An Integrated Study on the Differential Expression of the FOX Gene Family in Cancer and Their Response to Chemotherapy Drugs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Gene Correlation Analysis in Gene Expression Atlas
2.2. Other Gene Correlation Analysis Methods
3. Results
3.1. Identification of FOX Genes That Play Important Roles in Various Types of Cancer
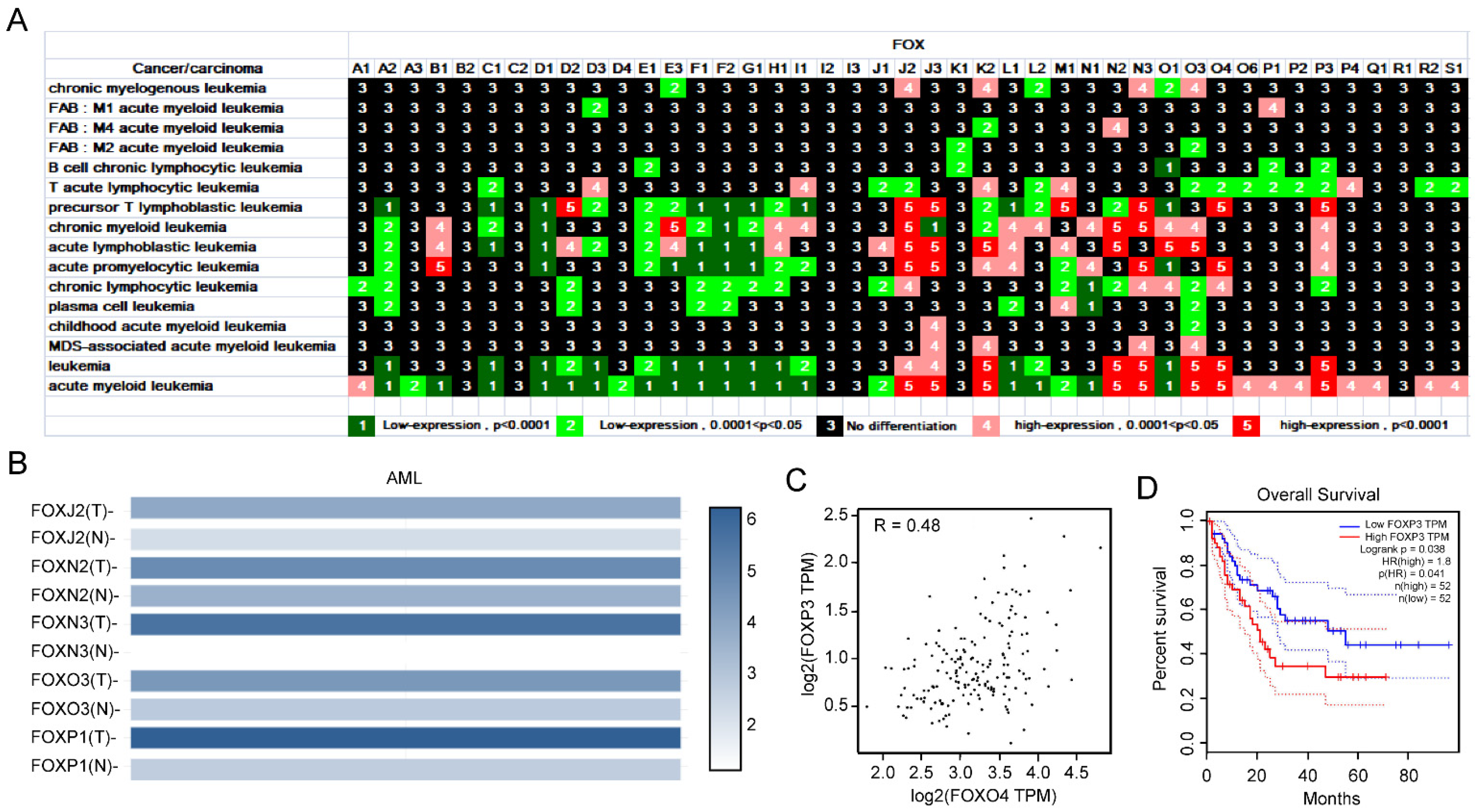
3.1.1. Expression Profile of FOX Genes in Leukemia and Identification of Several FOX Genes with High Expression in AML
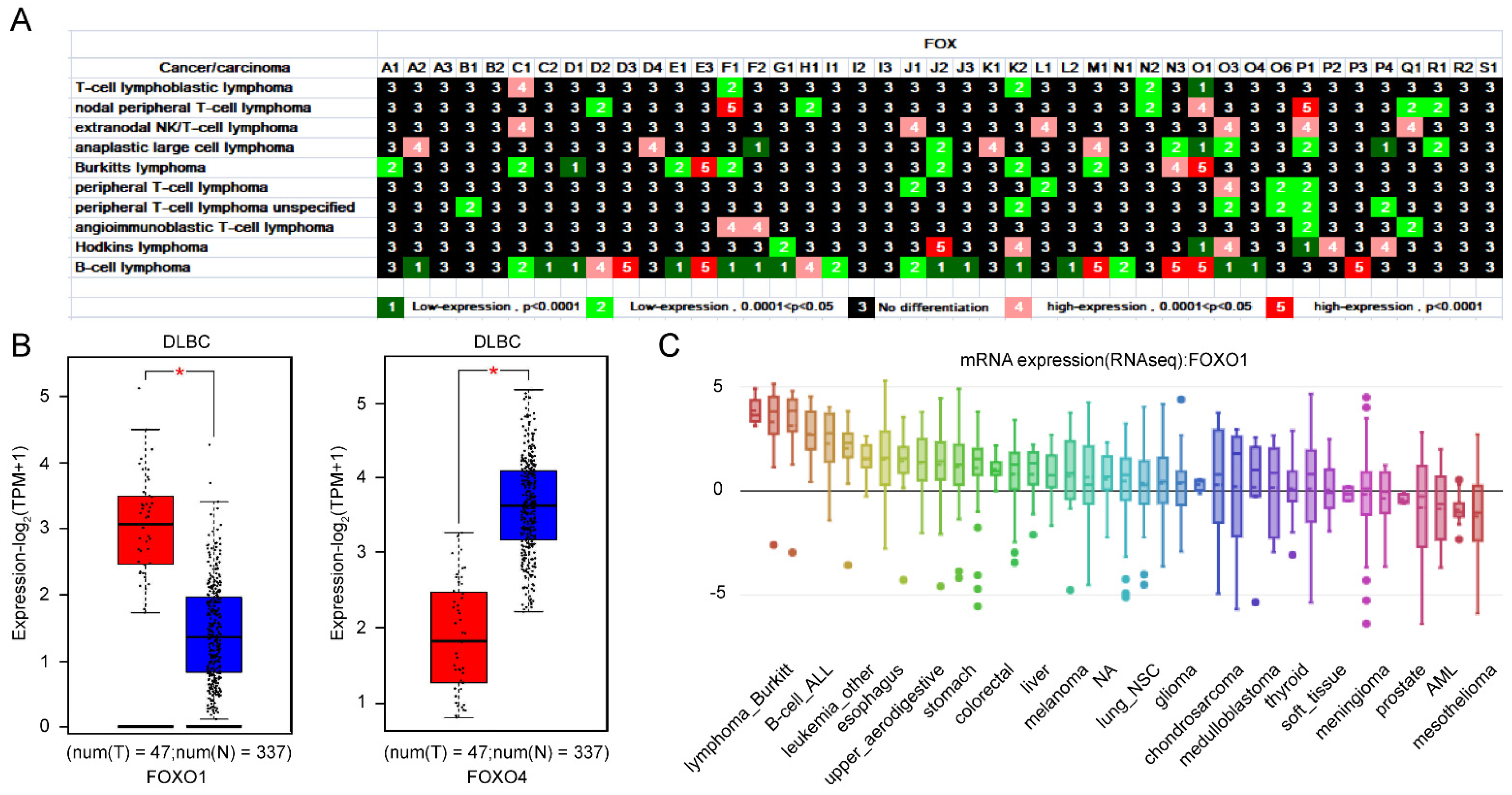
3.1.2. Importance of FOXO1 and FOXO4 from the Expression Profile of FOX Genes in Lymphoma
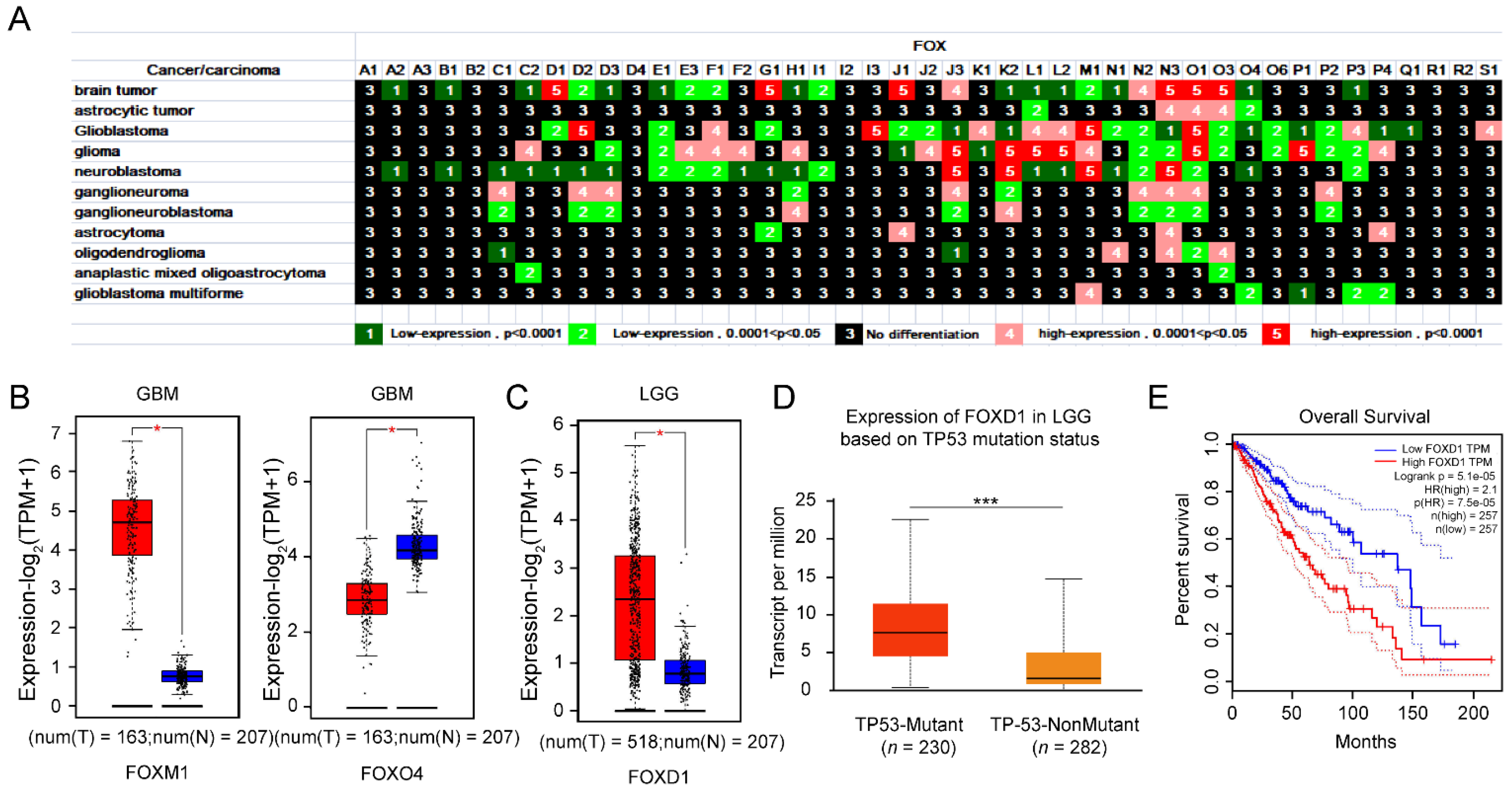
3.1.3. Upregulation of FOXD1 Based on TP53 Mutation Is a Poor Prognosis Factor for LGG Patients
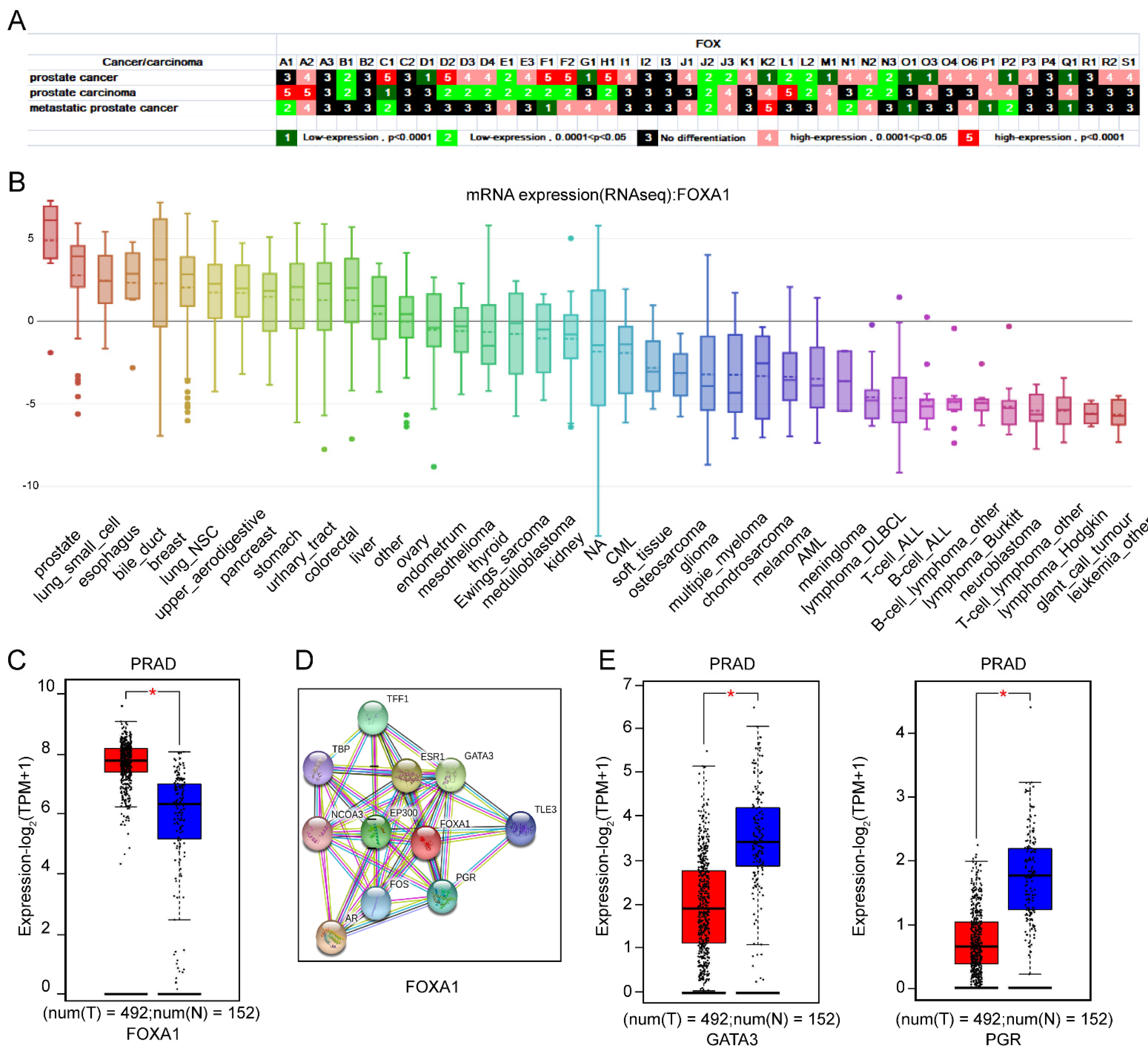
3.1.4. The Importance of FOXA1 in Prostate as Well as Its Interaction with GATA3 and PGR
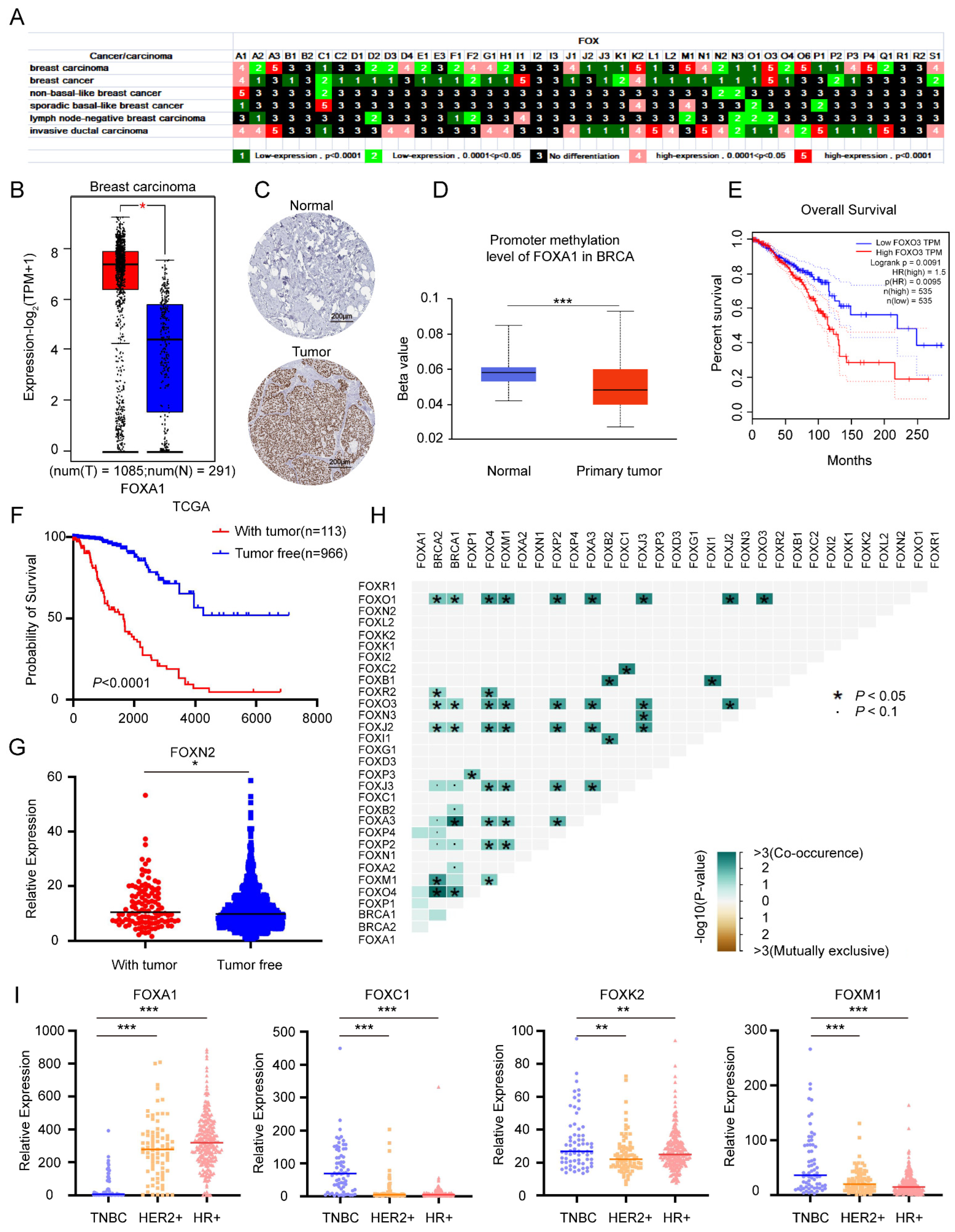
3.1.5. Multiple Roles of FOX Gene in Breast Cancer and Identification of Several FOX Genes with High Expression in TNBC
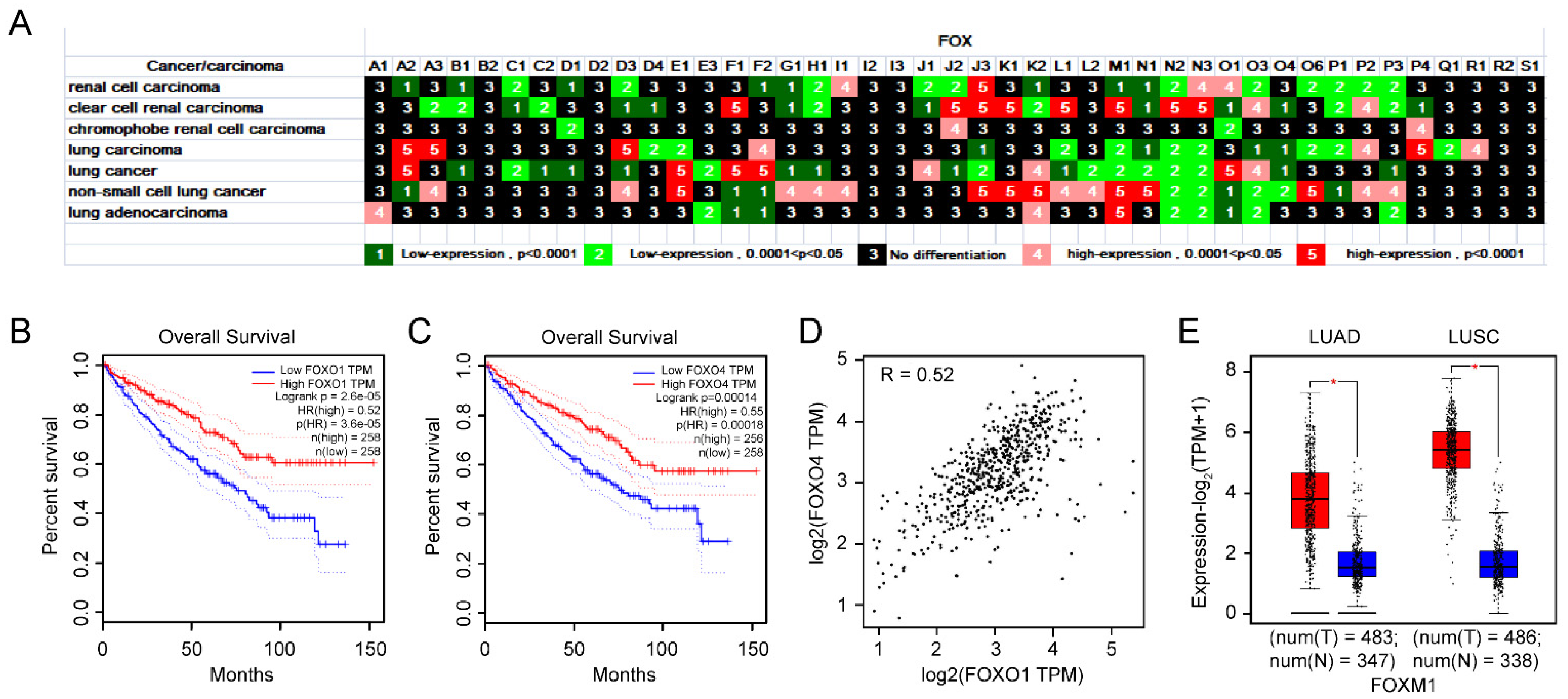
3.1.6. The Importance of FOXO1 and FOXO4 in KIRC from Expression, Prognosis, and Correlation Analysis
3.1.7. The Importance of FOXM1 and FOXP1 in PAAD Is Underlined by Their Expression and Prognosis
3.2. Integrative Analysis of FOX Genes Differential Expression and Response to Chemicals
3.2.1. Screening for FOX Genes Which Are Important for the Progression and Treatment of a Wide Range of Cancers
3.2.2. Evaluating the Potential of Anticancer Drug Therapy for a Specific Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kaestner, K.H.; Knochel, W.; Martinez, D.E. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000, 14, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.W.F.; Brosens, J.J.; Gomes, A.R.; Koo, C.-Y. Forkhead box proteins: Tuning forks for transcriptional harmony. Nat. Rev. Cancer 2013, 13, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Golson, M.L.; Kaestner, K.H. Fox transcription factors: From development to disease. Development 2016, 143, 4558–4570. [Google Scholar] [CrossRef] [PubMed]
