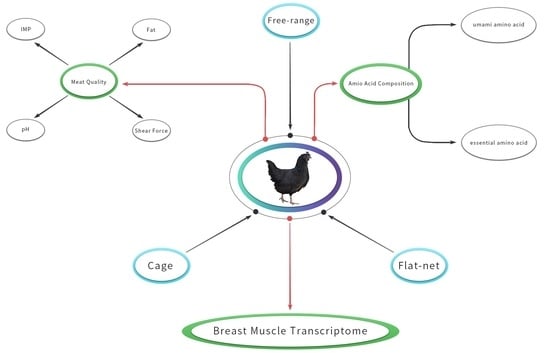
Effects of Different Rearing Systems on Lueyang Black-Bone Chickens: Meat Quality, Amino Acid Composition, and Breast Muscle Transcriptome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Approval Statements
2.2. Animals, Diets and Experimental Design
2.3. Measurement of Meat Quality Traits
2.4. Total RNA Extraction, cDNA Library Preparation, and Sequencing
2.5. Bioinformatic Analyses of DEGs
2.6. Real-Time Quantitative PCR (RT-qPCR) Verification
2.7. Statistical Analyses
3. Results
3.1. Meat Quality Parameters
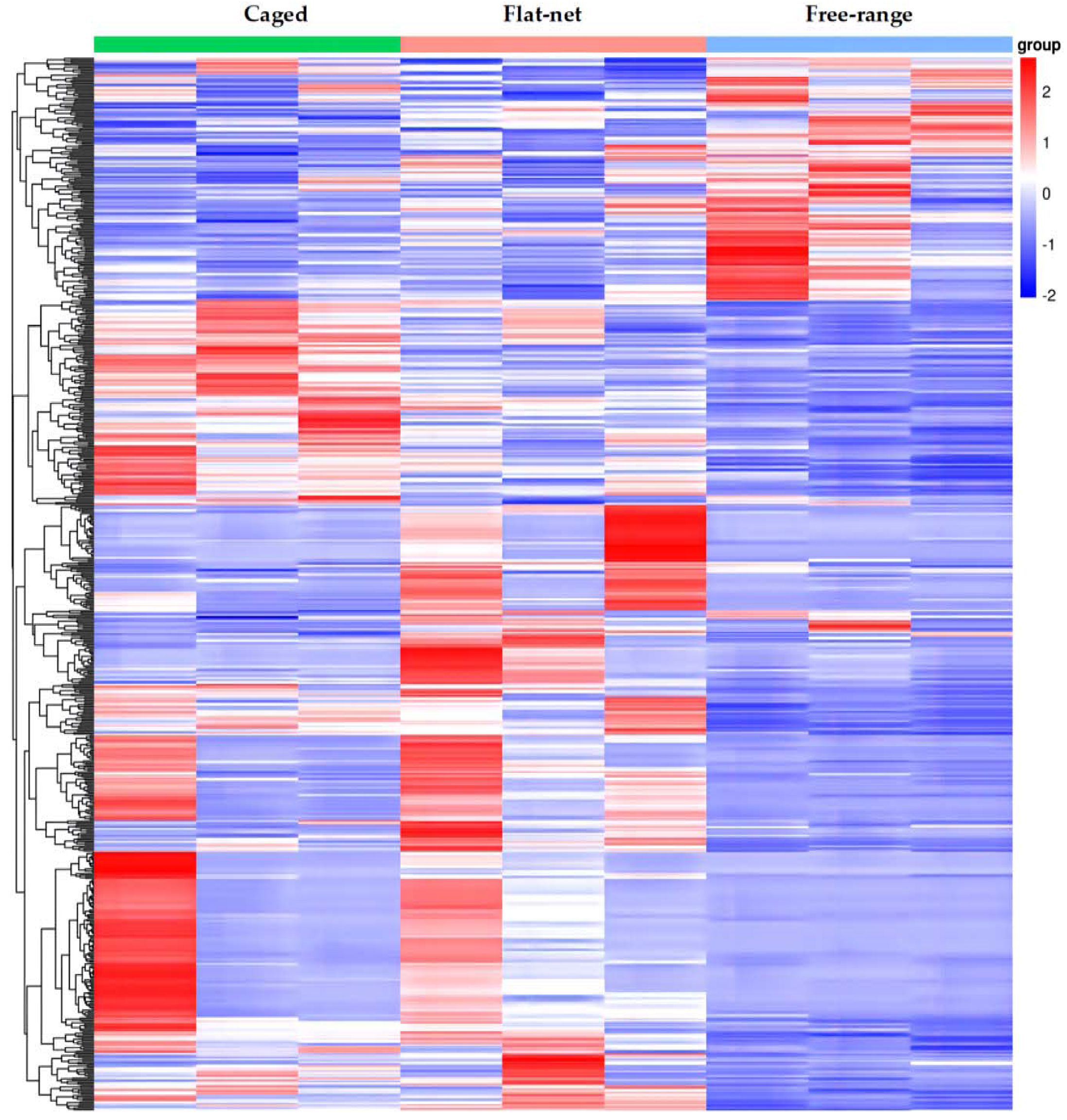
3.2. Sequencing of mRNA in Chicken Breast Muscle
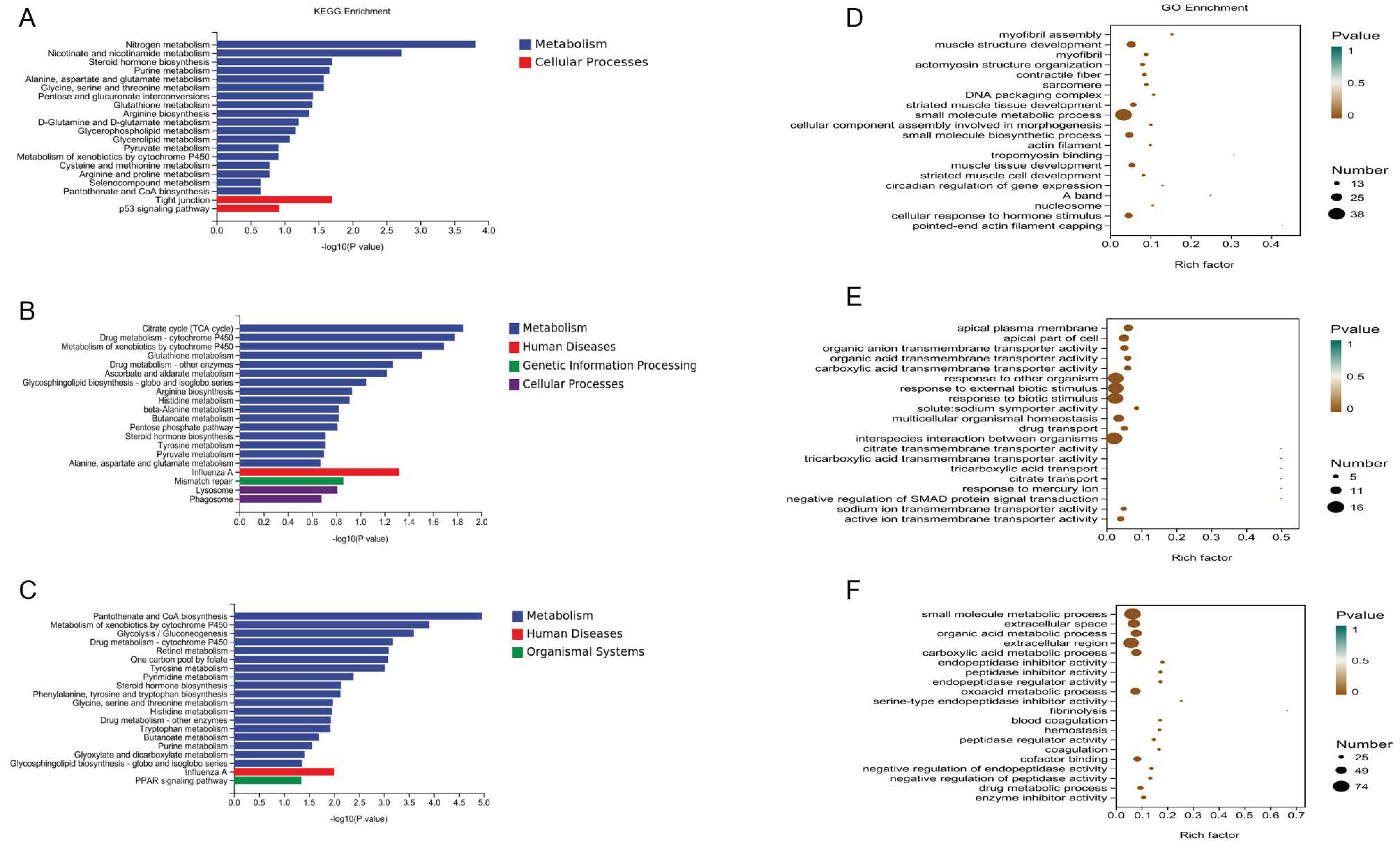
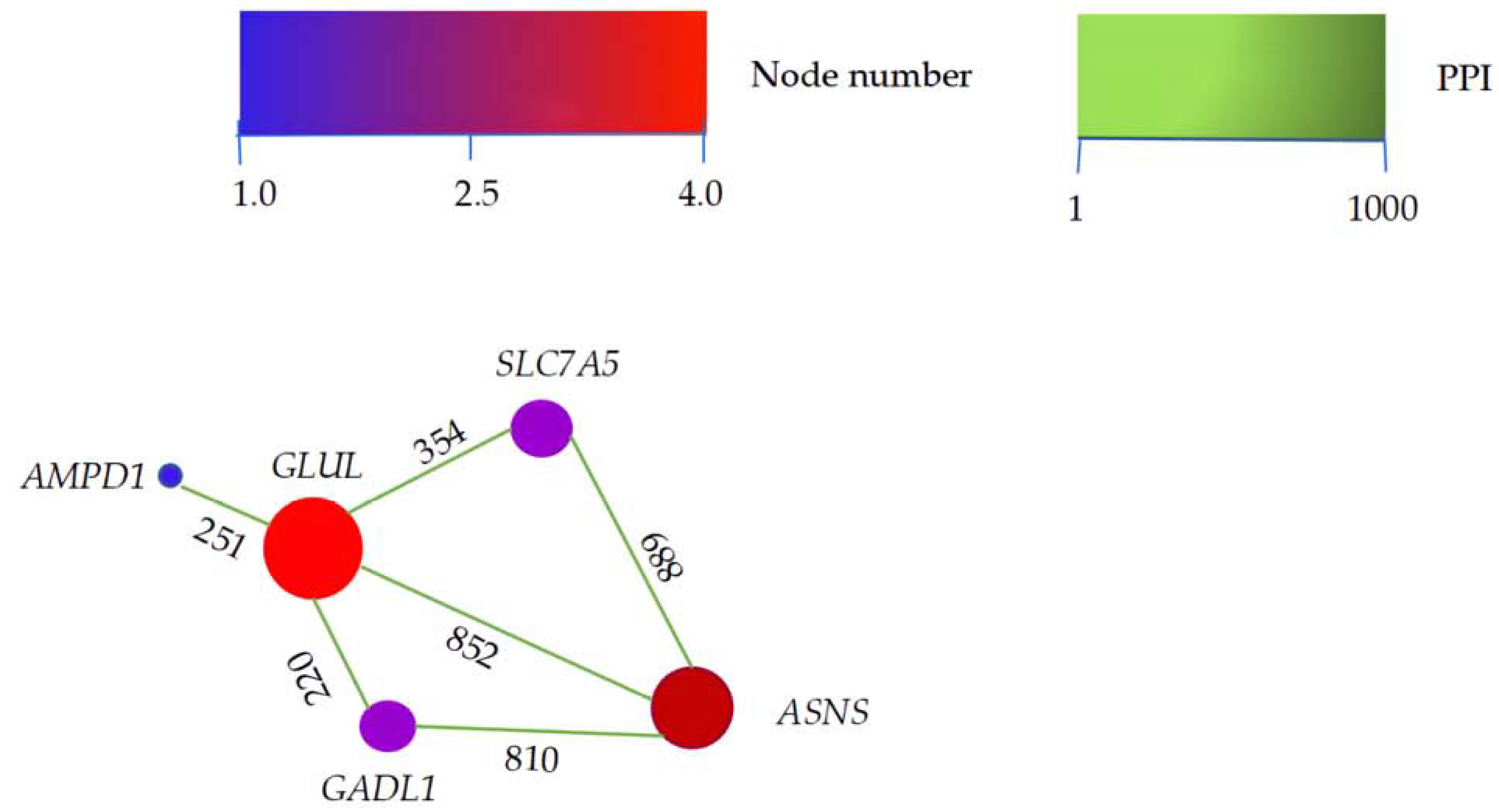
3.3. Enrichment and Interaction Analyses of DEGs
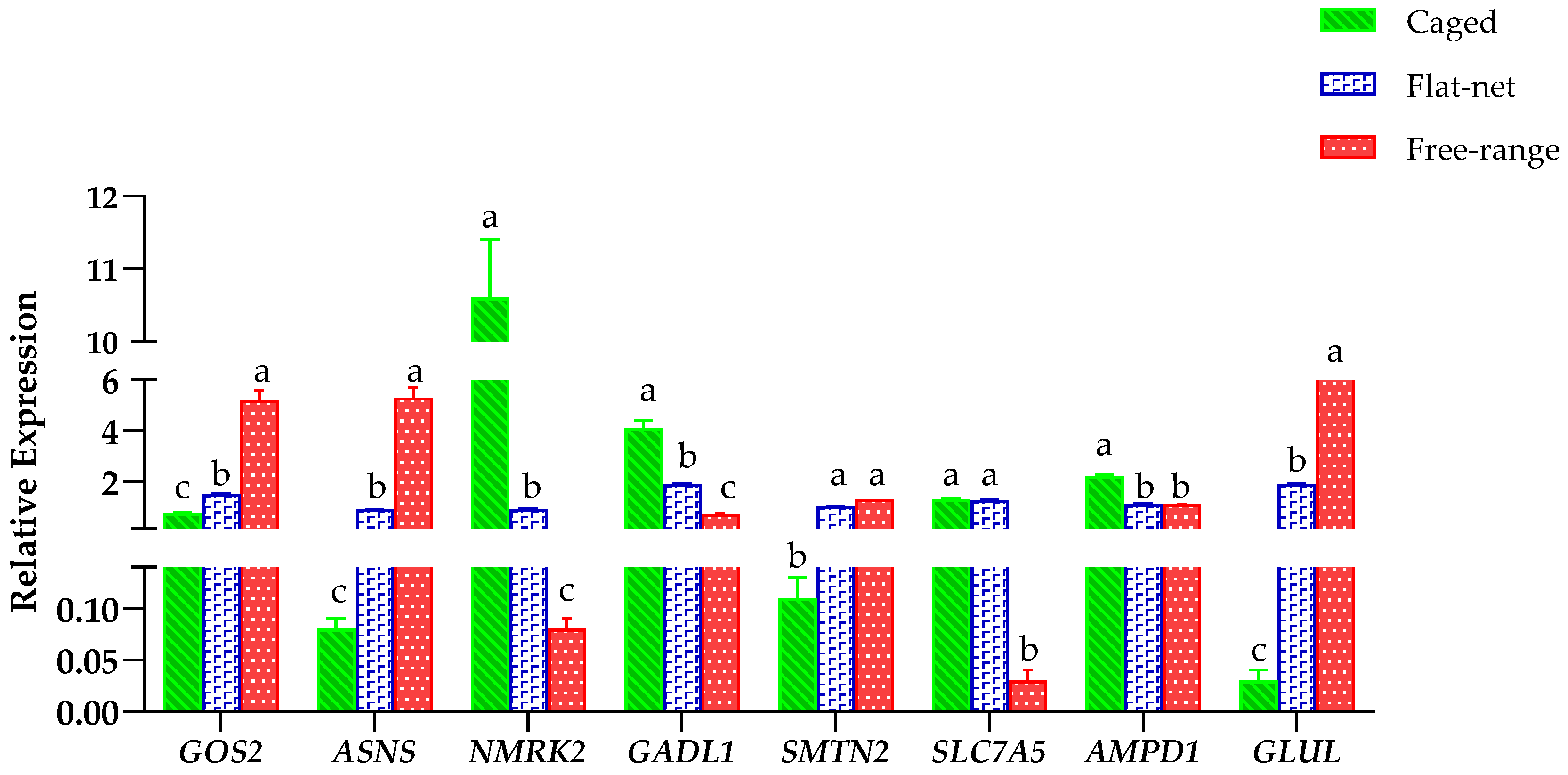
3.4. RT-qPCR Verification of Candidate Genes
4. Discussion
4.1. Meat Quality Parameters
4.2. Regulatory Pathways Affecting Muscle Development and Flavor
4.3. DEGs on Muscle Development and Flavor
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, T.; Liu, H.; Yang, L.-K.; Yin, Y.-J.; Lu, H.-Z.; Wang, L. The complete mitochondrial genome and molecular phylogeny of Lueyang black-bone chicken. Br. Poult. Sci. 2018, 59, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Wang, L.; Wang, S.; Chen, R.; Zhang, T.; Ma, H.; Lu, H.; Yuan, G. Transcriptomic analysis of thigh muscle of Lueyang black-bone chicken in free-range and caged feeding. Anim. Biotechnol. 2021, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dang, L.-P.; Liu, R.-F.; Zhao, W.-Y.; Zhou, W.-X.; Min, Y.-N.; Wang, Z.-P. Investigating structural impact of a valine to isoleucine substitution on anti-Müllerian hormone in silico and genetic association of the variant and AMH expression with egg production in chickens. J. Integr. Agric. 2020, 19, 1635–1643. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, F.; Cao, J.; Liu, X. Skin Transcriptome Profiles Associated with Skin Color in Chickens. PLoS ONE 2015, 10, e0127301. [Google Scholar] [CrossRef]
