Genome-Wide Identification and Characterization of the CCT Gene Family in Foxtail Millet (Setaria italica) Response to Diurnal Rhythm and Abiotic Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Identification, Chromosomal Location, and Phylogenetic Relationships of CCT Family Members in Foxtail Millet
2.2. Collinearity Analysis and Gene Duplication
2.3. Gene Structure and Conserved Sequence Analysis
2.4. Cis-Acting Element Analysis
2.5. Plant Materials and Treatments
2.6. Analysis of CCT Gene Expression in Millet and Quantitative Real-Time PCR
3. Results
3.1. Identification and Analysis of CCT Gene Family Members in Foxtail Millet
3.2. Evolutionary and Synteny Analysis of CCT Genes in Foxtail Millet and Other Species
3.3. Gene Structure and Conserved Motif Analysis
3.4. Analysis of Cis-Acting Elements in the SiCCT Gene Promoter Region
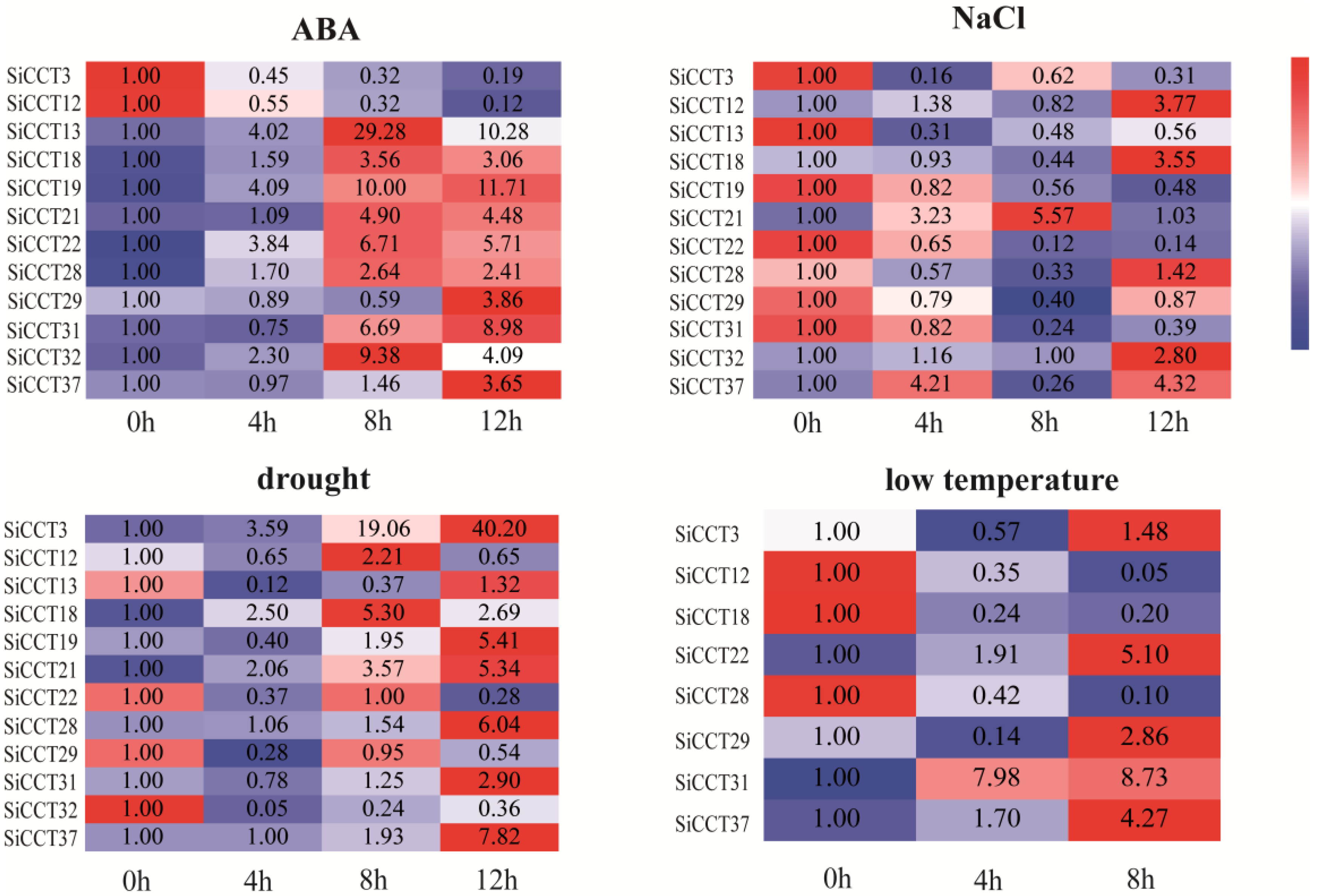
3.5. Expression Analysis of SiCCT Genes in Different Tissues and under Abiotic Stress and Exogenous Hormone Treatments
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Veatch-Blohm, M.E. Principles of plant genetics and breeding. Crop Sci. 2007, 47, 385. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Hossain, M.A.; da Silva, J.A.T.; Fujita, M. Plant Response and Tolerance to Abiotic Oxidative Stress: Antioxidant Defense Is a Key Factor. In Crop Stress and Its Management: Perspectives and Strategies; Venkateswarlu, B., Shanker, A., Shanker, C., Maheswari, M., Eds.; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Wong, C.E.; Teo, Z.W.N.; Shen, L.; Yu, H. Seeing the lights for leafy greens in indoor vertical farming. Trends Food Sci. Technol. 2020, 106, 48–63. [Google Scholar] [CrossRef]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef]
- Mizuno, T.; Nakamichi, N. Pseudo-response regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol. 2005, 46, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Mengarelli, D.A.; Zanor, M.I. Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max). Planta 2021, 253, 15. [Google Scholar] [CrossRef] [PubMed]
