Transcriptome Response of Differentiating Muscle Satellite Cells to Thermal Challenge in Commercial Turkey
Abstract
:1. Introduction
2. Materials and Methods
2.1. Turkey Myogenic Satellite Cells
2.2. RNA Isolation and Sequencing
2.3. RNAseq Data Analyses
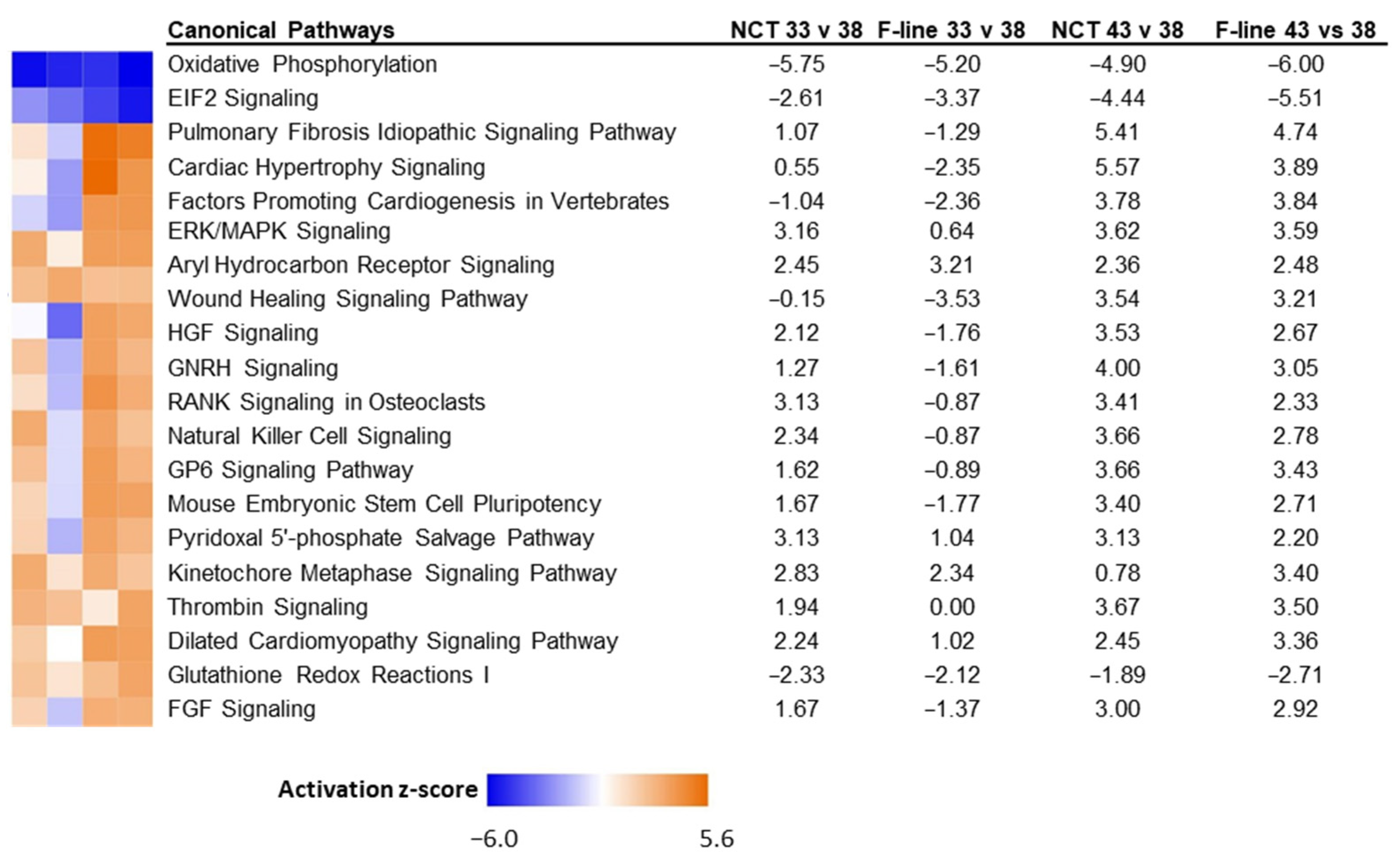
3. Results
3.1. Gene Expression
3.2. Differential Expression
3.2.1. Effect of Cold Treatment
3.2.2. Effect of Heat Treatment
3.3. Effects of Selection
3.3.1. Line Differences: NCT versus RBC2 SCs
3.3.2. Line Differences: NCT Versus F-Line SCs
4. Discussion
4.1. Effect of Cold Stress
4.2. Effect of Heat Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cardasis, C.A.; Cooper, G.W. An analysis of nuclear numbers in individual muscle fibers during differentiation and growth: A satellite cell-muscle fiber growth unit. J. Exp. Zool. 1975, 191, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, R. The satellite cell and muscle regeneration. In Myology, 2nd ed.; Engel, A.G., Franzini-Armstrong, C., Eds.; McGraw-Hill, Inc.: New York, NY, USA, 1994; Volume 1, pp. 97–118. [Google Scholar]
- Yablonka-Reuveni, Z.; Quinn, L.S.; Nameroff, M. Isolation and clonal analysis of satellite cells from chicken pectoralis muscle. Dev. Biol. 1987, 119, 252–259. [Google Scholar] [CrossRef] [Green Version]
- Hartley, R.S.; Bandman, E.; Yablonka-Reuveni, Z. Skeletal muscle satellite cells appear during late chicken embryogenesis. Dev. Biol. 1992, 153, 206–216. [Google Scholar] [CrossRef] [Green Version]
- Velleman, S.G.; Zhang, X.; Coy, C.S.; Song, Y.; McFarland, D.C. Changes in satellite cell proliferation and differentiation during turkey muscle development. Poult. Sci. 2010, 89, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Asakura, A.; Komaki, M.; Rudnicki, M. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation 2001, 68, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.J.; Velleman, S.G.; Cowieson, A.J.; Singh, M.; Muir, W.I. Influence of hatch time and access to feed on intramuscular adipose tissue deposition in broilers. Poult. Sci. 2016, 95, 1449–1456. [Google Scholar] [CrossRef]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef] [Green Version]
- Conboy, I.M.; Rando, T.A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 2002, 3, 397–409. [Google Scholar] [CrossRef] [Green Version]
- Fujimaki, S.; Machida, M.; Hidaka, R.; Asashima, M.; Takemasa, T.; Kuwabara, T. Intrinsic ability of adult stem cell in skeletal muscle: An effective and replenishable resource to the establishment of pluripotent stem cells. Stem Cells Int. 2013, 2013, 420164. [Google Scholar] [CrossRef]
- Pallafacchina, G.; Blaauw, B.; Schiaffino, S. Role of satellite cells in muscle growth and maintenance of muscle mass. Nutr. Metab. Cardiovasc. Dis. 2013, 23 (Suppl. 1), S12–S18. [Google Scholar] [CrossRef]