- Bai, H.; Yang, B.; Dong, Z.; Li, X.; Song, Q.; Jiang, Y.; Chang, G.; Chen, G. Research Note: Effects of cage and floor rearing systems on growth performance, carcass traits, and meat quality in small-sized meat ducks. Poult. Sci. 2021, 101, 101520. [Google Scholar] [CrossRef]
- Semwogerere, F.; Neethling, J.; Muchenje, V.; Hoffman, L. Effects of production systems on the carcass and meat quality characteristics of spent laying hens. Poult. Sci. 2018, 97, 1990–1997. [Google Scholar] [CrossRef]
- Vanhonacker, F.; Verbeke, W. Public and Consumer Policies for Higher Welfare Food Products: Challenges and Opportunities. J. Agric. Environ. Ethic 2013, 27, 153–171. [Google Scholar] [CrossRef]
- Wang, K.H.; Shi, S.R.; Dou, T.C.; Sun, H.J. Effect of a free-range raising system on growth performance, carcass yield, and meat quality of slow-growing chicken. Poult. Sci. 2009, 88, 2219–2223. [Google Scholar] [CrossRef]
- Lefaucheur, L.; Lebret, B. The rearing system modulates biochemical and histological differences in loin and ham muscles between Basque and Large White pigs. Animal 2020, 14, 1976–1986. [Google Scholar] [CrossRef]
- Bogosavljević-Bošković, S.; Rakonjac, S.; Dosković, V.; Petrovic, M. Broiler rearing systems: A review of major fattening results and meat quality traits. World′s Poult. Sci. J. 2012, 68, 217–228. [Google Scholar] [CrossRef]
- Huo, W.; Weng, K.; Gu, T.; Luo, X.; Zhang, Y.; Zhang, Y.; Xu, Q.; Chen, G. Effects of integrated rice-duck farming system on duck carcass traits, meat quality, amino acid, and fatty acid composition. Poult. Sci. 2021, 100, 101107. [Google Scholar] [CrossRef] [PubMed]
- San, J.; Du, Y.; Wu, G.; Xu, R.; Yang, J.; Hu, J. Transcriptome analysis identifies signaling pathways related to meat quality in broiler chickens–the extracellular matrix (ECM) receptor interaction signaling pathway. Poult. Sci. 2021, 100, 101135. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, X.; Chen, D.; Yu, B.; Yin, J.; Huang, Z. Effects of dietary apple polyphenol supplementation on carcass traits, meat quality, muscle amino acid and fatty acid composition in finishing pigs. Food Funct. 2019, 10, 7426–7434. [Google Scholar] [CrossRef] [PubMed]
