCluster Molecular Subtypes Based on the Expression of Innate Immune-Related Prognostic Signatures in Breast Cancer
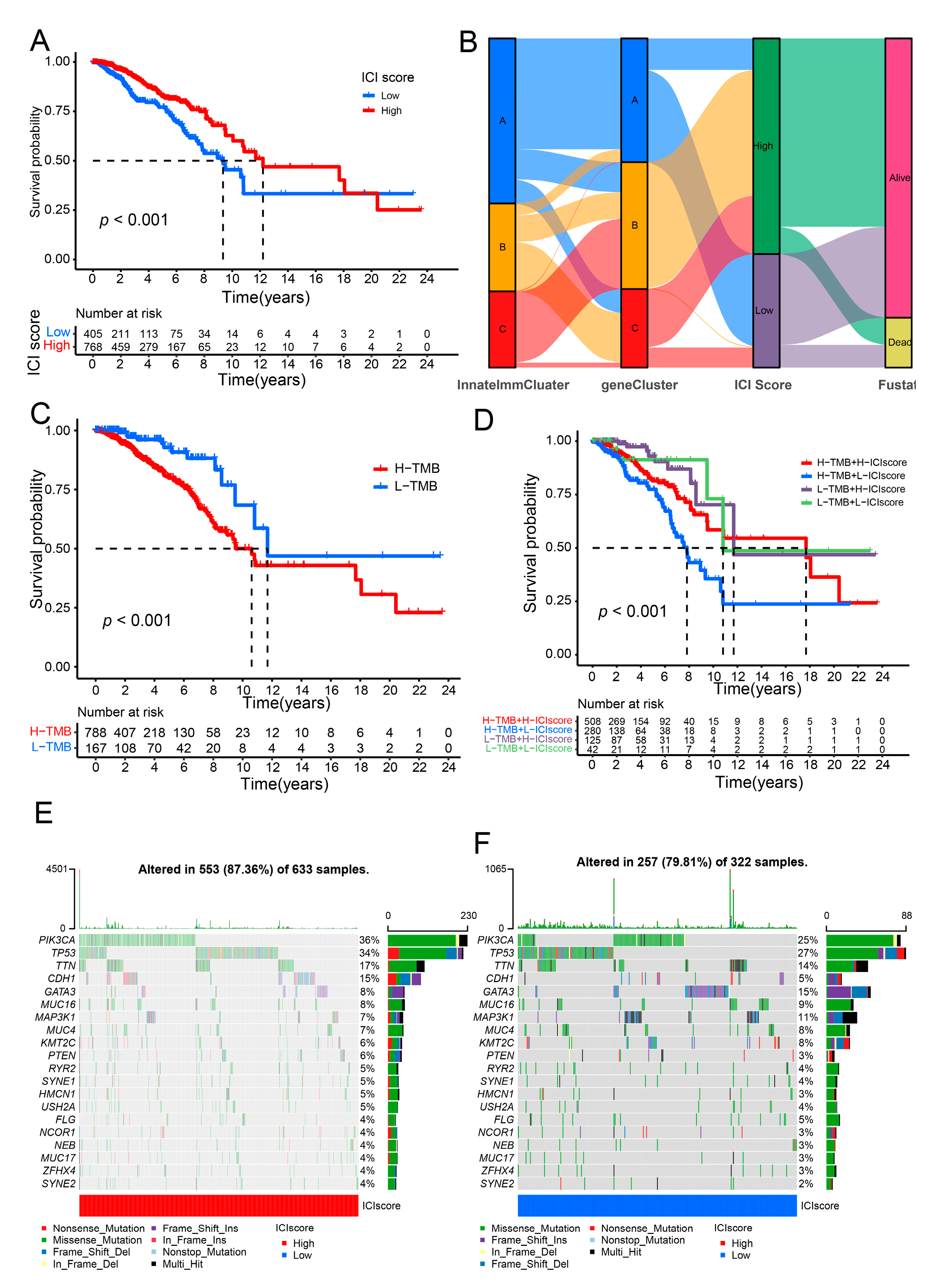
3.3. Calculated the ICI Score to Forecast the Prognosis for Breast Cancer Patients
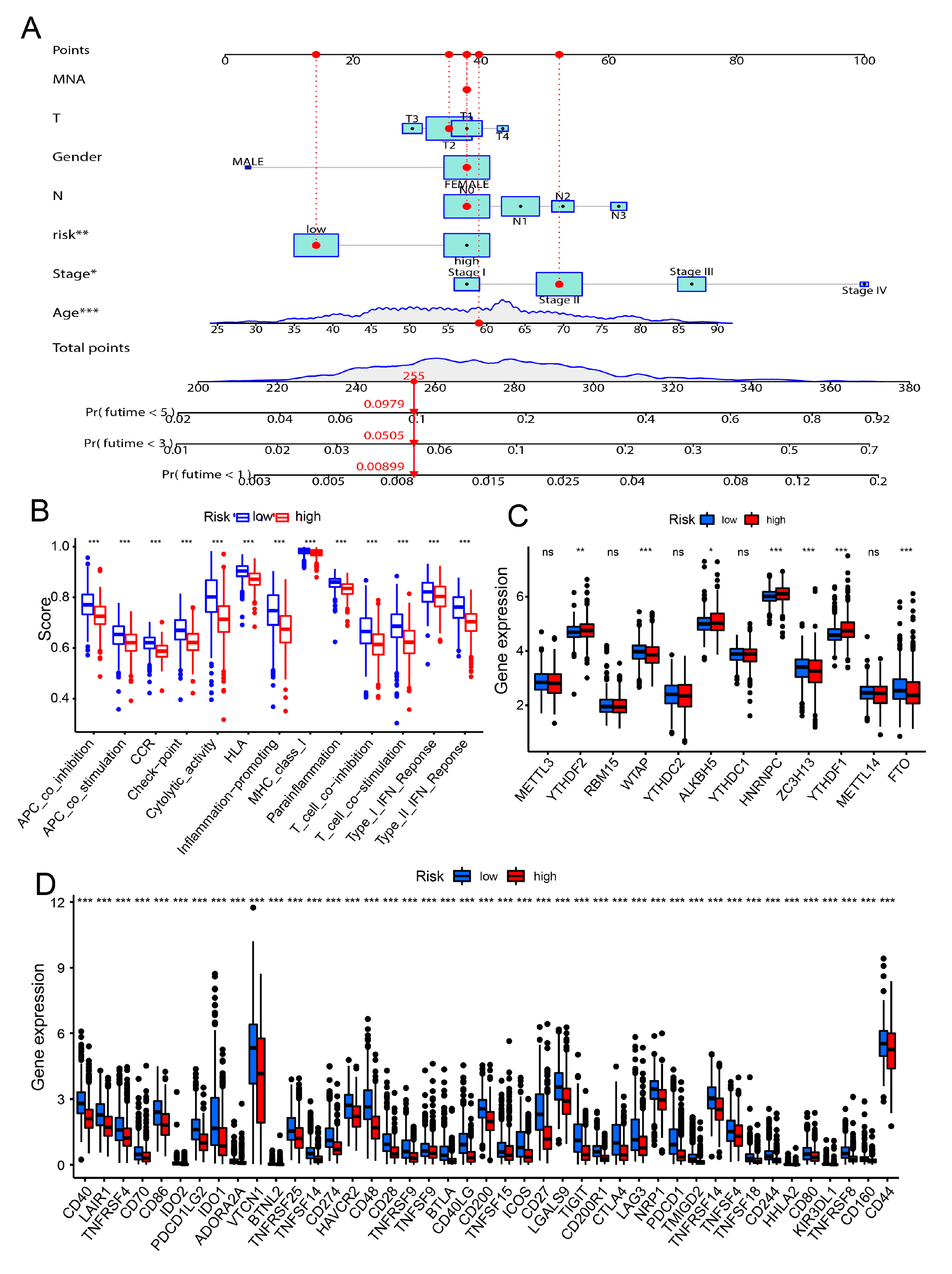
3.4. Constructed the Risk Model for the Prognostic in Breast Cancer Patients
3.5. Correlation of the Risk Model and Cancer Immunity
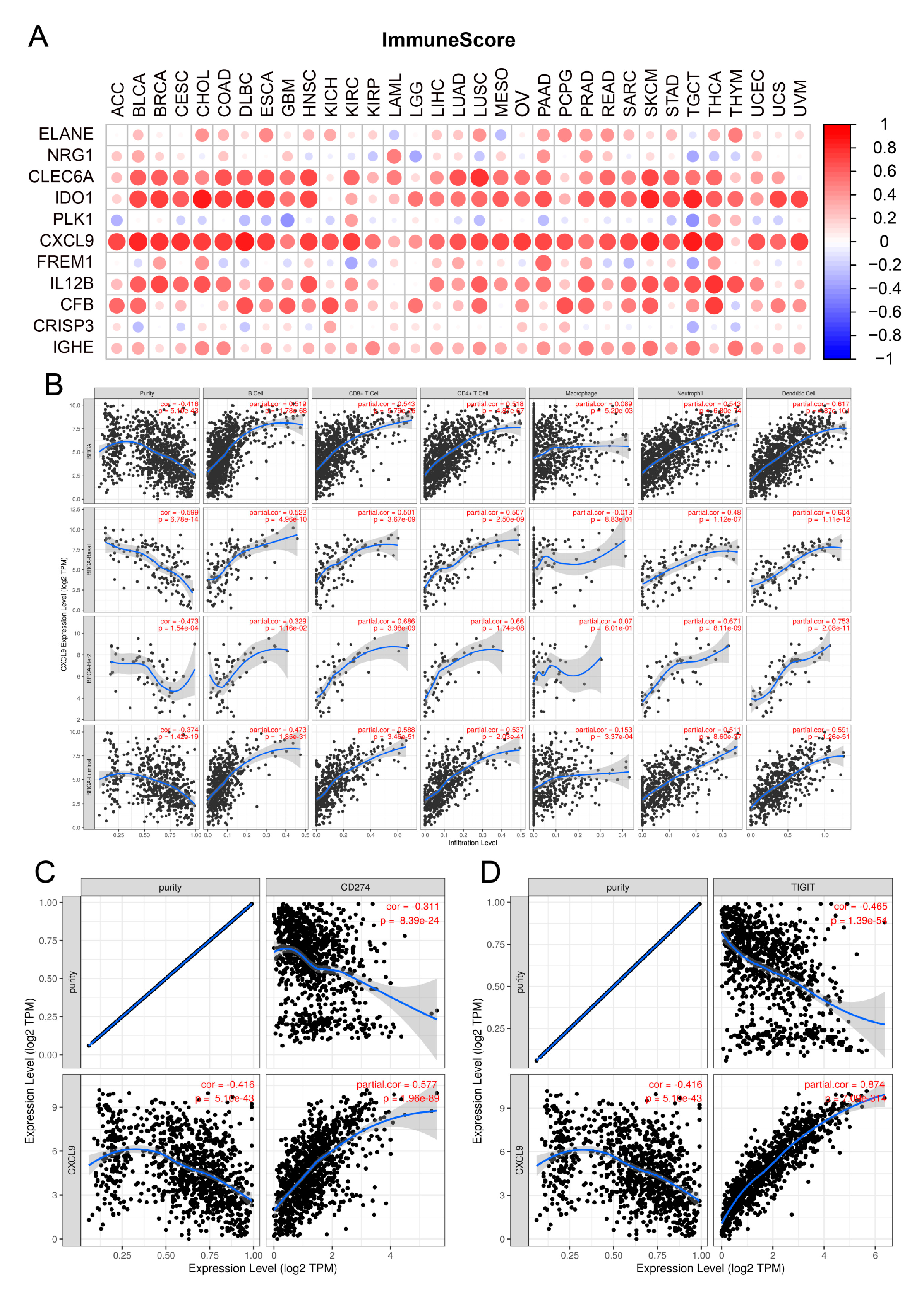
3.6. Identification of CXCL9 as the Key Innate Immune-Related Prognostic Biomarker for Breast Cancer through Pan Cancer Analysis and Immune Infiltration Analysis
3.7. Identification of Small Molecular Drug for Target CXCL9 through Molecular Docking
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Eliyatkin, N.; Yalcin, E.; Zengel, B.; Aktaş, S.; Vardar, E. Molecular Classification of Breast Carcinoma: From Traditional, Old-Fashioned Way to A New Age, and A New Way. J. Breast Health 2015, 11, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Sinn, H.P.; Kreipe, H. A Brief Overview of the WHO Classification of Breast Tumors, 4th Edition, Focusing on Issues and Updates from the 3rd Edition. Breast Care 2013, 8, 149–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viale, G. The current state of breast cancer classification. Ann. Oncol. 2012, 23 (Suppl. 10), x207–x210. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, Y.; Kiani, M.F.; Wang, B. Classification, Treatment Strategy, and Associated Drug Resistance in Breast Cancer. Clin. Breast Cancer 2016, 16, 335–343. [Google Scholar] [CrossRef]
- Mutebi, M.; Anderson, B.O.; Duggan, C.; Adebamowo, C.; Ms, G.A.; Ali, Z.; Bird, P.; Bourque, J.; DeBoer, R.; Gebrim, L.H.; et al. Breast cancer treatment: A phased approach to implementation. Cancer 2020, 126 (Suppl. 10), 2365–2378. [Google Scholar] [CrossRef]