- Zammit, P.S. All muscle satellite cells are equal, but are some more equal than others? J. Cell Sci. 2008, 121, 2975–2982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Wang, C.; Xiao, F.; Wang, H.; Wu, Z. JAK2/STAT2/STAT3 are required for myogenic differentiation. Mol. Basis Cell Dev. Biol. 2008, 283, 34029–34036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halevy, O.; Geyra, A.; Barak, M.; Uni, Z.; Sklan, D. Early posthatch starvation decreases satellite cell proliferation and skeletal muscle growth in chicks. J. Nutr. 2000, 130, 858–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mozdziak, P.E.; Walsh, T.J.; McCoy, D.W. The effect of early posthatch nutrition on satellite cell mitotic activity. Poult. Sci. 2002, 81, 1703–1708. [Google Scholar] [CrossRef]
- Piestun, Y.; Druyan, S.; Brake, J.; Yahav, S. Thermal manipulations during broiler incubation alter performance of broilers to 70 days of age. Poult. Sci. 2013, 92, 1155–1163. [Google Scholar] [CrossRef]
- Loyau, T.; Metayer-Coustard, S.; Berri, C.; Crochet, S.; Cailleau-Audouin, E.; Sannier, M.; Chartrin, P.; Praud, C.; Hennequet-Antier, C.; Rideau, N.; et al. Thermal manipulation during embryogenesis has long-term effects on muscle and liver metabolism in fast-growing chickens. PLoS ONE 2014, 9, e105339. [Google Scholar] [CrossRef]
- Myhre, K. Behavioral temperature regulation in neonate chick of Bantam hen (Gallus domesticus). Poult. Sci. 1978, 57, 1369–1375. [Google Scholar] [CrossRef]
- Modrey, P.; Nichelmann, M. Development of autonomic and behavioral thermoregulation in turkeys (Meleagris gallopavo). J. Biol. 1992, 17, 287–292. [Google Scholar]
- Shinder, D.; Rusal, M.; Tanny, J.; Druyan, S.; Yahav, S. Thermoregulatory response of chicks (Gallus domesticus) to low ambient temperatures at an early age. Poult. Sci. 2007, 86, 2200–2209. [Google Scholar] [CrossRef]
- Clark, D.L.; Coy, C.S.; Strasburg, G.M.; Reed, K.M.; Velleman, S.G. Temperature effect on proliferation and differentiation of satellite cells from turkeys with different growth rates. Poult. Sci. 2016, 95, 934–947. [Google Scholar] [CrossRef]
- Nestor, K.E. Genetics of growth and reproduction in the turkey. 5. Selection for increased body weight alone and in combination with increased egg production. Poult. Sci. 1977, 56, 337–347. [Google Scholar] [CrossRef]
- Nestor, K.E. Genetics of growth and reproduction in the turkey. 9. Long-term selection for increased 16-week body weight. Poult Sci. 1984, 63, 2114–2122. [Google Scholar] [CrossRef] [PubMed]
- Reed, K.M.; Mendoza, K.M.; Abrahante, J.E.; Barnes, N.E.; Velleman, S.G.; Strasburg, G.M. Response of turkey muscle satellite cells to thermal challenge. I. Transcriptome effects in proliferating cells. BMC Genom. 2017, 18, 352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, K.M.; Mendoza, K.M.; Strasburg, G.M.; Velleman, S.G. Response of turkey muscle satellite cells to thermal challenge. II. Transcriptome effects in differentiating cells. Front. Physiol. 2017, 8, 948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, K.M.; Mendoza, K.M.; Strasburg, G.M.; Velleman, S.G. Transcriptome response of proliferating muscle satellite cells to thermal challenge in commercial turkey. Front. Physiol. 2022, 13, 970243. [Google Scholar] [CrossRef] [PubMed]