- Benayoun, B.A.; Caburet, S.; Veitia, R.A. Forkhead transcription factors: Key players in health and disease. Trends Genet. 2011, 27, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci. 2016, 144, 194–201. [Google Scholar] [CrossRef]
- Katoh, M.; Igarashi, M.; Fukuda, H.; Nakagama, H.; Katoh, M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013, 328, 198–206. [Google Scholar] [CrossRef]
- de Brachène, A.C.; Demoulin, J.-B. FOXO transcription factors in cancer development and therapy. Cell Mol. Life Sci. 2016, 73, 1159–1172. [Google Scholar] [CrossRef] [PubMed]
- Kapushesky, M.; Emam, I.; Holloway, E.; Kurnosov, P.; Zorin, A.; Malone, J.; Rustici, G.; Williams, E.; Parkinson, H.; Brazma, A. Gene expression atlas at the European bioinformatics institute. Nucleic Acids Res. 2010, 38, D690–D698. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Colwill, K.; Gräslund, S. A roadmap to generate renewable protein binders to the human proteome. Nat. Methods 2011, 8, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Zhang, B.; Teng, X.; Hu, P.; Xu, S.; Zheng, Z.; Liu, R.; Tang, T.; Ye, F. Validating a targeted next-generation sequencing assay and profiling somatic variants in Chinese non-small cell lung cancer patients. Sci. Rep. 2020, 10, 2070. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H. Targeting forkhead box transcription factors FOXM1 and FOXO in leukemia (Review). Oncol. Rep. 2014, 32, 1327–1334. [Google Scholar] [CrossRef]
- Widick, P.; Winer, E.S. Leukocytosis and Leukemia. Prim Care 2016, 43, 575–587. [Google Scholar] [CrossRef]
- Rose-Inman, H.; Kuehl, D. Acute Leukemia. Hematol. Oncol. Clin. N. Am. 2017, 31, 1011–1028. [Google Scholar] [CrossRef]
- Hallek, M. Chronic lymphocytic leukemia: 2017 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2017, 92, 946–965. [Google Scholar] [CrossRef]
- Bennour, A.; Saad, A.; Sennana, H. Chronic myeloid leukemia: Relevance of cytogenetic and molecular assays. Crit. Rev. Oncol. Hematol. 2016, 97, 263–274. [Google Scholar] [CrossRef]