- Demirel, G.; Wachira, A.M.; Sinclair, L.A.; Wilkinson, R.G.; Wood, J.D.; Enser, M. Effects of dietary n-3 polyunsaturated fatty acids, breed and dietary vitamin E on the fatty acids of lamb muscle, liver and adipose tissue. Br. J. Nutr. 2004, 91, 551–565. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Lu, H.; Wang, L.; Yin, M.; Yang, L. Specific expression pattern of IMP metabolism related-genes in chicken muscle between cage and free-range conditions. PLoS ONE 2018, 13, e0201736. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Tong, H.B.; Wang, Q.; Lu, J.; Zou, J.M.; Chang, L.L.; Fu, S.Y. Effect of free-range days on a local chicken breed: Growth performance, carcass yield, meat quality, and lymphoid organ index. Poult. Sci. 2014, 93, 1883–1889. [Google Scholar] [CrossRef]
- Husak, R.L.; Sebranek, J.G.; Bregendahl, K. Processing, Products, and Food Safety A Survey of Commercially Available Broilers Marketed as Organic, Free-Range, and Conventional Broilers for Cooked Meat Yields, Meat Composition, and Relative Value. Poult. Sci. 2008, 87, 2367–2376. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Jiang, Z.; Lin, Y.; Zhou, G.; Chen, F.; Zheng, C. Effects of different rearing and feeding methods on meat quality and antioxidative properties in Chinese Yellow male broilers. Br. Poult. Sci. 2011, 52, 352–358. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.; Zhang, S.; Xie, J.; Shan, T. The Effect of Rearing Conditions on Carcass Traits, Meat Quality and the Compositions of Fatty Acid and Amino Acid of LTL in Heigai Pigs. Animals 2021, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jiang, W.; Tan, H.Z.; Xu, G.F.; Zhang, X.B.; Wei, S.; Wang, X.Q. Effects of outdoor access on growth performance, carcass composition, and meat characteristics of broiler chickens. Poult. Sci. 2013, 92, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Zhang, D.; Xu, G.; Li, K.; Wang, Q.; Zhang, Z.; Li, J.; Chen, Y.; Jia, Y.; Qu, L. Effects of different rearing systems on meat production traits and meat fiber microstructure of Beijing-you chicken. Anim. Sci. J. 2014, 86, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Q.; Lou, L.; Liu, J.; Chen, X.; Zhang, C.; Wang, H. High-resolution melting curve analysis of the ADSL and LPL genes and their correlation with meat quality and blood parameters in chickens. Genet. Mol. Res. 2015, 14, 2031–2040. [Google Scholar] [CrossRef]