- Strayer, C.; Oyama, T.; Schultz, T.F.; Raman, R.; Somers, D.E.; Más, P.; Panda, S.; Kreps, J.A.; Kay, S.A. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 2000, 289, 768–771. [Google Scholar] [CrossRef]
- Cockram, J.; Thiel, T.; Steuernagel, B.; Stein, N.; Taudien, S.; Bailey, P.C.; O’Sullivan, D.M. Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS ONE 2012, 7, e45307. [Google Scholar] [CrossRef]
- Robson, F.; Costa, M.M.; Hepworth, S.R.; Vizir, I.; Piñeiro, M.; Reeves, P.H.; Putterill, J.; Coupland, G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001, 28, 619–631. [Google Scholar] [CrossRef]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2484. [Google Scholar] [CrossRef]
- Miller, T.A.; Muslin, E.H.; Dorweiler, J.E. A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 2008, 227, 1377–1388. [Google Scholar] [CrossRef]
- Yan, L.; Loukoianov, A.; Blechl, A.; Tranquilli, G.; Ramakrishna, W.; SanMiguel, P.; Bennetzen, J.L.; Echenique, V.; Dubcovsky, J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 2004, 303, 1640–1644. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Shen, Y.; Chang, H.C.; Hou, Y.; Harris, A.; Ma, S.F.; McPartland, M.; Hymus, G.J.; Adam, L.; Marion, C.; et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010, 187, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, Y.; Nonoue, Y.; Yano, M.; Izawa, T. Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 2016, 86, 221–233. [Google Scholar] [CrossRef]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kita, M.; Ito, S.; Yamashino, T.; Mizuno, T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Alabadi, D. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef]
- Legnaioli, T.; Cuevas, J.; Mas, P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009, 28, 3745–3757. [Google Scholar] [CrossRef]
- Liu, T.; Carlsson, J.; Takeuchi, T.; Newton, L.; Farré, E.M. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 2013, 76, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Min, J.H.; Chung, J.S.; Lee, K.H.; Kim, C.S. The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 2015, 57, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Wang, L.; Wang, J.; Hu, Y.; Du, H.; Xu, C.; Xing, Y.; Li, X.; Xiao, J.; Zhang, Q. Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 2014, 164, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shen, J.; Xu, Y.; Li, X.; Xiao, J.; Xiong, L. Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J. Exp. Bot. 2016, 67, 5785–5798. [Google Scholar] [CrossRef]