- Armitage, J.O.; Gascoyne, R.D.; Lunning, M.A.; Cavalli, F. Non-Hodgkin lymphoma. Lancet 2017, 390, 298–310. [Google Scholar] [CrossRef]
- Jiang, M.; Bennani, N.N.; Feldman, A.L. Lymphoma classification update: T-cell lymphomas, Hodgkin lymphomas, and histiocytic/dendritic cell neoplasms. Expert Rev. Hematol. 2017, 10, 239–249. [Google Scholar] [CrossRef] [PubMed]
- DeWitt, J.C.; Mock, A.; Louis, D.N. The 2016 WHO classification of central nervous system tumors: What neurologists need to know. Curr. Opin. Neurol. 2017, 30, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Good, D.W.; Stewart, G.D.; Hammer, S.; Scanlan, P.; Shu, W.; Phipps, S.; Reuben, R.; McNeill, A.S. Elasticity as a biomarker for prostate cancer: A systematic review. BJU Int. 2014, 113, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Myatt, S.S.; Lam, E.W.F. The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer 2007, 7, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, C.E.; Baca, S.C.; Lawrence, M.S.; Demichelis, F.; Blattner, M.; Theurillat, J.-P.; White, T.A.; Stojanov, P.; Van Allen, E.; Stransky, N.; et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012, 44, 685–689. [Google Scholar] [CrossRef]
- Jin, H.-J.; Zhao, J.C.; Ogden, I.; Bergan, R.C.; Yu, J. Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer Res. 2013, 73, 3725–3736. [Google Scholar] [CrossRef]
- Jiang, X.; Chen, Y.; Du, E.; Yang, K.; Zhang, Z.; Qi, S.; Xu, Y. GATA3-driven expression of miR-503 inhibits prostate cancer progression by repressing ZNF217 expression. Cell. Signal. 2016, 28, 1216–1224. [Google Scholar] [CrossRef]
- Shafer, M.E.R.; Nguyen, A.H.T.; Tremblay, M.; Viala, S.; Béland, M.; Bertos, N.R.; Park, M.; Bouchard, M. Lineage Specification from Prostate Progenitor Cells Requires Gata3-Dependent Mitotic Spindle Orientation. Stem Cell Rep. 2017, 8, 1018–1031. [Google Scholar] [CrossRef]