- Bayraktar, S.; Batoo, S.; Okuno, S.; Glück, S. Immunotherapy in breast cancer. J. Carcinog. 2019, 18, 2. [Google Scholar] [CrossRef]
- Barzaman, K.; Moradi-Kalbolandi, S.; Hosseinzadeh, A.; Kazemi, M.H.; Khorramdelazad, H.; Safari, E.; Farahmand, L. Breast cancer immunotherapy: Current and novel approaches. Int. Immunopharmacol. 2021, 98, 107886. [Google Scholar] [CrossRef]
- Lee, A.H.; Sun, L.; Mochizuki, A.Y.; Reynoso, J.G.; Orpilla, J.; Chow, F.; Kienzler, J.C.; Everson, R.G.; Nathanson, D.A.; Bensinger, S.J.; et al. Neoadjuvant PD-1 blockade induces T cell and cDC1 activation but fails to overcome the immunosuppressive tumor associated macrophages in recurrent glioblastoma. Nat. Commun. 2021, 12, 6938. [Google Scholar] [CrossRef]
- Shen, M.; Smith, H.A.; Wei, Y.; Jiang, Y.-Z.; Zhao, S.; Wang, N.; Rowicki, M.; Tang, Y.; Hang, X.; Wu, S.; et al. Pharmacological disruption of the MTDH–SND1 complex enhances tumor antigen presentation and synergizes with anti-PD-1 therapy in metastatic breast cancer. Nat. Rev. Cancer 2021, 1–15. [Google Scholar] [CrossRef]
- Xia, Q.; Jia, J.; Hu, C.; Lu, J.; Li, J.; Xu, H.; Fang, J.; Feng, D.; Wang, L.; Chen, Y. Tumor-associated macrophages promote PD-L1 expression in tumor cells by regulating PKM2 nuclear translocation in pancreatic ductal adenocarcinoma. Oncogene 2021, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Dai, H.; Wang, H.; Han, W. Exploring innate immunity in cancer immunotherapy: Opportunities and challenges. Cell. Mol. Immunol. 2021, 18, 1607–1609. [Google Scholar] [CrossRef]