- Su, H.; Liang, J.; Abou-Elwafa, S.F.; Cheng, H.; Dou, D.; Ren, Z.; Xie, J.; Chen, Z.; Gao, F.; Ku, L.; et al. ZmCCT regulates photoperiod-dependent flowering and response to stresses in maize. BMC Plant Biol. 2021, 21, 453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, X.; Quan, Z.; Cheng, S.; Xu, X.; Pan, S.; Xie, M.; Zeng, P.; Yue, Z.; Wang, W.; et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 2012, 30, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Huang, X.; Zhi, H.; Zhao, Y.; Zhao, Q.; Li, W.; Chai, Y.; Yang, L.; Liu, K.; Lu, H.; et al. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 2013, 45, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Doust, A.N.; Kellogg, E.A.; Devos, K.M.; Bennetzen, J.L. Foxtail millet: A sequence-driven grass model system. Plant Physiol. 2009, 149, 137–141. [Google Scholar] [CrossRef]
- Santos, C.M.; Romeiro, D.; Silva, J.P.; Basso, M.F.; Molinari, H.B.C.; Centeno, D.C. An improved protocol for efficient transformation and regeneration of Setaria italica. Plant Cell Rep. 2020, 39, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Shalmani, A.; Jing, X.Q.; Shi, Y.; Muhammad, I.; Zhou, M.R.; Wei, X.Y.; Chen, Q.Q.; Li, W.Q.; Liu, W.T.; Chen, K.M. Characterization of B-BOX gene family and their expression profiles under hormonal, abiotic and metal stresses in Poaceae plants. BMC Genom. 2019, 20, 27. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.H. The Arabidopsis B-box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.R.; Arnon, D.S. The water culture method for growing plants without soil. Calif. Agric. Exp. Stn. Circ. 1950, 347, 32. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Huo, M.; Li, J.; Yu, J. Time-series transcriptomics reveals a drought-responsive temporal network and crosstalk between drought stress and the circadian clock in foxtail millet. Plant J. 2022, 110, 1213–1228. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Ren, D.; Huang, M.; Sun, K.; Feng, J.; Zhao, J.; Xiao, D.; Xie, W.; Liu, S.; Zhang, H.; et al. Strong photoperiod sensitivity is controlled by cooperation and competition among Hd1, Ghd7 and DTH8 in rice heading. New Phytol. 2021, 229, 1635–1649. [Google Scholar] [CrossRef]
