Homozygous Inversion on Chromosome 13 Involving SGCG Detected by Short Read Whole Genome Sequencing in a Patient Suffering from Limb-Girdle Muscular Dystrophy
Abstract
1. Introduction
2. Patient and Methods
2.1. Case Presentation
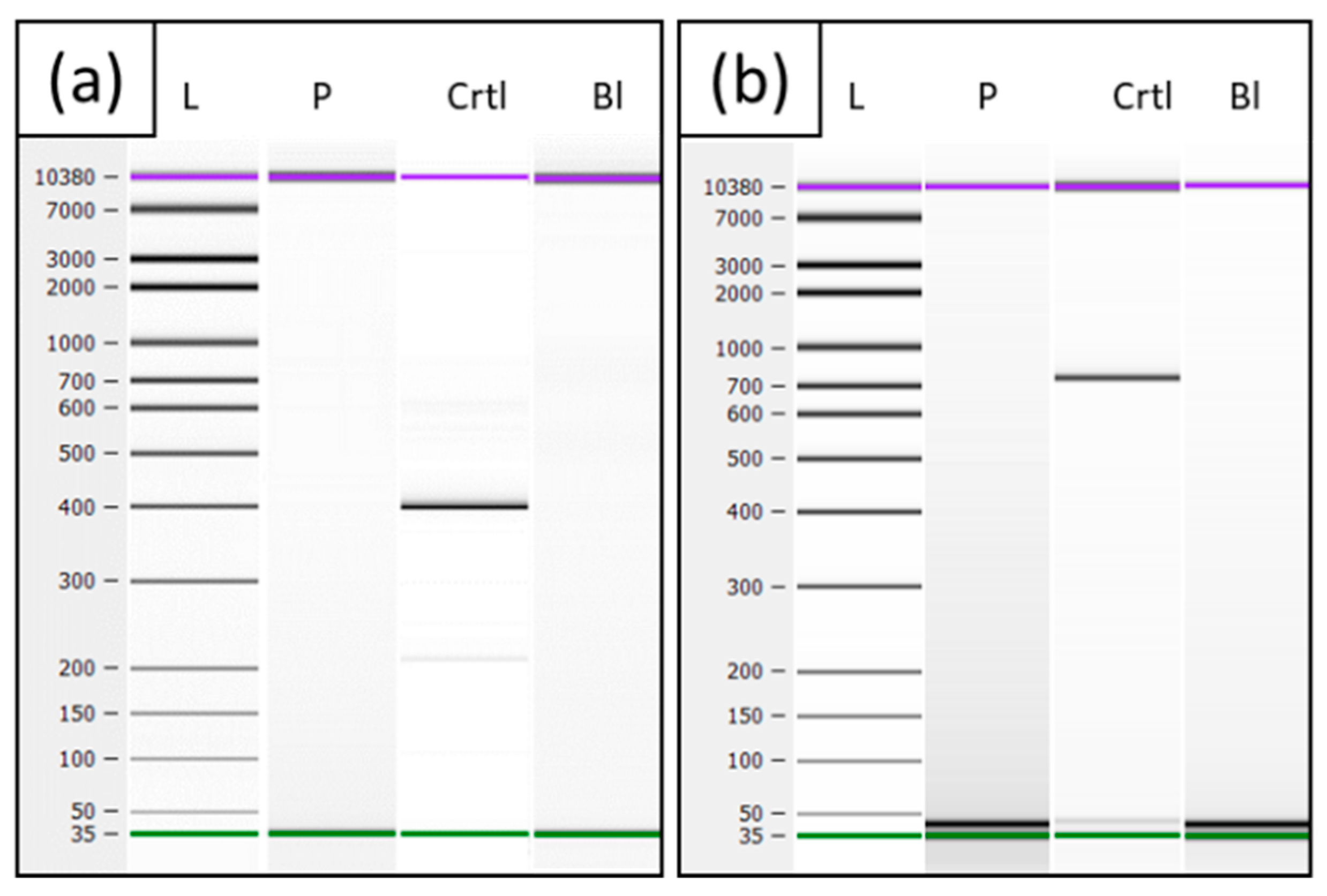
2.2. mRNA Analysis
2.3. Whole Genome Sequencing
2.4. Muscle Biopsy Analyses
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kirschner, J.; Lochmuller, H. Sarcoglycanopathies. Handb. Clin. Neurol. 2011, 101, 41–46. [Google Scholar] [CrossRef]
- Ginjaar, H.B.; van der Kooi, A.J.; Ceelie, H.; Kneppers, A.L.; van Meegen, M.; Barth, P.G.; Busch, H.F.; Wokke, J.H.; Anderson, L.V.; Bonnemann, C.G.; et al. Sarcoglycanopathies in Dutch patients with autosomal recessive limb girdle muscular dystrophy. J. Neurol. 2000, 247, 524–529. [Google Scholar] [CrossRef]
- Holt, K.H.; Campbell, K.P. Assembly of the sarcoglycan complex. Insights for muscular dystrophy. J. Biol. Chem. 1998, 273, 34667–34670. [Google Scholar] [CrossRef] [PubMed]
- Straub, V.; Murphy, A.; Udd, B.; LGMD Workshop Study Group. 229th ENMC International Workshop: Limb Girdle Muscular Dystrophies—Nomenclature and Reformed Classification, Naarden, The Netherlands, 17–19 March 2017. Neuromuscul. Disord. 2018, 28, 702–710. [Google Scholar] [CrossRef] [PubMed]
- McNally, E.M.; Passos-Bueno, M.R.; Bonnemann, C.G.; Vainzof, M.; Moreira, E.d.; Lidov, H.G.; Othmane, K.B.; Denton, P.H.; Vance, J.M.; Zatz, M.; et al. Mild and severe muscular dystrophy caused by a single gamma-sarcoglycan mutation. Am. J. Hum. Genet. 1996, 59, 1040–1047. [Google Scholar]
- Alonso-Perez, J.; Gonzalez-Quereda, L.; Bello, L.; Guglieri, M.; Straub, V.; Gallano, P.; Semplicini, C.; Pegoraro, E.; Zangaro, V.; Nascimento, A.; et al. New genotype-phenotype correlations in a large European cohort of patients with sarcoglycanopathy. Brain 2020, 143, 2696–2708. [Google Scholar] [CrossRef] [PubMed]
- Nigro, V.; Savarese, M. Next-generation sequencing approaches for the diagnosis of skeletal muscle disorders. Curr. Opin. Neurol. 2016, 29, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Ormond, K.E.; Wheeler, M.T.; Hudgins, L.; Klein, T.E.; Butte, A.J.; Altman, R.B.; Ashley, E.A.; Greely, H.T. Challenges in the clinical application of whole-genome sequencing. Lancet 2010, 375, 1749–1751. [Google Scholar] [CrossRef]
- Zhou, B.; Ho, S.S.; Zhang, X.L.; Pattni, R.; Haraksingh, R.R.; Urban, A.E. Whole-genome sequencing analysis of CNV using low-coverage and paired-end strategies is efficient and outperforms array-based CNV analysis. J. Med. Genet. 2018, 55, 735–743. [Google Scholar] [CrossRef]