- Hoa, V.B.; Cho, S.-H.; Seong, P.-N.; Kang, S.M.; Kim, Y.-S.; Moon, S.-S.; Choi, Y.-M.; Kim, J.-H.; Seol, K.-H. Quality characteristics, fatty acid profiles, flavor compounds and eating quality of cull sow meat in comparison with commercial pork. Asian-Australas. J. Anim. Sci. 2020, 33, 640–650. [Google Scholar] [CrossRef] [Green Version]
- Kouba, M.; Benatmane, F.; Blochet, J.; Mourot, J. Effect of a linseed diet on lipid oxidation, fatty acid composition of muscle, perirenal fat, and raw and cooked rabbit meat. Meat Sci. 2008, 80, 829–834. [Google Scholar] [CrossRef]
- Zhao, C.J.; Schieber, A.; Gänzle, M.G. Formation of taste-active amino acids, amino acid derivatives and peptides in food fermentations—A review. Food Res. Int. 2016, 89, 39–47. [Google Scholar] [CrossRef]
- Walker, M.C.; A van der Donk, W. The many roles of glutamate in metabolism. J. Ind. Microbiol. Biotechnol. 2016, 43, 419–430. [Google Scholar] [CrossRef]
- Lorenzo, J.M.; Franco, D. Fat effect on physico-chemical, microbial and textural changes through the manufactured of dry-cured foal sausage Lipolysis, proteolysis and sensory properties. Meat Sci. 2012, 92, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Wang, Y.; Long, K.; Jiang, A.; Jin, L. LncRNA Regulatory Mechanism for LncRNAs in Skeletal Muscle Development and Progress on Its Research in Domestic Animals. Yi Chuan 2018, 40, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Chal, J.; Pourquié, O. Making muscle: Skeletal myogenesis in vivo and in vitro. Development 2017, 144, 2104–2122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaumatin, F.; O’prey, J.; Barthet, V.J.A.; Zunino, B.; Parvy, J.-P.; Bachmann, A.M.; O’Prey, M.; Kania, Z.; Sierra Gonzalez, P.; Macintosh, R.; et al. Supplemental Information MTORC1 Activation Requires DRAM-1 by Facilitating Lysosomal Amino Acid Efflux. Mol. Cell 2019, 76, 163–176.e8. [Google Scholar] [CrossRef] [Green Version]