- Liu, Y.; Zeng, G. Cancer and innate immune system interactions: Translational potentials for cancer immunotherapy. J. Immunother. 2012, 35, 299–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pennemann, F.L.; Mussabekova, A.; Urban, C.; Stukalov, A.; Andersen, L.L.; Grass, V.; Lavacca, T.M.; Holze, C.; Oubraham, L.; Benamrouche, Y.; et al. Cross-species analysis of viral nucleic acid interacting proteins identifies TAOKs as innate immune regulators. Nat. Commun. 2021, 12, 7009. [Google Scholar] [CrossRef]
- Xu, B.; Tian, L.; Chen, J.; Wang, J.; Ma, R.; Dong, W.; Li, A.; Zhang, J.; Chiocca, E.A.; Kaur, B.; et al. An oncolytic virus expressing a full-length antibody enhances antitumor innate immune response to glioblastoma. Nat. Commun. 2021, 12, 5908. [Google Scholar] [CrossRef] [PubMed]
- Hai, J.; Zhu, C.-Q.; Wang, T.; Organ, S.L.; Shepherd, F.A.; Tsao, M.-S. TRIM14 is a Putative Tumor Suppressor and Regulator of Innate Immune Response in Non-Small Cell Lung Cancer. Sci. Rep. 2017, 7, 39692. [Google Scholar] [CrossRef] [Green Version]
- Koelwyn, G.J.; Newman, A.A.C.; Afonso, M.S.; Van Solingen, C.; Corr, E.M.; Brown, E.J.; Albers, K.B.; Yamaguchi, N.; Narke, D.; Schlegel, M.; et al. Myocardial infarction accelerates breast cancer via innate immune reprogramming. Nat. Med. 2020, 26, 1452–1458. [Google Scholar] [CrossRef]
- Ritzmann, F.; Jungnickel, C.; Vella, G.; Kamyschnikow, A.; Herr, C.; Li, D.; Menger, M.M.; Angenendt, A.; Hoth, M.; Lis, A.; et al. IL-17C-mediated innate inflammation decreases the response to PD-1 blockade in a model of Kras-driven lung cancer. Sci. Rep. 2019, 9, 10353. [Google Scholar] [CrossRef]
- Brägelmann, J.; Lorenz, C.; Borchmann, S.; Nishii, K.; Wegner, J.; Meder, L.; Ostendorp, J.; Ast, D.F.; Heimsoeth, A.; Nakasuka, T.; et al. MAPK-pathway inhibition mediates inflammatory reprogramming and sensitizes tumors to targeted activation of innate immunity sensor RIG-I. Nat. Commun. 2021, 12, 5505. [Google Scholar] [CrossRef]