- Jin, M.; Liu, X.; Jia, W.; Liu, H.; Li, W.; Peng, Y.; Du, Y.; Wang, Y.; Yin, Y.; Zhang, X.; et al. ZmCOL3, a CCT gene represses flowering in maize by interfering with the circadian clock and activating expression of ZmCCT. J. Integr. Plant Biol. 2018, 60, 465–480. [Google Scholar] [CrossRef]
- Zhang, J.; Fan, X.; Hu, Y.; Zhou, X.; He, Q.; Liang, L.; Xing, Y. Global analysis of CCT family knockout mutants identifies four genes involved in regulating heading date in rice. J. Integr. Plant Biol. 2021, 63, 913–923. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 1, 10. [Google Scholar]
- Murat, F.; Xu, J.H.; Tannier, E.; Abrouk, M.; Guilhot, N.; Pont, C.; Messing, J.; Salse, J. Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res. 2010, 20, 1545–1557. [Google Scholar] [CrossRef]
- Cui, L.; Wall, P.K.; Leebens-Mack, J.H.; Lindsay, B.G.; Soltis, D.E.; Doyle, J.J.; Soltis, P.S.; Carlson, J.E.; Arumuganathan, K.; Barakat, A.; et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006, 16, 738–749. [Google Scholar] [CrossRef]
- McKain, M.R.; Tang, H.; McNeal, J.R.; Ayyampalayam, S.; Davis, J.I.; de Pamphilis, C.W.; Givnish, T.J.; Pires, J.C.; Stevenson, D.W.; Leebens-Mack, J.H. A Phylogenomic Assessment of Ancient Polyploidy and Genome Evolution across the Poales. Genome Biol. Evol. 2016, 8, 1150–1164. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.F.; Liu, Y.L.; Jin, G.H.; Liu, J.X.; Wu, H.; He, J.; Guo, Z.H.; Li, D.Z. The Pharus latifolius genome bridges the gap of early grass evolution. Plant Cell 2021, 33, 846–864. [Google Scholar] [CrossRef]
- Li, Y.P.; Xu, M.L. CCT family genes in cereal crops: A current overview. Crop J. 2017, 5, 449–458. [Google Scholar] [CrossRef]
- Shibaya, T.; Nonoue, Y.; Ono, N.; Yamanouchi, U.; Hori, K.; Yano, M. Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions. Theor. Appl. Genet. 2011, 123, 1133–1143. [Google Scholar] [CrossRef]