- Mahmassani, Z.S.; Mckenzie, A.I.; Petrocelli, J.J.; de Hart, N.M.; Fix, D.K.; Kelly, J.J.; Baird, L.M.; Howard, M.T.; Drummond, M.J. Reduced Physical Activity Alters the Leucine-Stimulated Translatome in Aged Skeletal Muscle. J. Gerontol. A Biol. Sci. Med. Sci. 2021, 76, 2112–2121. [Google Scholar] [CrossRef]
- Deloux, R.; Tannous, C.; Ferry, A.; Li, Z.; Mericskay, M.; di Felice, V. Aged Nicotinamide Riboside Kinase 2 Deficient Mice Present an Altered Response to Endurance Exercise Training. Front. Physiol. 2018, 9, 1290. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Ge, X.; Ding, H.; Jiang, H.; Christensen, B.M.; Li, J. Role of Glutamate Decarboxylase-like Protein 1 (GADL1) in Taurine Biosynthesis*. J. Biol. Chem. 2012, 287, 40898–40906. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Zhang, X.; Yue, B.; Ge, W.; Zhang, M.; Ma, C.; Kong, M. Effects of pantothenic acid on growth performance, slaughter performance, lipid metabolism, and antioxidant function of Wulong geese aged one to four weeks. Anim. Nutr. 2016, 2, 312–317. [Google Scholar] [CrossRef]
- Li, Y.; Wang, M.; Li, Q.; Gao, Y.; Li, Q.; Li, J.; Cao, Y. Transcriptome profiling of longissimus lumborum in Holstein bulls and steers with different beef qualities. PLoS ONE 2020, 15, e0235218. [Google Scholar] [CrossRef]
- Hu, J.; Yu, P.; Ding, X.; Xu, M.; Guo, B.; Xu, Y. NU SC College of Animal Science and Technology, Nanjing Agricultural University, Nanjing. Gene 2015, 574, 204–209. [Google Scholar] [CrossRef]
- Páez-Pérez, E.D.; Llamas-García, M.L.; Benítez-Cardoza, C.G.; Montero-Morán, G.M.; Lara-González, S. Bioinformatic Analysis and Biophysical Characterization Reveal Structural Disorder in G0S2 Protein. ACS Omega 2020, 5, 25841–25847. [Google Scholar] [CrossRef] [PubMed]
- Rha, G.B.; Chi, Y.; Guerin, T.M.; Smart, E.J.; Yang, X.; Lu, X.; Lombe, M. The G 0 / G 1 Switch Gene 2 Regulates Adipose Lipolysis through Association with Adipose Triglyceride Lipase. Cell Metab. 2010, 11, 194–205. [Google Scholar] [CrossRef] [Green Version]