- Velleman, S.G.; Liu, X.; Nestor, K.E.; McFarland, D.C. Heterogeneity in growth and differentiation characteristics in male and female satellite cells isolated from turkey lines with different growth rates. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2000, 125, 503–509. [Google Scholar] [CrossRef]
- Nestor, K.E.; McCartney, M.G.; Bachev, N. Relative contributions of genetics and environment to turkey improvement. Poult. Sci. 1969, 48, 1944–1949. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucl. Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Defer, N.; Best-Belpomme, M.; Hanoune, J. Tissue specificity and physiological relevance of various isoforms of adenylyl cyclase. Am. J. Physiol. Ren. Physiol. 2000, 279, 400–416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Abreu, J.G.; Yokota, C.; MacDonald, B.T.; Singh, S.; Coburn, K.L.; Cheong, S.M.; Zhang, M.M.; Ye, Q.Z.; Hang, H.C.; et al. Tiki1 is required for head formation via Wnt cleavage-oxidation and inactivation. Cell 2012, 149, 1565–1577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pingault, V.; Zerad, L.; Bertani-Torres, W.; Bondurand, N. SOX10: 20 years of phenotypic plurality and current understanding of its developmental function. J. Med. Genet. 2022, 59, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.M.; Nijstad, N.; Franceschini, L. Regulation of microsomal triglyceride transfer protein. Clin. Lipidol. 2011, 6, 293–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, B.L.; Hong, W. ADAMTS: A novel family of proteases with an ADAM protease domain and thrombospondin 1 repeats. FEBS Lett. 1999, 445, 223–225. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Denard, B.; Lee, C.E.; Han, S.; Ye, J.S.; Ye, J. Inverting the topology of a transmembrane protein by regulating the translocation of the first transmembrane helix. Mol. Cell. 2016, 63, 567–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilliland, D.G.; Griffin, J.D. The roles of FLT3 in hematopoiesis and leukemia. Blood 2002, 100, 1532–1542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Cabrera, P.J.; Shi, T.; Yun, J.; McCune, D.F.; Rorabaugh, B.R.; Perez, D.M. Differential regulation of the cell cycle by alpha1-adrenergic receptor subtypes. Endocrinology 2004, 145, 5157–5167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulero, J.J.; Boyle, B.J.; Bradley, S.; Bright, J.M.; Nelken, S.T.; Ho, T.T.; Mize, N.K.; Childs, J.D.; Ballinger, D.G.; Ford, J.E.; et al. Three new human members of the lipid transfer/lipopolysaccharide binding protein family (LT/LBP). Immunogenetics 2002, 54, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Chou, K.B.; Takeda, J.; Sachais, B.S.; Krause, J.E. Molecular cloning, structural characterization and functional expression of the human substance P receptor. Biochem. Biophys. Res. Commun. 1991, 179, 1232–1240. [Google Scholar] [CrossRef]