- Yang, M.; Li, J.C.; Tao, C.; Wu, S.; Liu, B.; Shu, Q.; Li, B.; Zhu, R. PAQR6 Upregulation Is Associated with AR Signaling and Unfavorite Prognosis in Prostate Cancers. Biomolecules 2021, 11, 1383. [Google Scholar] [CrossRef]
- Anastasiadi, Z.; Lianos, G.D.; Ignatiadou, E.; Harissis, H.V.; Mitsis, M. Breast cancer in young women: An overview. Updat. Surg. 2017, 69, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Merino Bonilla, J.A.; Torres Tabanera, M.; Ros Mendoza, L.H. Breast cancer in the 21st century: From early detection to new therapies. Radiologia 2017, 59, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zou, L.; Lu, W.-Q.; Zhang, Y.; Shen, A.-G. Foxo3a expression is a prognostic marker in breast cancer. PLoS ONE 2013, 8, e70746. [Google Scholar] [CrossRef]
- Arruabarrena-Aristorena, A.; Maag, J.L.V.; Kittane, S.; Cai, Y.; Karthaus, W.R.; Ladewig, E.; Park, J.; Kannan, S.; Ferrando, L.; Cocco, E.; et al. FOXA1 Mutations Reveal Distinct Chromatin Profiles and Influence Therapeutic Response in Breast Cancer. Cancer Cell 2020, 38, 534–550.e539. [Google Scholar] [CrossRef]
- Bell, R.; Barraclough, R.; Vasieva, O. Gene Expression Meta-Analysis of Potential Metastatic Breast Cancer Markers. Curr. Mol. Med. 2017, 17, 200–210. [Google Scholar] [CrossRef]
- Tung, N.M.; Garber, J.E. BRCA1/2 testing: Therapeutic implications for breast cancer management. Br. J. Cancer 2018, 119, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Vagia, E.; Mahalingam, D.; Cristofanilli, M. The Landscape of Targeted Therapies in TNBC. Cancers 2020, 12, 916. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Jia, Z.; Wan, F.; Zhu, Y.; Shi, G.; Zhang, H.; Dai, B.; Ye, D. Forkhead-box series expression network is associated with outcome of clear-cell renal cell carcinoma. Oncol. Lett. 2018, 15, 8669–8680. [Google Scholar] [CrossRef]
- Nasim, F.; Sabath, B.F.; Eapen, G.A. Lung Cancer. Med. Clin. N. Am. 2019, 103, 463–473. [Google Scholar] [CrossRef]