- Lomelino, C.L.; Andring, J.T.; McKenna, R.; Kilberg, M.S. Asparagine synthetase: Function, structure, and role in disease. J. Biol. Chem. 2017, 292, 19952–19958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keogh, K.; Kenny, D.A.; Cormican, P.; Kelly, A.K.; Waters, S.M. Effect of dietary restriction and subsequent re-alimentation on the transcriptional profile of hepatic tissue in cattle. BMC Genom. 2016, 17, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Mullins, Y.; Keogh, K.; Blackshields, G.; Kenny, D.A.; Kelly, A.K.; Waters, S.M. Transcriptome assisted label free proteomics of hepatic tissue in response to both dietary restriction and compensatory growth in cattle. J. Proteom. 2020, 232, 104048. [Google Scholar] [CrossRef] [PubMed]
- Murali, M.; MacDonald, J.A. Smoothelins and the Control of Muscle Contractility. Adv. Pharmacol. 2018, 81, 39–78. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Huang, H.; Vandekeere, S.; Kalucka, J.; Lange, C.; Wyns, S.; Souffreau, J.; Schoonjans, L.; Lamers, W.H.; Wu, Y.; et al. Role of Glutamine Synthetase in Angiogenesis beyond Glutamine Synthesis. Nature 2018, 561, 63–69. [Google Scholar]
| Gene | Accession Number | Primers | Sequence (5′-3′) | Product Length (bp) |
|---|---|---|---|---|
| β-actin | NM_L08165.1 | F | TGTGATGGACTCTGGTGATGGT | 164 |
| R | TCTCGGCTGTGGTGGTGAAG | |||
| COS2 | XM_015299248.3 | F | CGTTCTTCGGCGTGGTCAT | 155 |
| R | CGACTTCTTGCTCTGCTCCA | |||
| ASNS | XM_015281395.3 | F | AAGGTGCTGACTGATGATGGA | 270 |
| R | CGGATGTTGCTCTTCACTGTT | |||
| NMRK2 | XM_015299836.3 | F | CTCTTCGACCTCCGCTACTT | 154 |
| R | CGCTGTCCTCCATCTCCTT | |||
| GADL1 | XM_025148022.2 | F | TGGCTAGATACCTTGTGGAAGA | 244 |
| R | ACCTGGCGGAAGAAGTTGA | |||
| SMTNL2 | XM_040650160.1 | F | CTCTGTCACTAAGCCTGTCTCT | 157 |
| R | CACCAGTTCTCGCCTCCTT | |||
| SLC7A5 | NM_001030579.2 | F | TGCTCTACGCCTTCTCCAAT | 104 |
| R | ACGCAGCCACATCATACCA | |||
| AMPD1 | XM_004935004.4 | F | AGCCTCGCCTCTCAATCTATG | 280 |
| R | GATTCATCATCCACGCTGTCAA | |||
| GLUL | NM_205493.1 | F | CGTGCCTGTATGCTGGTGTGAA | 274 |