- Lawrence, E.J.; Arpag, G.; Norris, S.R.; Zanic, M. Human CLASP2 specifically regulates microtubule catastrophe and rescue. Mol. Biol. Cell. 2018, 29, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.K.; Sowa, H.; Hinoi, E.; Ferron, M.; Ahn, J.D.; Confavreux, C.; Dacquin, R.; Mee, P.J.; McKee, M.D.; Jung, D.Y.; et al. Endocrine regulation of energy metabolism by the skeleton. Cell 2007, 130, 456–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.P.; Lo, C.Y.; Tseng, H.M.; Chao, C.C. Knockdown of protein kinase CK2 blocked gene expression mediated by brain-derived neurotrophic factor-induced serum response element. Chin. J. Physiol. 2019, 62, 63–69. [Google Scholar]
- Stewart, A.; Guan, H.; Yang, K. BMP-3 promotes mesenchymal stem cell proliferation through the TGF-beta/activin signaling pathway. J. Cell. Physiol. 2010, 223, 658–666. [Google Scholar] [PubMed]
- Clevers, H.; Nusse, R. Wnt/β-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunk, C.; Shams, M.; Nijjar, S.; Rhaman, M.; Qiu, Y.; Bussolati, B.; Ahmed, A. Angiopoietin-1 and angiopoietin-2 activate trophoblast Tie-2 to promote growth and migration during placental development. Am. J. Pathol. 2000, 156, 2185–2199. [Google Scholar] [CrossRef] [Green Version]
- Smyth, I.; Du, X.; Taylor, M.S.; Justice, M.J.; Beutler, B.; Jackson, I.J. The extracellular matrix gene Frem1 is essential for the normal adhesion of the embryonic epidermis. Proc. Natl. Acad. Sci. USA 2004, 101, 13560–13565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Cazarín, M.L.; Smith, J.L.; Olszewski, K.A.; McCune, D.F.; Simmerman, L.A.; Hadley, R.W.; Kraner, S.D.; Piascik, M.T. The alpha1D-adrenergic receptor is expressed intracellularly and coupled to increases in intracellular calcium and reactive oxygen species in human aortic smooth muscle cells. J. Mol. Signal. 2008, 3, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, K.M.; Madsen, L.; Prag, S.; Johnsen, A.H.; Semple, C.A.; Hendil, K.B.; Hartmann-Petersen, R. Thioredoxin Txnl1/TRP32 is a redox-active cofactor of the 26 S proteasome. J. Biol. Chem. 2009, 284, 15246–15254. [Google Scholar] [CrossRef] [Green Version]
- Schultz, E.; Lipton, B.H. Skeletal muscle satellite cells: Changes in proliferation potential as a function of age. Mech. Ageing Dev. 1982, 20, 377–383. [Google Scholar] [CrossRef]
- Tanaka, S.; Terada, K.; Nohno, T. Canonical Wnt signaling is involved in switching from cell proliferation to myogenic differentiation of mouse myoblast cells. J. Mol. Sign. 2011, 6, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Shang, R.; Bi, P. Feedback regulation of Notch signaling and myogenesis connected by MyoD–Dll1 axis. PLoS Genet. 2021, 17, e1009729. [Google Scholar] [CrossRef] [PubMed]
- Holst, D.; Luquet, S.; Kristiansen, K.; Grimaldi, P.A. Roles of peroxisome proliferator-activated receptors delta and gamma in myoblast transdifferentiation. Exp. Cell Res. 2003, 288, 168–176. [Google Scholar] [CrossRef]