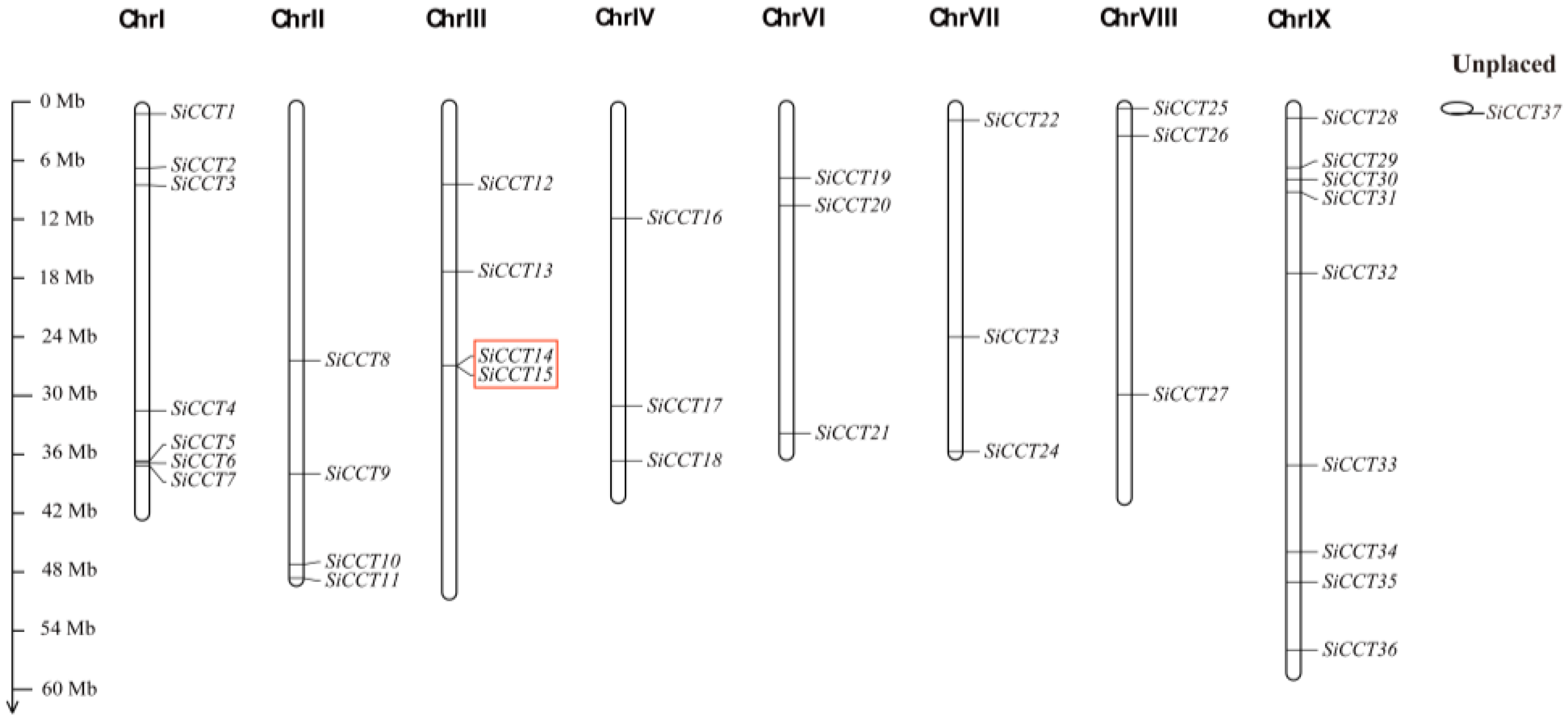
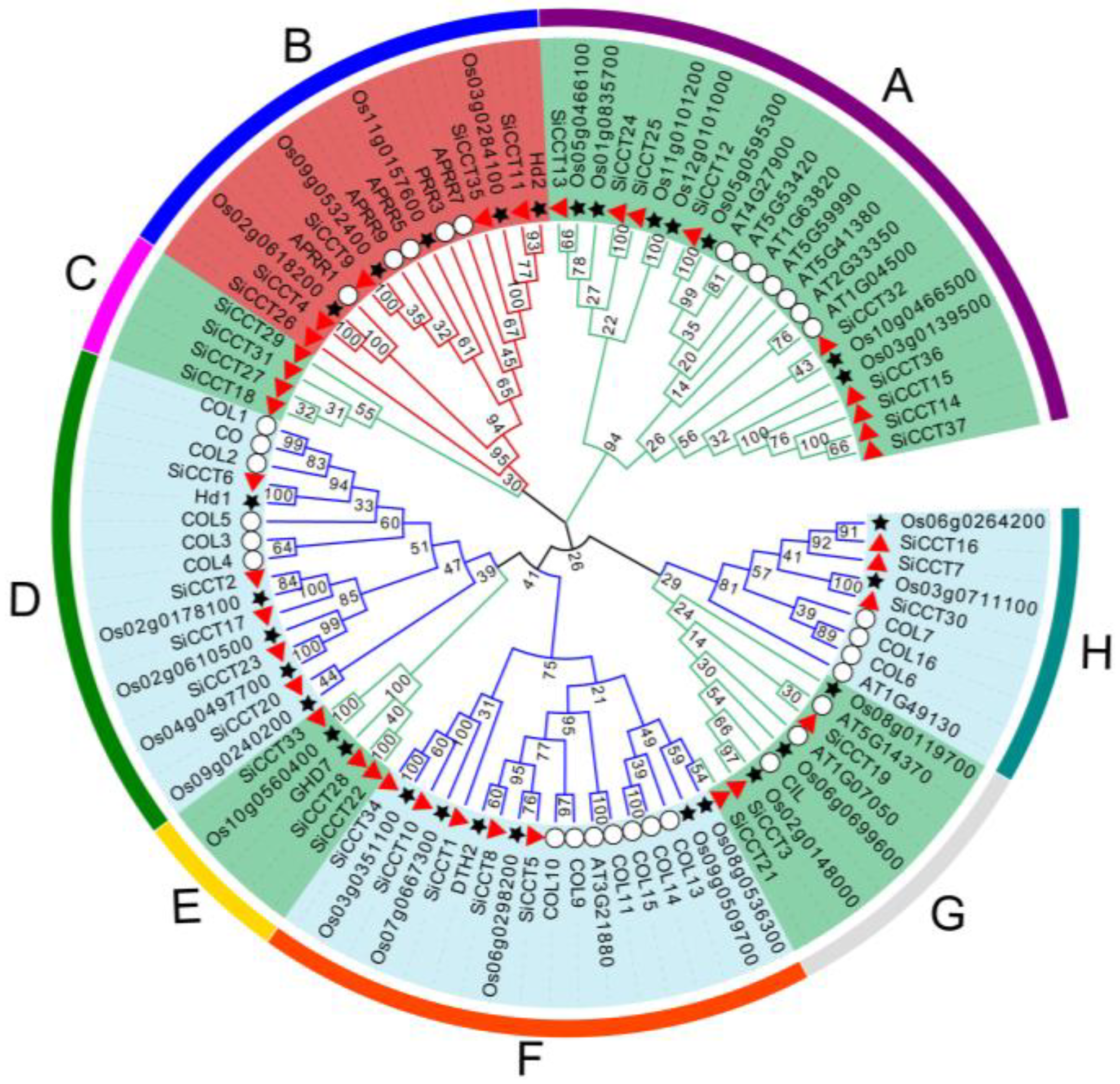



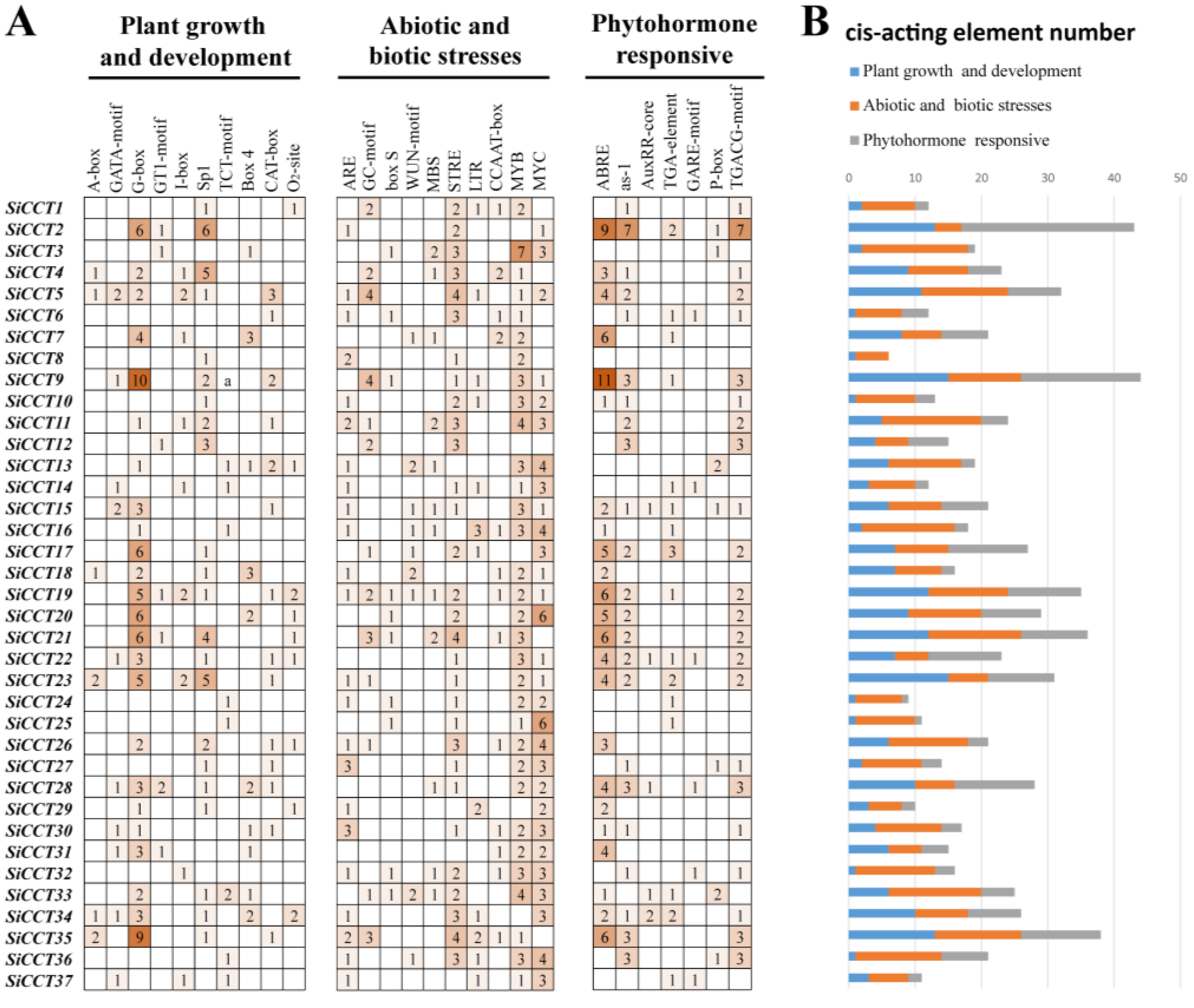
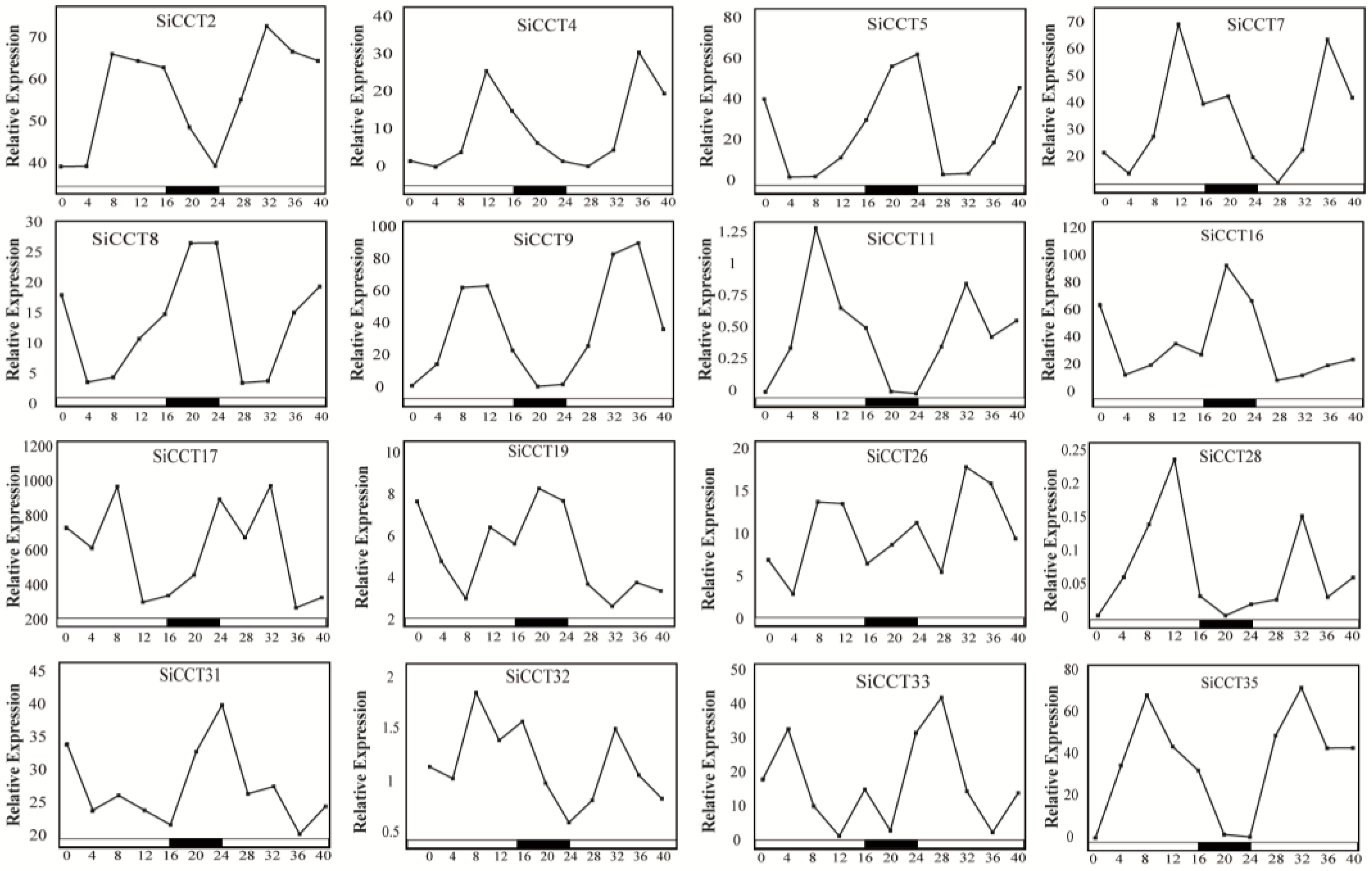
| Gene Name | Transcript ID | Alias | Chr | Protein Length (aa) | MW (D) | pI | Location |
|---|---|---|---|---|---|---|---|
| SETIT_019213mg | KQL27760 | SiCCT1 | Ⅰ | 432 | 46,243.27 | 5.60 | Nucleus |
| SETIT_017487mg | KQL28473 | SiCCT2 | Ⅰ | 386 | 39,459.15 | 6.32 | Nucleus |
| SETIT_017125mg | KQL28750 | SiCCT3 | Ⅰ | 453 | 47,725.85 | 8.14 | Nucleus |
| SETIT_016922mg | KQL30521 | SiCCT4 | Ⅰ | 518 | 57,741.59 | 5.59 | Nucleus/Mitochondrion |
| SETIT_017374mg | KQL31286 | SiCCT5 | Ⅰ | 407 | 44,610.57 | 4.99 | Nucleus |
| SETIT_019803mg | KQL31326 | SiCCT6 | Ⅰ | 355 | 39,793.63 | 8.00 | Nucleus |
| SETIT_017124mg | KQL31374 | SiCCT7 | Ⅰ | 440 | 47,328.18 | 6.54 | Nucleus |
| SETIT_030034mg | KQL24119 | SiCCT8 | Ⅱ | 406 | 43,630.40 | 5.20 | Nucleus |
| SETIT_029202mg | KQL25444 | SiCCT9 | Ⅱ | 630 | 70,287.20 | 6.22 | Nucleus |
| SETIT_030140mg | KQL27073 | SiCCT10 | Ⅱ | 384 | 40,363.07 | 6.10 | Nucleus |
| SETIT_033274mg | KQL27318 | SiCCT11 | Ⅱ | 661 | 71,442.89 | 8.74 | Nucleus |
| SETIT_022659mg | KQL13967 | SiCCT12 | Ⅲ | 325 | 35,522.48 | 4.93 | Nucleus |
| SETIT_024852mg | KQL15112 | SiCCT13 | Ⅲ | 319 | 35,116.48 | 5.75 | Nucleus |
| SETIT_024038mg | KQL16011 | SiCCT14 | Ⅲ | 331 | 36,436.76 | 4.58 | Nucleus |
| SETIT_024806mg | KQL16012 | SiCCT15 | Ⅲ | 268 | 29,552.18 | 4.78 | Nucleus |
| SETIT_006432mg | KQL10228 | SiCCT16 | Ⅳ | 445 | 47,590.03 | 5.09 | Nucleus |
| SETIT_006690mg | KQL11132 | SiCCT17 | Ⅳ | 372 | 39,574.38 | 6.13 | Nucleus |
| SETIT_006750mg | KQL11783 | SiCCT18 | Ⅳ | 357 | 37,898.91 | 5.01 | Nucleus |
| SETIT_014122mg | KQL01011 | SiCCT19 | Ⅵ | 313 | 33,098.76 | 7.65 | Nucleus |
| SETIT_014224mg | KQL01176 | SiCCT20 | Ⅵ | 281 | 28,425.62 | 6.42 | Nucleus |
| SETIT_014037mg | KQL02690 | SiCCT21 | Ⅵ | 235 | 26,147.14 | 9.58 | Nucleus |
| SETIT_011920mg | KQK96183 | SiCCT22 | Ⅶ | 217 | 23,959.73 | 6.75 | Nucleus |
| SETIT_010592mg | KQK97912 | SiCCT23 | Ⅶ | 326 | 34,393.37 | 5.15 | Nucleus |