- Muliaditan, T.; Caron, J.; Okesola, M.; Opzoomer, J.; Kosti, P.; Georgouli, M.; Gordon, P.; Lall, S.; Kuzeva, D.M.; Pedro, L.; et al. Macrophages are exploited from an innate wound healing response to facilitate cancer metastasis. Nat. Commun. 2018, 9, 2951. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; He, H.; Wang, H. Oncolytic herpes simplex virus and immunotherapy. BMC Immunol. 2018, 19, 40. [Google Scholar] [CrossRef] [Green Version]
- Chiba, S.; Baghdadi, M.; Akiba, H.; Yoshiyama, H.; Kinoshita, I.; Dosaka-Akita, H.; Fujioka, Y.; Ohba, Y.; Gorman, J.V.; Colgan, J.D.; et al. Tumor-infiltrating DCs suppress nucleic acid–mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat. Immunol. 2012, 13, 832–842. [Google Scholar] [CrossRef] [PubMed]
- Pol, J.; Kroemer, G. Anti-CTLA-4 immunotherapy: Uncoupling toxicity and efficacy. Cell Res. 2018, 28, 501–502. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, W.; Huang, Y.; Cui, R.; Li, X.; Li, B. Evolving Roles for Targeting CTLA-4 in Cancer Immunotherapy. Cell. Physiol. Biochem. 2018, 47, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, N.; Tardiel-Cyril, D.R.; Davtyan, A.; Generali, D.; Roudi, R.; Li, Y. CTLA-4 in Regulatory T Cells for Cancer Immunotherapy. Cancers 2021, 13, 1440. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, M.; Nie, H.; Yuan, Y. PD-1 and PD-L1 in cancer immunotherapy: Clinical implications and future considerations. Hum. Vaccines Immunother. 2019, 15, 1111–1122. [Google Scholar] [CrossRef]
- Gong, J.; Chehrazi-Raffle, A.; Reddi, S.; Salgia, R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: A comprehensive review of registration trials and future considerations. J. Immunother. Cancer 2018, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Chauvin, J.-M.; Zarour, H.M. TIGIT in cancer immunotherapy. J. Immunother. Cancer 2020, 8, e000957. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.-S.; Ko, M.; Choi, D.-S.; Kim, J.H.; Lee, D.-H.; Kang, S.-H.; Kim, I.; Lee, H.J.; Choi, E.K.; Kim, K.-P.; et al. CD226hiCD8+ T Cells Are a Prerequisite for Anti-TIGIT Immunotherapy. Cancer Immunol. Res. 2020, 8, 912–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, R.J.; Comps-Agrar, L.; Hackney, J.; Yu, X.; Huseni, M.; Yang, Y.; Park, S.; Javinal, V.; Chiu, H.; Irving, B.; et al. The Immunoreceptor TIGIT Regulates Antitumor and Antiviral CD8 + T Cell Effector Function. Cancer Cell 2014, 26, 923–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef] [Green Version]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Kursa, M.; Rudnicki, W. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Shlien, A.; Malkin, D. Copy number variations and cancer. Genome Med. 2009, 1, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Walker, L.C.; Pearson, J.F.; Wiggins, G.A.R.; Giles, G.G.; Hopper, J.L.; Southey, M.C. Increased genomic burden of germline copy number variants is associated with early onset breast cancer: Australian breast cancer family registry. Breast Cancer Res. 2017, 19, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Hu, Y.; Luo, R.; Zhao, Y.; Pan, H.; Ji, L.; Zhou, T.; Zhang, L.; Long, H.; Fu, J.; et al. Multi-region exome sequencing reveals the intratumoral heterogeneity of surgically resected small cell lung cancer. Nat. Commun. 2021, 12, 5431. [Google Scholar] [CrossRef] [PubMed]
- Saito, R.; Smith, C.C.; Utsumi, T.; Bixby, L.M.; Kardos, J.; Wobker, S.E.; Stewart, K.G.; Chai, S.; Manocha, U.; Byrd, K.M.; et al. Molecular Subtype-Specific Immunocompetent Models of High-Grade Urothelial Carcinoma Reveal Differential Neoantigen Expression and Response to Immunotherapy. Cancer Res. 2018, 78, 3954–3968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.; Liang, Y.; Hu, Z.; Li, H.; Yang, J.; Hsu, E.J.; Zhu, J.; Zhou, J.; Fu, Y.-X. Next generation of tumor-activating type I IFN enhances anti-tumor immune responses to overcome therapy resistance. Nat. Commun. 2021, 12, 5866. [Google Scholar] [CrossRef]