- Xu, J.; Strasburg, G.M.; Reed, K.M.; Velleman, S.G. Effect of temperature and selection for growth on intracellular lipid accumulation and adipogenic gene expression in turkey pectoralis major muscle satellite cells. Front. Physiol. 2021, 12, 667814. [Google Scholar] [CrossRef] [PubMed]
- Rı́os, R.; Carneiro, I.; Arce, V.M.; Devesa, J. Myostatin is an inhibitor of myogenic differentiation. Am. J. Physiol. Cell Physiol. 2002, 282, C993–C999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.-H.; Choi, Y.M.; Suh, Y.; Shin, S.; Lee, J.; Hwang, S.; Lee, K. Research note: Association of temporal expression of myostatin with hypertrophic muscle growth in different Japanese quail lines. Poult. Sci. 2020, 99, 2926–2930. [Google Scholar] [CrossRef]
- Buckingham, M. Muscle differentiation: Which myogenic factors make muscle? Curr. Biol. 1994, 4, 61–63. [Google Scholar] [CrossRef]
- Lazure, F.; Blackburn, D.M.; Corchado, A.H.; Sahinyan, K.; Karam, N.; Sharanek, A.; Nguyen, D.; Lepper, C.; Najafabadi, H.S.; Perkins, T.J.; et al. Myf6/MRF4 is a myogenic niche regulator required for the maintenance of the muscle stem cell pool. EMBO Rep. 2020, 21, e49499. [Google Scholar] [CrossRef]
- Schiroli, D.; Cirrincione, S.; Donini, S.; Peracchi, A. Strict reaction and substrate specificity of AGXT2L1, the human O-phosphoethanolamine phospho-lyase. IUBMB Life 2013, 65, 645–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Q.; Kang, J.; Dai, J.; Tang, M.; Wang, Q.; Zhang, H.; Guo, W.; Sun, R.; Yu, H. AGXT2L1 is down-regulated in heptocellular carcinoma and associated with abnormal lipogenesis. J. Clin. Pathol. 2016, 69, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zheng, L.; Lu, K.; Gao, Y.; Guo, L.; Xu, W.; Wang, X. Downregulation of cohesin loading factor nipped-B-like protein (NIPBL) induces cell cycle arrest, apoptosis, and autophagy of breast cancer cell lines. Med. Sci. Monit. 2017, 23, 4817–4825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Strasburg, G.M.; Reed, K.M.; Velleman, S.G. Thermal stress affects proliferation and differentiation of turkey satellite cells through the mTOR/S6K pathway in a growth-dependent manner. PLoS ONE 2022, 17, e0262576. [Google Scholar] [CrossRef]
- Xu, J.; Strasburg, G.M.; Reed, K.M.; Velleman, S.G. Temperature and growth selection effects on proliferation, differentiation, and adipogenic potential of turkey myogenic satellite cells through frizzled-7-mediated Wnt planar cell polarity pathway. Front. Physiol. 2022, 13, 892887. [Google Scholar] [CrossRef] [PubMed]
- Thoma, A.; Lightfoot, A.P. NF-kB and inflammatory cytokine signaling: Role in skeletal muscle atrophy. Adv. Exp. Med. Biol. 2018, 1088, 267–279. [Google Scholar] [PubMed]