- Pierrou, S.; Hellqvist, M.; Samuelsson, L.; Enerbäck, S.; Carlsson, P. Cloning and characterization of seven human forkhead proteins: Binding site specificity and DNA bending. EMBO J. 1994, 13, 5002–5012. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Mo, W.; Zhang, R.; Li, Y. The clinical and prognostic significance of FOXN3 downregulation in acute myeloid leukaemia. Int. J. Lab. Hematol. 2020, 42, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Jia, M.Y.; Fang, W.Y.; Chen, X.J.; Mu, L.L.; Wang, Z.Y.; Shen, Y.; Xiang, R.F.; Wang, L.N.; Wang, L.; et al. FLT3 inhibition upregulates HDAC8 via FOXO to inactivate p53 and promote maintenance of FLT3-ITD+ acute myeloid leukemia. Blood 2020, 135, 1472–1483. [Google Scholar] [CrossRef] [PubMed]
- Miyazato, P.; Matsuoka, M. Human T-cell leukemia virus type 1 and Foxp3 expression: Viral strategy in vivo. Int. Immunol. 2014, 26, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Pei, S.N.; Qi, J.; Zeng, Z.; Iyer, S.P.; Lin, P.; Tung, C.H.; Zu, Y. Oligonucleotide aptamer-drug conjugates for targeted therapy of acute myeloid leukemia. Biomaterials 2015, 67, 42–51. [Google Scholar] [CrossRef]
- Gehringer, F.; Weissinger, S.E.; Swier, L.J.; Möller, P.; Wirth, T.; Ushmorov, A. FOXO1 Confers Maintenance of the Dark Zone Proliferation and Survival Program and Can Be Pharmacologically Targeted in Burkitt Lymphoma. Cancers 2019, 11, 1427. [Google Scholar] [CrossRef]
- Sehn, L.H.; Salles, G. Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2021, 384, 842–858. [Google Scholar] [CrossRef] [PubMed]
- Roberto, M.P.; Varano, G.; Vinas-Castells, R.; Holmes, A.B.; Kumar, R.; Pasqualucci, L.; Farinha, P.; Scott, D.W.; Dominguez-Sola, D. Mutations in the transcription factor FOXO1 mimic positive selection signals to promote germinal center B cell expansion and lymphomagenesis. Immunity 2021, 54, 1807–1824.e1814. [Google Scholar] [CrossRef]
- Langendonk, M.; de Jong, M.R.W.; Smit, N.; Seiler, J.; Reitsma, B.; Ammatuna, E.; Glaudemans, A.; van den Berg, A.; Huls, G.A.; Visser, L.; et al. Identification of the estrogen receptor beta as a possible new tamoxifen-sensitive target in diffuse large B-cell lymphoma. Blood Cancer J. 2022, 12, 36. [Google Scholar] [CrossRef] [PubMed]