| R | GCCTCCTCGATGTGCTTGAGAC |
| Parameters | Caged | Flat-Net | Free-Range | SEM | p-Value | ||
|---|---|---|---|---|---|---|---|
| Caged vs. Flat-Net | Caged vs. Free-Rang | Flat-Net vs. Free-Range | |||||
| pH | 5.31 c | 5.52 b | 5.67 a | 0.17 | 0.007 | 0.000 | 0.016 |
| Shear force (kg) | 18.98 b | 21.97 b | 30.68 a | 5.75 | 0.146 | 0.000 | 0.001 |
| Crude protein (g/100 g) | 26.52 a | 25.68 b | 26.00 ab | 0.16 | 0.036 | 0.173 | 0.394 |
| Fat (g/100 g) | 0.58 a | 0.43 b | 0.33 c | 0.06 | 0.036 | 0.004 | 0.041 |
| IMP (mg/g) | 1.92 b | 2.29 a | 2.40 a | 0.06 | 0.001 | 0.000 | 0.228 |
| Palmitic acid (%) # | 25.88 b | 25.27 b | 27.87 a | 0.39 | 0.418 | 0.017 | 0.003 |
| Stearic acid (%) # | 11.70 | 12.57 | 11.96 | 0.30 | 0.413 | 0.811 | 0.539 |
| Myristic acid (%) # | 1.12 | 1.53 | 1.23 | 0.17 | 0.346 | 0.804 | 0.482 |
| Oleic acid (%) * | 40.30 bc | 42.57 a | 38.93 c | 0.51 | 0.031 | 0.171 | 0.002 |
| Linoleic acid (%) * | 19.13 b | 18.39 b | 21.72 a | 0.53 | 0.536 | 0.038 | 0.009 |
| Palmitoleic acid (%) * | 2.97 ab | 3.60 a | 2.58 b | 0.18 | 0.130 | 0.342 | 0.021 |
| Itemsmg/g | Caged | Flat-Net | Free-Rang | SEM | p-Value | ||
|---|---|---|---|---|---|---|---|
| Caged vs. Flat-Net | Caged vs. Free-Rang | Flat-Net vs. Free-Range | |||||
| Essential amino acid | |||||||
| Threonine (Thr) | 1.08 a | 1.09 a | 1.04 b | 0.007 | 0.399 | 0.005 | 0.001 |
| Valine (Val) | 1.21 a | 1.20 a | 1.09 b | 0.067 | 0.631 | 0.000 | 0.000 |
| Methionine (Met) | 0.72 a | 0.73 a | 0.68 b | 0.007 | 0.745 | 0.000 | 0.000 |
| Isoleucine (Ile) | 1.16 a | 1.15 a | 1.05 b | 0.015 | 0.726 | 0.000 | 0.001 |
| Leucine (Leu) | 1.99 a | 1.99 a | 1.85 b | 0.079 | 0.902 | 0.000 | 0.000 |
| Phenylalanine (Phe) | 0.96 | 0.94 | 0.94 | 0.006 | 0.265 | 0.092 | 0.530 |
| Lysine (Lys) | 2.18 a | 2.18 a | 2.05 b | 0.019 | 0.957 | 0.001 | 0.001 |
| Total essential amino acid | 9.29 a | 9.27 a | 8.70 b | 0.083 | 0.875 | 0.000 | 0.000 |
| Dispensable amino acid | |||||||
| Aspartic (Asp) # | 2.35 a | 2.36 a | 2.25 b | 0.016 | 0.656 | 0.001 | 0.001 |
| Serine (Ser) | 0.95 ab | 0.96 a | 0.93 b | 0.007 | 0.470 | 0.067 | 0.016 |