| SETIT_011862mg | KQL00040 | SiCCT24 | Ⅶ | 202 | 21,587.85 | 5.61 | Nucleus |
| SETIT_027626mg | KQK93230 | SiCCT25 | Ⅷ | 236 | 25,887.70 | 6.50 | Nucleus |
| SETIT_026170mg | KQK93667 | SiCCT26 | Ⅷ | 566 | 62,003.23 | 7.40 | Nucleus |
| SETIT_027518mg | KQK95022 | SiCCT27 | Ⅷ | 160 | 17,635.30 | 10.18 | Nucleus |
| SETIT_039184mg | KQK86275 | SiCCT28 | Ⅸ | 248 | 27,292.51 | 6.86 | Nucleus |
| SETIT_036747mg | KQK87242 | SiCCT29 | Ⅸ | 308 | 32,998.37 | 8.66 | Nucleus |
| SETIT_035937mg | KQK87465 | SiCCT30 | Ⅸ | 406 | 44,451.22 | 6.65 | Nucleus |
| SETIT_036492mg | KQK87704 | SiCCT31 | Ⅸ | 339 | 35,998.10 | 4.77 | Nucleus |
| SETIT_035901mg | KQK88797 | SiCCT32 | Ⅸ | 412 | 43,116.93 | 4.37 | Nucleus |
| SETIT_036910mg | KQK89929 | SiCCT33 | Ⅸ | 286 | 29,324.74 | 7.14 | Nucleus |
| SETIT_034611mg | KQK90887 | SiCCT34 | Ⅸ | 648 | 69,184.42 | 8.49 | Nucleus |
| SETIT_034368mg | KQK91381 | SiCCT35 | Ⅸ | 760 | 82,668.80 | 6.07 | Nucleus |
| SETIT_039219mg | KQK92647 | SiCCT36 | Ⅸ | 365 | 39,857.30 | 4.67 | Nucleus |
| SETIT_020907mg | KQK85264 | SiCCT37 | Unplaced | 353 | 38,888.61 | 4.75 | Nucleus |
| Type | Locus 1 | Locus 2 | Ka | Ks | Ka/Ks | T (MYA) | Period |
|---|---|---|---|---|---|---|---|
| Millet | SiCCT32 | SiCCT36 | 1.04842 | 0.879676 | 1.19183 | 67.67 | Intermediate ancestor |
| Millet | SiCCT2 | SiCCT17 | 0.242598 | 1.71138 | 0.141756 | 131.64 | Poaceae ancestor |
| Millet | SiCCT7 | SiCCT16 | 0.218529 | 1.39626 | 0.15651 | 107.40 | Poaceae ancestor |
| Millet | SiCCT10 | SiCCT34 | 0.332322 | 2.14794 | 0.154716 | 165.23 | Poaceae ancestor |
| Millet | SiCCT24 | SiCCT25 | 0.00952499 | 0.0261684 | 0.363988 | 2.01 | Millet |
| Millet | SiCCT11 | SiCCT35 | 0.974077 | 1.10846 | 0.878766 | 85.27 | Intermediate ancestor |
| Sorghum | EES15491 | EES07834 | 0.0549948 | 0.12169 | 0.451925 | 9.36 | Sorghum |
| Sorghum | KXG22181 | EES03844 | 0.362499 | 2.54405 | 0.142489 | 195.70 | Poaceae ancestor |
| Sorghum | EES15491 | EES03844 | 0.879346 | 0.771143 | 1.14032 | 59.32 | Intermediate ancestor |
| Sorghum | KXG40217 | EER94114 | 0.952785 | 1.15926 | 0.821889 | 89.17 | Intermediate ancestor |
| Sorghum | EES05406 | EES12446 | 0.144635 | 1.20479 | 0.12005 | 92.68 | Intermediate ancestor |