- Xu, J.; Strasburg, G.M.; Reed, K.M.; Velleman, S.G. Response of turkey pectoralis major muscle satellite cells to hot and cold thermal stress: Effect of growth selection on satellite cell proliferation and differentiation. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2021, 252, 110823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.W.; Lv, Z.H.; Li, J.L.; Li, S.; Xu, W.; Wang, X.L. Effects of cold stress on nitric oxide in duodenum of chicks. Poult. Sci. 2011, 90, 1555–1561. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.Q.; Zhang, Z.W.; Qu, J.P.; Yao, H.D.; Li, M.; Li, S.; Xu, S.W. Cold stress induces antioxidants and Hsps in chicken immune organs. Cell Stress Chaperones 2014, 9, 635–648. [Google Scholar] [CrossRef] [Green Version]
- Barnes, N.E.; Mendoza, K.M.; Strasburg, G.M.; Velleman, S.G.; Reed, K.M. Thermal challenge alters the transcriptional profile of the breast muscle in turkey poults. Poult. Sci. 2019, 98, 74–91. [Google Scholar] [CrossRef]
- Ratner, C.; Shin, J.H.; Dwibedi, C.; Tremaroli, V.; Bjerregaard, A.; Hartmann, B.; Bäckhed, F.; Leinninger, G.; Seeley, R.J.; Holst, B. Anorexia and fat aversion induced by vertical sleeve gastrectomy is attenuated in neurotensin receptor 1–deficient mice. Endocrinology 2021, 162, bqab130. [Google Scholar] [CrossRef]
- Saini, A.; Al-Shanti, N.; Stewart, C. C2 skeletal myoblast survival, death, proliferation and differentiation: Regulation by Adra1d. Cell Physiol. Biochem. 2010, 25, 253–262. [Google Scholar] [CrossRef]
- Mimori-Kiyosue, Y.; Grigoriev, I.; Lansbergen, G.; Sasaki, H.; Matsui, C.; Severin, F.; Galjart, N.; Grosveld, F.; Vorobjev, I.; Tsukita, S.; et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 2005, 168, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Papah, M.B.; Brannick, E.M.; Schmidt, C.J.; Abasht, B. Evidence and role of phlebitis and lipid infiltration in the onset and pathogenesis of wooden breast disease in modern broiler chickens. Avian Pathol. 2017, 46, 623–643. [Google Scholar] [CrossRef] [PubMed]
- Velleman, S.G.; Clark, D.L.; Tonniges, J.R. The effect of the wooden breast myopathy on sarcomere structure and organization. Avian Dis. 2018, 62, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Daughtry, M.R.; Berio, E.; Shen, Z.; Suess, E.J.R.; Shah, N.; Geiger, A.E.; Berguson, E.R.; Dalloul, R.A.; Persia, M.E.; Shi, H.; et al. Satellite cell-mediated breast muscle regeneration decreases with broiler size. Poult. Sci. 2017, 96, 3457–3464. [Google Scholar] [CrossRef]
- Velleman, S.G. Why breast muscle satellite cell heterogeneity is an issue of importance for the poultry industry: An opinion paper. Front. Physiol. 2022, 13, 987883. [Google Scholar] [CrossRef]


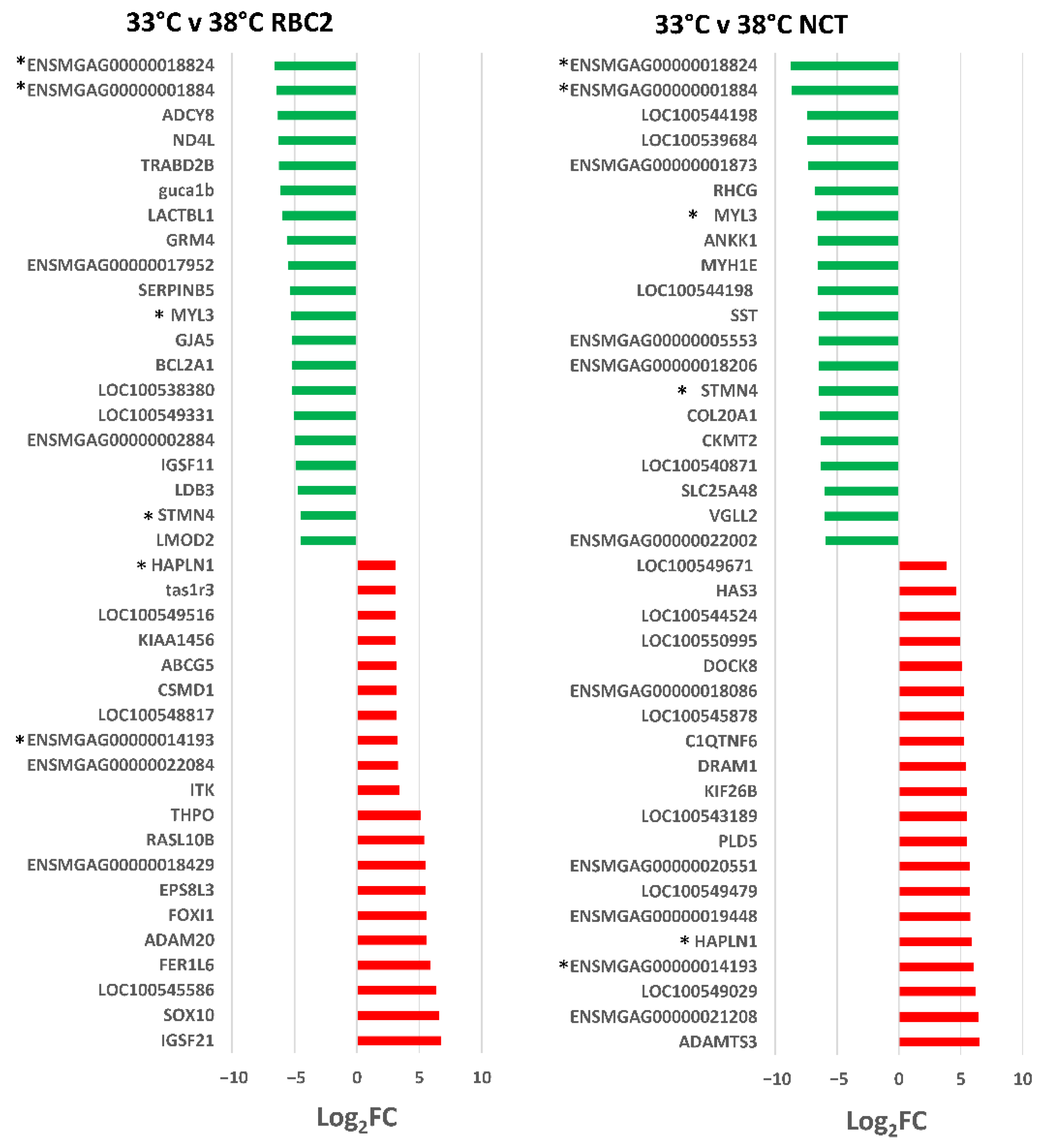
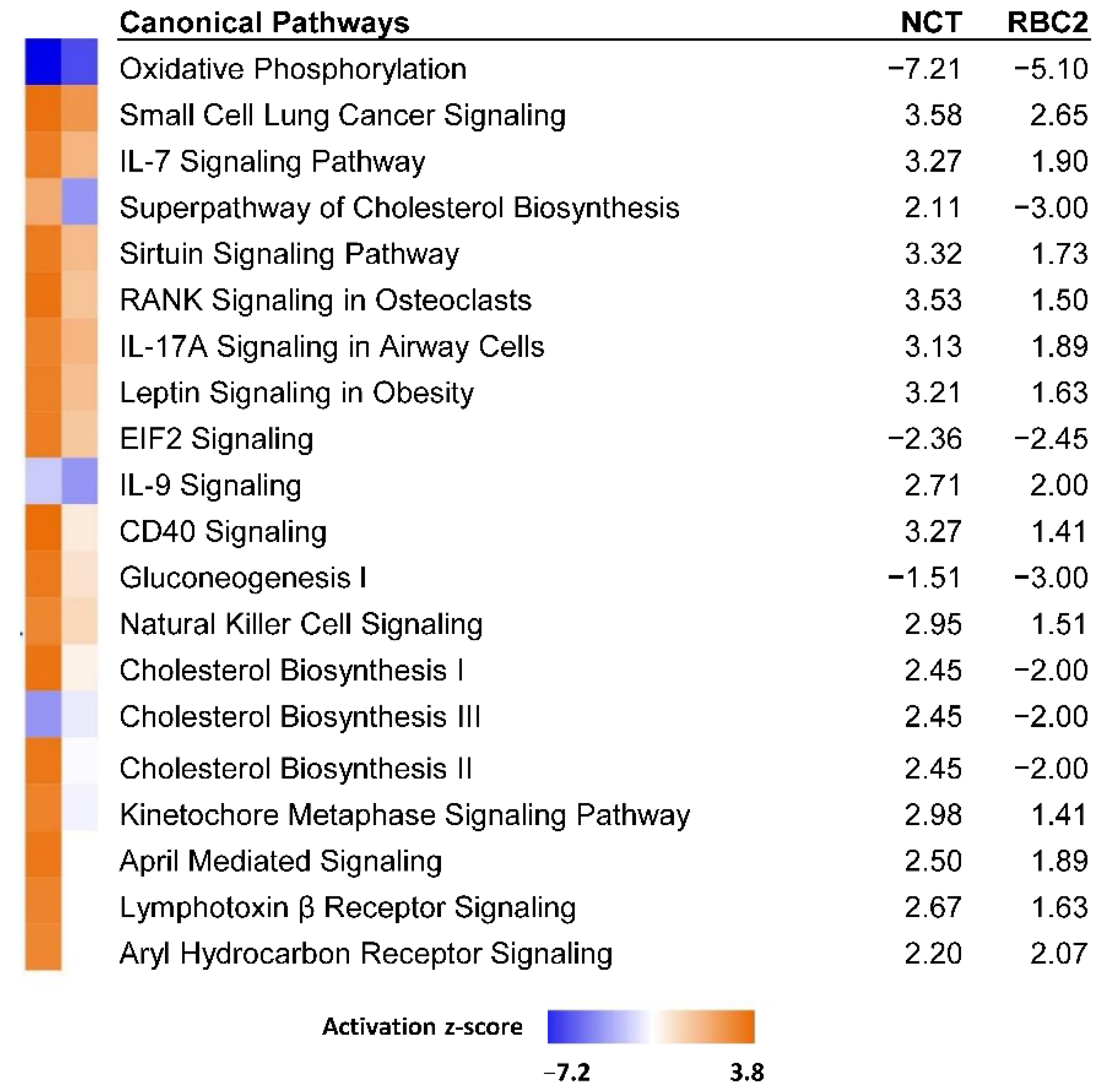

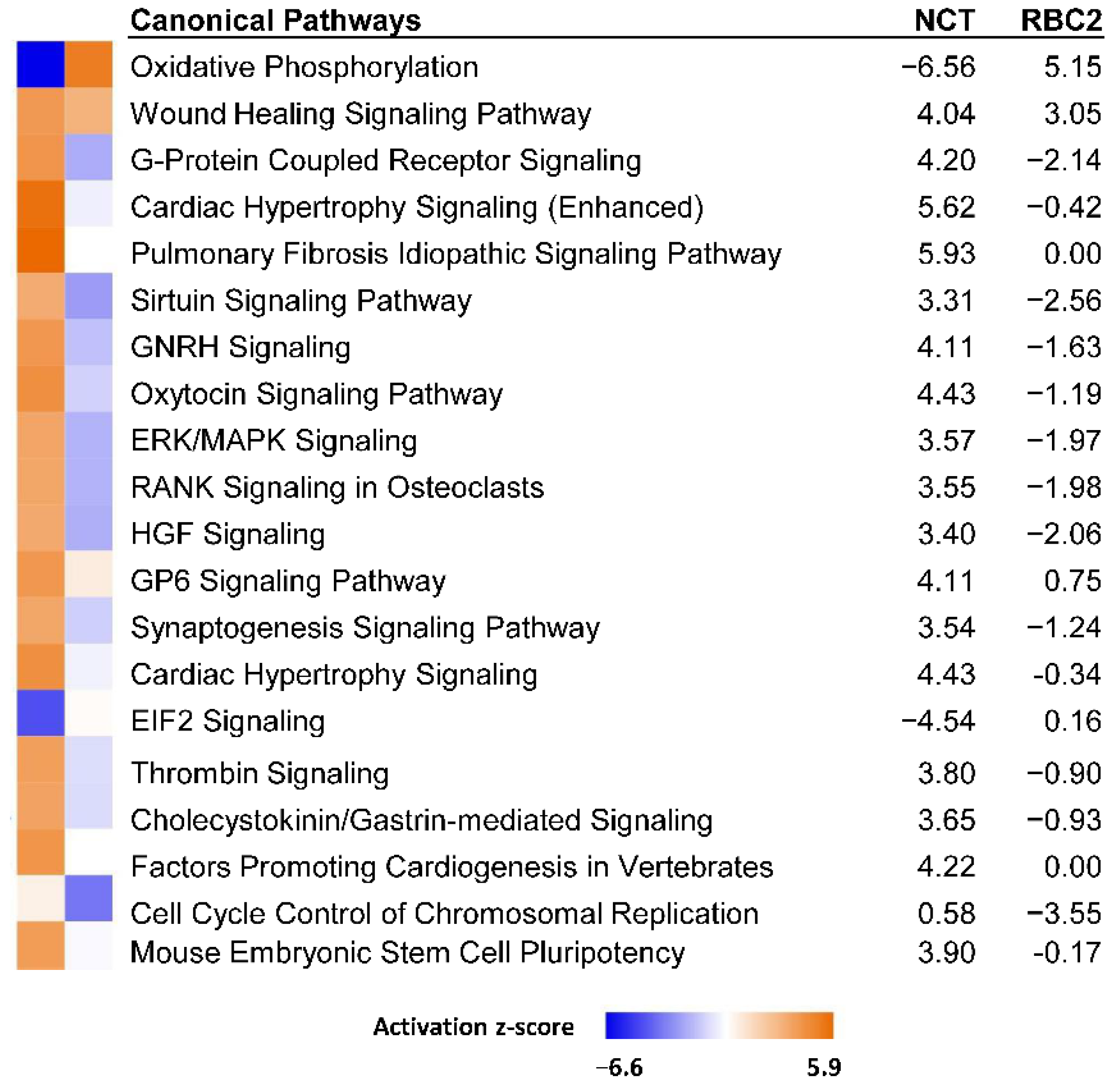
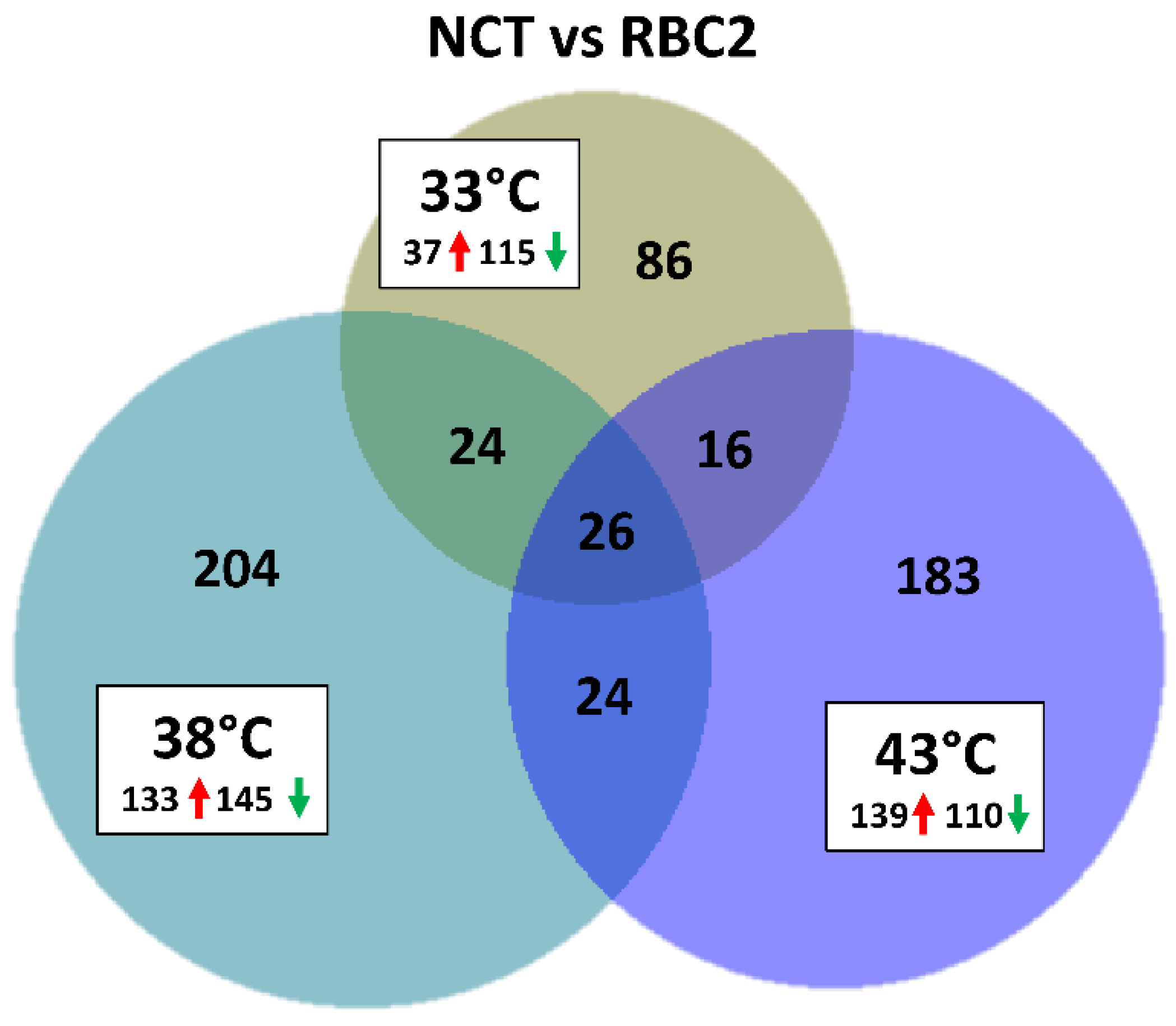

| Line | Temp °C | Replicate | PE Reads | Median Read Quality R1 | Median Read Quality R2 | Observed Genes | Mean Observed Genes | Group Observed Genes | % Expressed Genes |
|---|---|---|---|---|---|---|---|---|---|
| RBC2 | 33 | 1 | 22,142,413 | 36.2 | 35.8 | 13,926 | 13,917.5 | 14,459 | 0.805 |
| 2 | 24,577,416 | 36.3 | 35.8 | 13,909 | |||||
| 38 | 1 | 19,480,565 | 36.2 | 35.8 | 13,638 | 13,732.5 | 14,295 | 0.795 | |