| Glutamate (Glu) # | 3.65 b | 3.73 ab | 3.75 a | 0.010 | 0.152 | 0.046 | 0.717 |
| Glycine (Gly) | 1.11 | 1.10 | 1.07 | 0.013 | 0.874 | 0.235 | 0.299 |
| Alanine (Ala) | 1.46 a | 1.46 a | 1.35 b | 0.013 | 0.941 | 0.000 | 0.000 |
| Tyrosine (Tyr) | 0.87 a | 0.88 a | 0.85 b | 0.006 | 0.793 | 0.049 | 0.012 |
| Histidine (His) | 0.81 | 0.81 | 0.82 | 0.011 | 0.781 | 0.824 | 0.618 |
| Arginine (Arg) | 1.62 | 1.61 | 1.58 | 0.010 | 0.667 | 0.113 | 0.233 |
| Proline (Pro) | 0.76 ab | 078 a | 0.73 b | 0.009 | 0.395 | 0.222 | 0.048 |
| Cystine (Cys) | 0.22 | 0.22 | 0.22 | 0.003 | 0.396 | 0.830 | 0.292 |
| Total dispensable amino acid | 13.89 | 13.70 | 13.43 | 0.102 | 0.466 | 0.073 | 0.256 |
| Total umami amino acid | 15.35 a | 15.17 ab | 14.66 b | 0.119 | 0.489 | 0.016 | 0.063 |
| Total amino acid | 23.20 a | 23.18 a | 22.13 b | 0.162 | 0.954 | 0.002 | 0.002 |
| Gene | Flat-Net vs. Caged | Flat-Net vs. Free-Range | Caged vs. Free-Range | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Differentiate | Log2 FC | p | Differentiate | Log2 FC | p | Differentiate | Log2 FC | p | |
| GOS2 | Up | 1.01 | * | Down | −1.38 | * | Down | −2.38 | * |
| GADL1 | Down | −1.24 | * | Up | 2.32 | * | Up | 3.56 | * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Zhang, J.; Cao, C.; Cai, Y.; Li, Y.; Song, Y.; Bao, X.; Zhang, J. Effects of Different Rearing Systems on Lueyang Black-Bone Chickens: Meat Quality, Amino Acid Composition, and Breast Muscle Transcriptome. Genes 2022, 13, 1898. https://doi.org/10.3390/genes13101898
Zhang S, Zhang J, Cao C, Cai Y, Li Y, Song Y, Bao X, Zhang J. Effects of Different Rearing Systems on Lueyang Black-Bone Chickens: Meat Quality, Amino Acid Composition, and Breast Muscle Transcriptome. Genes. 2022; 13(10):1898. https://doi.org/10.3390/genes13101898
Chicago/Turabian StyleZhang, Shuya, Jiqiao Zhang, Chang Cao, Yingjie Cai, Yuxiao Li, Yiping Song, Xiuyu Bao, and Jianqin Zhang. 2022. "Effects of Different Rearing Systems on Lueyang Black-Bone Chickens: Meat Quality, Amino Acid Composition, and Breast Muscle Transcriptome" Genes 13, no. 10: 1898. https://doi.org/10.3390/genes13101898