- Weedon, M.N.; Cebola, I.; Patch, A.M.; Flanagan, S.E.; de Franco, E.; Caswell, R.; Rodriguez-Segui, S.A.; Shaw-Smith, C.; Cho, C.H.; Allen, H.L.; et al. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat. Genet. 2014, 46, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Herdewyn, S.; Zhao, H.; Moisse, M.; Race, V.; Matthijs, G.; Reumers, J.; Kusters, B.; Schelhaas, H.J.; van den Berg, L.H.; Goris, A.; et al. Whole-genome sequencing reveals a coding non-pathogenic variant tagging a non-coding pathogenic hexanucleotide repeat expansion in C9orf72 as cause of amyotrophic lateral sclerosis. Hum. Mol. Genet. 2012, 21, 2412–2419. [Google Scholar] [CrossRef]
- Schluth-Bolard, C.; Diguet, F.; Chatron, N.; Rollat-Farnier, P.A.; Bardel, C.; Afenjar, A.; Amblard, F.; Amiel, J.; Blesson, S.; Callier, P.; et al. Whole genome paired-end sequencing elucidates functional and phenotypic consequences of balanced chromosomal rearrangement in patients with developmental disorders. J. Med. Genet. 2019, 56, 526–535. [Google Scholar] [CrossRef]
- Taylor, J.C.; Martin, H.C.; Lise, S.; Broxholme, J.; Cazier, J.B.; Rimmer, A.; Kanapin, A.; Lunter, G.; Fiddy, S.; Allan, C.; et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat. Genet. 2015, 47, 717–726. [Google Scholar] [CrossRef]
- Wolf, B.; Kuonen, P.; Dandekar, T.; Atlan, D. DNAseq Workflow in a Diagnostic Context and an Example of a User Friendly Implementation. Biomed. Res. Int. 2015, 2015, 403497. [Google Scholar] [CrossRef]
- Kohler, S.; Gargano, M.; Matentzoglu, N.; Carmody, L.C.; Lewis-Smith, D.; Vasilevsky, N.A.; Danis, D.; Balagura, G.; Baynam, G.; Brower, A.M.; et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 2021, 49, D1207. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Bondeson, M.L.; Dahl, N.; Malmgren, H.; Kleijer, W.J.; Tonnesen, T.; Carlberg, B.M.; Pettersson, U. Inversion of the IDS gene resulting from recombination with IDS-related sequences is a common cause of the Hunter syndrome. Hum. Mol. Genet. 1995, 4, 615–621. [Google Scholar] [CrossRef]
- Lakich, D.; Kazazian, H.H., Jr.; Antonarakis, S.E.; Gitschier, J. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat. Genet. 1993, 5, 236–241. [Google Scholar] [CrossRef]
- Cagliani, R.; Sironi, M.; Ciafaloni, E.; Bardoni, A.; Fortunato, F.; Prelle, A.; Serafini, M.; Bresolin, N.; Comi, G.P. An intragenic deletion/inversion event in the DMD gene determines a novel exon creation and results in a BMD phenotype. Hum. Genet. 2004, 115, 13–18. [Google Scholar] [CrossRef]