| 2 | 21,879,398 | 36.2 | 35.8 | 13,827 | |||||
| 43 | 1 | 20,058,265 | 36.3 | 35.8 | 13,589 | 13,612.5 | 14,184 | 0.789 | |
| 2 | 21,823,851 | 36.2 | 35.8 | 13,636 | |||||
| NCT | 33 | 1 | 25,893,254 | 36.2 | 35.9 | 13,816 | 13,763.0 | 14,309 | 0.796 |
| 2 | 21,843,150 | 36.2 | 35.8 | 13,710 | |||||
| 38 | 1 | 19,234,626 | 36.2 | 35.8 | 13,537 | 13,569.0 | 14,143 | 0.787 | |
| 2 | 24,062,238 | 36.3 | 35.8 | 13,601 | |||||
| 43 | 1 | 27,102,323 | 36.2 | 35.9 | 14,000 | 13,917.5 | 14,495 | 0.807 | |
| 2 | 25,216,803 | 36.3 | 35.8 | 13,835 | |||||
| Mean | 22,776,191.8 | 36.23 | 35.82 | 13,752.0 | 13,752.00 | 14,314.2 | 0.7966 |
| Comparison | Groups | Total Expressed Genes | Shared Genes | Unique Genes by Group | FDR < 0.05 | |Log2FC| > 1.0 | |Log2FC| > 2.0 |
|---|---|---|---|---|---|---|---|
| Cold (33 °C) | 33R vs. 38R | 14,132 | 13,224 | 527/381 | 3621 | 814 | 244 |
| 33N vs. 38N | 14,013 | 13,045 | 540/428 | 5600 | 1914 | 456 | |
| Hot (43 °C) | 43R vs. 38R | 14,072 | 13,075 | 467/530 | 6248 | 2043 | 525 |
| 43N vs. 38N | 14,124 | 13,106 | 651/367 | 4137 | 1795 | 395 | |
| Line | |||||||
| 38N vs. 38R | 14,027 | 13,051 | 422/554 | 3777 | 1357 | 278 | |
| 33N vs. 33R | 14,129 | 13,207 | 378/544 | 2919 | 501 | 152 | |
| 43N vs. 43R | 14,150 | 13,149 | 608/393 | 4574 | 1365 | 249 |
| Gene ID | Line | Change | Log2FC | FDR p-Value Correction |
|---|---|---|---|---|
| MYOD1, myogenic differentiation 1 | RBC2 | ↑ | 3.521 | 8.59 × 10−157 |
| NCT | ↑ | 1.873 | 2.72 × 10−13 | |
| PAX7, Paired box 7 | RBC2 | ↑ | 0.808 | 5.22 × 10−08 |
| NCT | ↑ | 0.946 | 6.71 × 10−03 | |
| MYOG, myogenin | RBC2 | ↑ | 8.387 | 0.00 |
| NCT | ↑ | 6.695 | 1.77 × 10−113 | |
| DLL1 delta like canonical Notch ligand 1 | RBC2 | ↑ | 3.835 | 1.31 × 10−46 |
| NCT | ↑ | 3.524 | 1.41 × 10−18 | |
| MSTN, myostatin | RBC2 | ↑ | 1.595 | 1.77 × 10−04 |
| NCT | ↑ | 2.519 | 6.23 × 10−08 | |
| MYF6, myogenic factor 6 | RBC2 | ↓ | −3.104 | 0.00 |
| NCT | ↓ | −2.502 | 3.09 × 10−18 | |
| PPARγ, peroxisome proliferator activated receptor γ | RBC2 | ↓ | −2.333 | 2.03 × 10−108 |
| NCT | ↓ | −2.228 | 1.96 × 10−13 | |
| LIF, LIF interleukin 6 family cytokine | RBC2 | ↓ | −3.416 | 4.24 × 10−58 |
| NCT | ↓ | −2.651 | 1.88 × 10−08 |
| Gallus gallus (18109) | Turkey DEGs | Expected | Fold Enrichment | p-Value | |
|---|---|---|---|---|---|
| Control 38 °C (3777 DEGs, 2915 uniquely mapped) | |||||
| striated muscle cell development (GO:0055002) | 45 | 27 | 7.24 | 3.73 | 2.37 × 10−03 |
| myofibril assembly (GO:0030239) | 44 | 26 | 7.08 | 3.67 | 4.76 × 10−03 |
| negative regulation of protein depolymerization (GO:1901880) | 48 | 25 | 7.73 | 3.24 | 4.15 × 10−02 |
| cellular component assembly involved in morphogenesis (GO:0010927) | 58 | 29 | 9.34 | 3.11 | 1.52 × 10−02 |
| regulation of protein depolymerization (GO:1901879) | 60 | 29 | 9.66 | 3.00 | 3.33 × 10−02 |
| regulation of protein-containing complex disassembly (GO:0043244) | 77 | 37 | 12.39 | 2.99 | 1.70 × 10−03 |