| Sorghum | EES05581 | EER88191 | 0.997347 | 1.00659 | 0.990813 | 77.43 | Intermediate ancestor |
| Sorghum | OQU84486 | EER90145 | 1.01935 | 0.960586 | 1.06118 | 73.89 | Intermediate ancestor |
| Sorghum | EES07342 | EER89633 | 0.148272 | 0.900118 | 0.164726 | 69.24 | Intermediate ancestor |
| Sorghum | OQU85473 | EER88227 | 0.996734 | 1.01133 | 0.985566 | 77.79 | Intermediate ancestor |
| Sorghum | EER94860 | EER99873 | 0.347808 | 1.79793 | 0.19345 | 138.30 | Poaceae ancestor |
| Maize | Zm00001d025770 | Zm00001d003162 | 0.0261931 | 0.239901 | 0.109183 | 18.45 | Intermediate ancestor |
| Maize | Zm00001d021291 | Zm00001d006212 | 0.0625971 | 0.129561 | 0.483147 | 9.97 | Maize |
| Maize | Zm00001d022500 | Zm00001d007107 | 0.176657 | 0.333117 | 0.530314 | 25.62 | Panicoideae ancestor |
| Maize | Zm00001d029149 | Zm00001d007107 | 0.641455 | 0.939475 | 0.68278 | 72.27 | Intermediate ancestor |
| Maize | Zm00001d042958 | Zm00001d012441 | 0.0781679 | 0.396518 | 0.197136 | 30.50 | Panicoideae ancestor |
| Maize | Zm00001d013443 | Zm00001d033719 | 0.0617102 | 0.38183 | 0.161617 | 29.37 | Panicoideae ancestor |
| Maize | Zm00001d027598 | Zm00001d014074 | 1.01132 | 0.971378 | 1.04112 | 74.72 | Intermediate ancestor |
| Maize | Zm00001d032768 | Zm00001d014074 | 1.10514 | 0.795633 | 1.38901 | 61.20 | Intermediate ancestor |
| Maize | Zm00001d048369 | Zm00001d014074 | 0.5782 | 0.687273 | 0.841296 | 52.87 | Intermediate ancestor |
| Maize | Zm00001d036494 | Zm00001d014656 | 1.02009 | 0.946121 | 1.07818 | 72.78 | Intermediate ancestor |
| Maize | Zm00001d015468 | Zm00001d046925 | 0.510484 | 1.31613 | 0.387869 | 101.24 | Poaceae ancestor |
| Maize | Zm00001d025770 | Zm00001d017176 | 0.157993 | 1.59228 | 0.0992244 | 122.48 | Poaceae ancestor |
| Maize | Zm00001d051047 | Zm00001d017176 | 0.0802383 | 0.492435 | 0.162942 | 37.88 | Panicoideae ancestor |
| Maize | Zm00001d017241 | Zm00001d051114 | 0.0547288 | 0.209798 | 0.260864 | 16.14 | Panicoideae ancestor |
| Maize | Zm00001d051684 | Zm00001d017885 | 0.995396 | 1.01465 | 0.981028 | 78.05 | Intermediate ancestor |