- Robertson, E.; Perry, C.; Doherty, R.; Madhusudan, S. Transcriptomic profiling of Forkhead box transcription factors in adult glioblastoma multiforme. Cancer Genom. Proteom. 2015, 12, 103–112. [Google Scholar]
- Li, C.; Guan, X.; Jing, H.; Xiao, X.; Jin, H.; Xiong, J.; Ai, S.; Wang, Y.; Su, T.; Sun, G.; et al. Circular RNA circBFAR promotes glioblastoma progression by regulating a miR-548b/FoxM1 axis. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2022, 36, e22183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Han, X.; Xu, X.; Zhou, Z.; Chen, X.; Tang, Y.; Cheng, J.; Moazzam, N.F.; Liu, F.; Xu, J.; et al. FoxM1 drives ADAM17/EGFR activation loop to promote mesenchymal transition in glioblastoma. Cell Death Dis. 2018, 9, 469. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Sun, L.A.; Jiang, X.C.; Han, Y.L.; Wang, L.; Niu, W.H.; Fei, M.X.; Zhaba, W.D.; Zheng, L.R.; Zhou, M.L. FOXO4 expression associates with glioblastoma development and FOXO4 expression inhibits cell malignant phenotypes in vitro and in vivo. Life Sci. 2020, 247, 117436. [Google Scholar] [CrossRef]
- Lopes, D.V.; de Fraga Dias, A.; Silva, L.F.L.; Scholl, J.N.; Sévigny, J.; Battastini, A.M.O.; Figueiró, F. Influence of NSAIDs and methotrexate on CD73 expression and glioma cell growth. Purinergic Signal. 2021, 17, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Baca, S.C.; Takeda, D.Y.; Seo, J.H.; Hwang, J.; Ku, S.Y.; Arafeh, R.; Arnoff, T.; Agarwal, S.; Bell, C.; O’Connor, E.; et al. Reprogramming of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer. Nat. Commun. 2021, 12, 1979. [Google Scholar] [CrossRef] [PubMed]
- Seachrist, D.D.; Anstine, L.J.; Keri, R.A. FOXA1: A Pioneer of Nuclear Receptor Action in Breast Cancer. Cancers 2021, 13, 5205. [Google Scholar] [CrossRef]