| actomyosin structure organization (GO:0031032) | 79 | 36 | 12.72 | 2.83 | 8.95 × 10−03 |
| muscle cell development (GO:0055001) | 84 | 38 | 13.52 | 2.81 | 4.57 × 10−03 |
| negative regulation of supramolecular fiber organization (GO:1902904) | 77 | 34 | 12.39 | 2.74 | 2.50 × 10−02 |
| striated muscle cell differentiation (GO:0051146) | 102 | 44 | 16.42 | 2.68 | 1.80 × 10−03 |
| Cold 33 °C (2919 DEGs, 2343 uniquely mapped) | |||||
| striated muscle cell development (GO:0055002) | 45 | 21 | 5.82 | 3.61 | 4.06 × 10−02 |
| stem cell development (GO:0048864) | 57 | 24 | 7.37 | 3.25 | 3.79 × 10−02 |
| striated muscle cell differentiation (GO:0051146) | 102 | 42 | 13.20 | 3.18 | 3.00 × 10−05 |
| muscle cell development (GO:0055001) | 84 | 34 | 10.87 | 3.13 | 1.12 × 10−03 |
| regulation of axonogenesis (GO:0050770) | 68 | 27 | 8.80 | 3.07 | 2.52 × 10−02 |
| ameboidal-type cell migration (GO:0001667) | 91 | 36 | 11.77 | 3.06 | 7.85 × 10−04 |
| muscle cell differentiation (GO:0042692) | 129 | 50 | 16.69 | 3.00 | 7.37 × 10−06 |
| mesenchymal cell differentiation (GO:0048762) | 103 | 38 | 13.33 | 2.85 | 1.67 × 10−03 |
| stem cell differentiation (GO:0048863) | 87 | 32 | 11.26 | 2.84 | 1.56 × 10−02 |
| muscle structure development (GO:0061061) | 201 | 73 | 26.01 | 2.81 | 1.28 × 10−08 |
| Hot 43 °C (4573 DEGs, 3885 uniquely mapped) | |||||
| negative regulation of cellular amide metabolic process (GO:0034249) | 99 | 47 | 19.45 | 2.42 | 1.27 × 10−02 |
| negative regulation of translation (GO:0017148) | 95 | 45 | 18.67 | 2.41 | 2.43 × 10−02 |
| translation (GO:0006412) | 290 | 133 | 56.98 | 2.33 | 6.42 × 10−11 |
| peptide biosynthetic process (GO:0043043) | 298 | 133 | 58.55 | 2.27 | 3.35 × 10−10 |
| peptide metabolic process (GO:0006518) | 377 | 156 | 74.07 | 2.11 | 3.04 × 10−10 |
| amide biosynthetic process (GO:0043604) | 374 | 147 | 73.48 | 2.00 | 3.76 × 10−08 |
| regulation of translation (GO:0006417) | 198 | 76 | 38.90 | 1.95 | 1.08 × 10−02 |
| positive regulation of catabolic process (GO:0009896) | 197 | 74 | 38.71 | 1.91 | 2.34 × 10−02 |
| regulation of cellular amide metabolic process (GO:0034248) | 213 | 80 | 41.85 | 1.91 | 1.08 × 10−02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reed, K.M.; Mendoza, K.M.; Xu, J.; Strasburg, G.M.; Velleman, S.G. Transcriptome Response of Differentiating Muscle Satellite Cells to Thermal Challenge in Commercial Turkey. Genes 2022, 13, 1857. https://doi.org/10.3390/genes13101857
Reed KM, Mendoza KM, Xu J, Strasburg GM, Velleman SG. Transcriptome Response of Differentiating Muscle Satellite Cells to Thermal Challenge in Commercial Turkey. Genes. 2022; 13(10):1857. https://doi.org/10.3390/genes13101857
Chicago/Turabian StyleReed, Kent M., Kristelle M. Mendoza, Jiahui Xu, Gale M. Strasburg, and Sandra G. Velleman. 2022. "Transcriptome Response of Differentiating Muscle Satellite Cells to Thermal Challenge in Commercial Turkey" Genes 13, no. 10: 1857. https://doi.org/10.3390/genes13101857