- Flanigan, K.M.; Dunn, D.; Larsen, C.A.; Medne, L.; Bonnemann, C.B.; Weiss, R.B. Becker muscular dystrophy due to an inversion of exons 23 and 24 of the DMD gene. Muscle Nerve 2011, 44, 822–825. [Google Scholar] [CrossRef]
- Madden, H.R.; Fletcher, S.; Davis, M.R.; Wilton, S.D. Characterization of a complex Duchenne muscular dystrophy-causing dystrophin gene inversion and restoration of the reading frame by induced exon skipping. Hum. Mutat. 2009, 30, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Sun, C.; Zhang, S.; Liu, Y.; Yu, M.; Zheng, Y.; Meng, L.; Acharya, A.; Cornejo-Sanchez, D.M.; Wang, G.; et al. Long-read whole-genome sequencing for the genetic diagnosis of dystrophinopathies. Ann. Clin. Transl. Neurol. 2020, 7, 2041–2046. [Google Scholar] [CrossRef]
- Puig, M.; Casillas, S.; Villatoro, S.; Caceres, M. Human inversions and their functional consequences. Brief. Funct. Genom. 2015, 14, 369–379. [Google Scholar] [CrossRef]
- Boulling, A.; Wicht, L.; Schorderet, D.F. Identification of HMX1 target genes: A predictive promoter model approach. Mol. Vis. 2013, 19, 1779–1794. [Google Scholar]
- Lee, J.; Han, K.; Meyer, T.J.; Kim, H.S.; Batzer, M.A. Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons. PLoS ONE 2008, 3, e4047. [Google Scholar] [CrossRef]
- Kim, S.; Cho, C.S.; Han, K.; Lee, J. Structural Variation of Alu Element and Human Disease. Genom. Inform. 2016, 14, 70–77. [Google Scholar] [CrossRef]
- Purandare, S.M.; Patel, P.I. Recombination hot spots and human disease. Genome Res. 1997, 7, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Loda, A.; Heard, E. Xist RNA in action: Past, present, and future. PLoS Genet. 2019, 15, e1008333. [Google Scholar] [CrossRef] [PubMed]
- St Laurent, G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef]
- Zaum, A.K.; Nanda, I.; Kress, W.; Rost, S. Detection of pericentric inversion with breakpoint in DMD by whole genome sequencing. Mol. Genet. Genom. Med. 2022, e2028. [Google Scholar] [CrossRef] [PubMed]
- Spiegler, S.; Rath, M.; Hoffjan, S.; Dammann, P.; Sure, U.; Pagenstecher, A.; Strom, T.; Felbor, U. First large genomic inversion in familial cerebral cavernous malformation identified by whole genome sequencing. Neurogenetics 2018, 19, 55–59. [Google Scholar] [CrossRef] [PubMed]




| Primer Name | Sequence | Used Transcript (RefSeq) |
|---|---|---|
| SGCG-cDNA-Ex1-F | 5′-ATTCGCCAGTGTGCTTTTCT-3′ | NM_000231.2 |