- Wen, W.; Chen, Z.; Bao, J.; Long, Q.; Shu, X.O.; Zheng, W.; Guo, X. Genetic variations of DNA bindings of FOXA1 and co-factors in breast cancer susceptibility. Nat. Commun. 2021, 12, 5318. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Liang, X.; Zhao, B.; Chen, M.; Liu, R.; Sun, S.; Yue, X.; Wang, S. Ultrasound assisted gene and photodynamic synergistic therapy with multifunctional FOXA1-siRNA loaded porphyrin microbubbles for enhancing therapeutic efficacy for breast cancer. Biomaterials 2018, 173, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, H.; Sun, Z.; Meng, X.; Ma, Z.; Wang, Z. Effects of etoposide combined with cisplatin on prognosis of patients with castration-resistant prostate cancer who failed castration treatment. Am. J. Transl. Res. 2022, 14, 1705–1713. [Google Scholar]
- Koushki, M.; Khedri, A.; Aberomand, M.; Akbari Baghbani, K.; Mohammadzadeh, G. Synergistic anti-cancer effects of silibinin-etoposide combination against human breast carcinoma MCF-7 and MDA-MB-231 cell lines. Iran. J. Basic Med. Sci. 2021, 24, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Dogra, A.; Mehta, A.; Doval, D.C. Are Basal-Like and Non-Basal-Like Triple-Negative Breast Cancers Really Different? J. Oncol. 2020, 2020, 4061063. [Google Scholar] [CrossRef] [PubMed]
- Yang, V.; Gouveia, M.J.; Santos, J.; Koksch, B.; Amorim, I.; Gärtner, F.; Vale, N. Breast cancer: Insights in disease and influence of drug methotrexate. RSC Med. Chem. 2020, 11, 646–664. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wu, J.Q.; Yu, X.F.; Yang, X.S.; Yang, Y. Trichostatin A inhibits proliferation of triple negative breast cancer cells by inducing cell cycle arrest and apoptosis. Neoplasma 2018, 65, 898–906. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.M.; Kimryn Rathmell, W.; Beckermann, K.E. Modeling clear cell renal cell carcinoma and therapeutic implications. Oncogene 2020, 39, 3413–3426. [Google Scholar] [CrossRef] [PubMed]
- Bychkov, A.; Saenko, V.; Nakashima, M.; Mitsutake, N.; Rogounovitch, T.; Nikitski, A.; Orim, F.; Yamashita, S. Patterns of FOXE1 expression in papillary thyroid carcinoma by immunohistochemistry. Thyroid 2013, 23, 817–828. [Google Scholar] [CrossRef]