- Huang, L.; Xu, H.; Peng, G. TLR-mediated metabolic reprogramming in the tumor microenvironment: Potential novel strategies for cancer immunotherapy. Cell. Mol. Immunol. 2018, 15, 428–437. [Google Scholar] [CrossRef] [Green Version]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef] [Green Version]
- Sha, D.; Jin, Z.; Budczies, J.; Kluck, K.; Stenzinger, A.; Sinicrope, F.A. Tumor Mutational Burden as a Predictive Biomarker in Solid Tumors. Cancer Discov. 2020, 10, 1808–1825. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Xia, L.; Xia, Z.; Chen, L. Comprehensive Analysis of Innate Immunophenotyping Based on Immune Score Predicting Immune Alterations and Prognosis in Breast Cancer Patients. Genes 2022, 13, 88. https://doi.org/10.3390/genes13010088
Liu W, Xia L, Xia Z, Chen L. Comprehensive Analysis of Innate Immunophenotyping Based on Immune Score Predicting Immune Alterations and Prognosis in Breast Cancer Patients. Genes. 2022; 13(1):88. https://doi.org/10.3390/genes13010088
Chicago/Turabian StyleLiu, Weiguang, Lingling Xia, Zhengmiao Xia, and Liming Chen. 2022. "Comprehensive Analysis of Innate Immunophenotyping Based on Immune Score Predicting Immune Alterations and Prognosis in Breast Cancer Patients" Genes 13, no. 1: 88. https://doi.org/10.3390/genes13010088
APA StyleLiu, W., Xia, L., Xia, Z., & Chen, L. (2022). Comprehensive Analysis of Innate Immunophenotyping Based on Immune Score Predicting Immune Alterations and Prognosis in Breast Cancer Patients. Genes, 13(1), 88. https://doi.org/10.3390/genes13010088