| SGCG-cDNA-Ex4-R | 5′-TGACCTCCCCTTCTGAGTTG-3′ | NM_000231.2 |
| DYSF-cDNA-Ex10-F | 5′-GGGCACCATTTACAGAGAGC-3′ | NM_003494.3 |
| DYSF-cDNA-Ex15-R | 5′-TTCGCACATGGAGGGAAA-3′ | NM_003494.3 |
| SACS-cDNA-Ex1-F | 5′-TGGAGTACACATTTAAAGGAAGCA-3′ | NM_014363.5 |
| SACS-cDNA-Ex5-R | 5′-AAGAATCTGTCCTCCTTCTGGA-3′ | NM_014363.5 |
| LINC00621-cDNA-Ex2-F | 5′-CCAAGTCTGGTCAACCCAAT-3′ | XR_001732839.1 |
| LINC00621-cDNA-Ex4-R | 5′-CTTTTTCCCATGTGAGGACAC-3′ | XR_001732839.1 |
| SGCG-Int2-F | 5′-GGTATCTAATTCAATCAGCACTTTTT-3′ | NM_000231.2 |
| SGCG-Int2-R | 5′-GCATGACACATACATGCCTGA-3′ | NM_000231.2 |
| LINC00621-Upstream-F | 5′-TTGAAGTAAATGTTTTCTAACCTGTCA-3′ | XR_001732839.1 |
| LINC00621-Upstream-R | 5′-TGATTTCAGTTTTATGTGGCATT-3′ | XR_001732839.1 |
| SGCG-Int1-g.23186799-F | 5′-AAGTATCCCCCATCCTCACA-3′ | NM_000231.2 |
| SGCG-Int1-g.23186799-R | 5′-GGCCAGCATCACTATCCAAG-3′ | NM_000231.2 |
| SGCG-Int1-g.23200194-F | 5′-AGCCTCACCAGACTGAAGGA-3′ | NM_000231.2 |
| SGCG-Int1-g.23200194-R | 5′-CCTCCAGGGTTTCAAGTGAG-3′ | NM_000231.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pluta, N.; Hoffjan, S.; Zimmer, F.; Köhler, C.; Lücke, T.; Mohr, J.; Vorgerd, M.; Nguyen, H.H.P.; Atlan, D.; Wolf, B.; et al. Homozygous Inversion on Chromosome 13 Involving SGCG Detected by Short Read Whole Genome Sequencing in a Patient Suffering from Limb-Girdle Muscular Dystrophy. Genes 2022, 13, 1752. https://doi.org/10.3390/genes13101752
Pluta N, Hoffjan S, Zimmer F, Köhler C, Lücke T, Mohr J, Vorgerd M, Nguyen HHP, Atlan D, Wolf B, et al. Homozygous Inversion on Chromosome 13 Involving SGCG Detected by Short Read Whole Genome Sequencing in a Patient Suffering from Limb-Girdle Muscular Dystrophy. Genes. 2022; 13(10):1752. https://doi.org/10.3390/genes13101752
Chicago/Turabian StylePluta, Natalie, Sabine Hoffjan, Frederic Zimmer, Cornelia Köhler, Thomas Lücke, Jennifer Mohr, Matthias Vorgerd, Hoa Huu Phuc Nguyen, David Atlan, Beat Wolf, and et al. 2022. "Homozygous Inversion on Chromosome 13 Involving SGCG Detected by Short Read Whole Genome Sequencing in a Patient Suffering from Limb-Girdle Muscular Dystrophy" Genes 13, no. 10: 1752. https://doi.org/10.3390/genes13101752
APA StylePluta, N., Hoffjan, S., Zimmer, F., Köhler, C., Lücke, T., Mohr, J., Vorgerd, M., Nguyen, H. H. P., Atlan, D., Wolf, B., Zaum, A.-K., & Rost, S. (2022). Homozygous Inversion on Chromosome 13 Involving SGCG Detected by Short Read Whole Genome Sequencing in a Patient Suffering from Limb-Girdle Muscular Dystrophy. Genes, 13(10), 1752. https://doi.org/10.3390/genes13101752