| Maize | Zm00001d037327 | Zm00001d017939 | 0.857576 | 1.41978 | 0.604022 | 109.21 | Poaceae ancestor |
| Maize | Zm00001d045661 | Zm00001d017939 | 0.289121 | 1.6396 | 0.176336 | 126.12 | Poaceae ancestor |
| Maize | Zm00001d021291 | Zm00001d052781 | 1.01446 | 0.944687 | 1.07386 | 72.67 | Intermediate ancestor |
| Maize | Zm00001d025770 | Zm00001d051047 | 0.169024 | 1.43393 | 0.117874 | 110.30 | Poaceae ancestor |
| Maize | Zm00001d027598 | Zm00001d048369 | 1.007 | 0.96874 | 1.0395 | 74.52 | Intermediate ancestor |
| Maize | Zm00001d035134 | Zm00001d049651 | 0.985493 | 1.03346 | 0.953586 | 79.50 | Intermediate ancestor |
| Maize | Zm00001d045661 | Zm00001d037327 | 0.0757818 | 0.430623 | 0.175982 | 33.12 | Poaceae ancestor |
| Maize | Zm00001d029149 | Zm00001d022500 | 0.838386 | 0.587664 | 1.42664 | 45.20 | Poaceae ancestor |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Yu, S.; Zhang, Q.; Wang, Z.; Liu, M.; Zhang, A.; Dong, X.; Fan, J.; Zhu, Y.; Ruan, Y.; et al. Genome-Wide Identification and Characterization of the CCT Gene Family in Foxtail Millet (Setaria italica) Response to Diurnal Rhythm and Abiotic Stress. Genes 2022, 13, 1829. https://doi.org/10.3390/genes13101829
Li Y, Yu S, Zhang Q, Wang Z, Liu M, Zhang A, Dong X, Fan J, Zhu Y, Ruan Y, et al. Genome-Wide Identification and Characterization of the CCT Gene Family in Foxtail Millet (Setaria italica) Response to Diurnal Rhythm and Abiotic Stress. Genes. 2022; 13(10):1829. https://doi.org/10.3390/genes13101829
Chicago/Turabian StyleLi, Yuntong, Shumin Yu, Qiyuan Zhang, Ziwei Wang, Meiling Liu, Ao Zhang, Xiaomei Dong, Jinjuan Fan, Yanshu Zhu, Yanye Ruan, and et al. 2022. "Genome-Wide Identification and Characterization of the CCT Gene Family in Foxtail Millet (Setaria italica) Response to Diurnal Rhythm and Abiotic Stress" Genes 13, no. 10: 1829. https://doi.org/10.3390/genes13101829
APA StyleLi, Y., Yu, S., Zhang, Q., Wang, Z., Liu, M., Zhang, A., Dong, X., Fan, J., Zhu, Y., Ruan, Y., & Li, C. (2022). Genome-Wide Identification and Characterization of the CCT Gene Family in Foxtail Millet (Setaria italica) Response to Diurnal Rhythm and Abiotic Stress. Genes, 13(10), 1829. https://doi.org/10.3390/genes13101829