- Fan, S.; Liao, Y.; Qiu, W.; Li, L.; Li, D.; Cao, X.; Ai, B. Targeting Toll-like receptor 4 with CLI-095 (TAK-242) enhances the antimetastatic effect of the estrogen receptor antagonist fulvestrant on non-small cell lung cancer. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2020, 22, 2074–2086. [Google Scholar] [CrossRef] [PubMed]
- Tempero, M.A.; Malafa, M.P.; Al-Hawary, M.; Behrman, S.W.; Benson, A.B.; Cardin, D.B.; Chiorean, E.G.; Chung, V.; Czito, B.; Del Chiaro, M.; et al. Pancreatic Adenocarcinoma, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. JNCCN 2021, 19, 439–457. [Google Scholar] [CrossRef]
- Huang, C.; Du, J.; Xie, K. FOXM1 and its oncogenic signaling in pancreatic cancer pathogenesis. Biochim. Biophys. Acta 2014, 1845, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wu, F.; Yang, J. A novel long non-coding RNA TTN-AS1/microRNA-589-5p/FOXP1 positive feedback loop increases the proliferation, migration and invasion of pancreatic cancer cell lines. Oncol. Lett. 2021, 22, 794. [Google Scholar] [CrossRef] [PubMed]
- Hoca, M.; Becer, E.; Kabadayı, H.; Yücecan, S.; Vatansever, H.S. The Effect of Resveratrol and Quercetin on Epithelial-Mesenchymal Transition in Pancreatic Cancer Stem Cell. Nutr. Cancer 2020, 72, 1231–1242. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, H.; Fan, X.; Zhang, Y.; Zhao, N.; Zhao, X.; Yin, K.; Zhang, Y. An Integrated Study on the Differential Expression of the FOX Gene Family in Cancer and Their Response to Chemotherapy Drugs. Genes 2022, 13, 1754. https://doi.org/10.3390/genes13101754
Yin H, Fan X, Zhang Y, Zhao N, Zhao X, Yin K, Zhang Y. An Integrated Study on the Differential Expression of the FOX Gene Family in Cancer and Their Response to Chemotherapy Drugs. Genes. 2022; 13(10):1754. https://doi.org/10.3390/genes13101754
Chicago/Turabian StyleYin, Haimeng, Xing Fan, Yanqiao Zhang, Nan Zhao, Xiaoyi Zhao, Kehan Yin, and Yali Zhang. 2022. "An Integrated Study on the Differential Expression of the FOX Gene Family in Cancer and Their Response to Chemotherapy Drugs" Genes 13, no. 10: 1754. https://doi.org/10.3390/genes13101754







