A Single Nucleotide Polymorphism Translates into a Radical Amino Acid Substitution at the Ligand-Binding Site in Fasciola hepatica Carboxylesterase B
Abstract
:1. Introduction
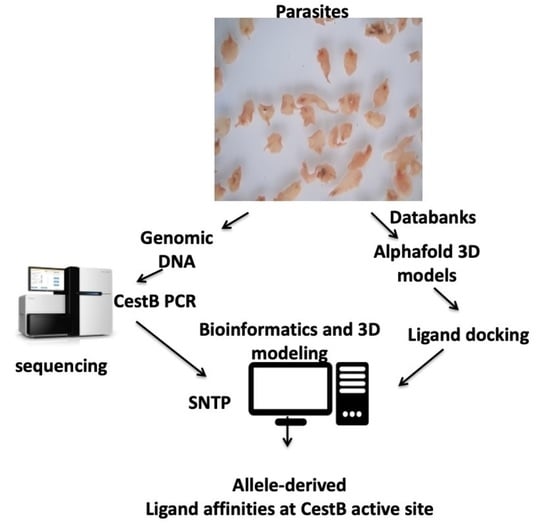
2. Materials and Methods
2.1. Protein Sequences
2.2. Protein 3D Modeling
2.3. Parasite Material
2.4. DNA Extraction
2.5. PCR Conditions
2.6. Sanger Amplicon Sequencing
3. Results
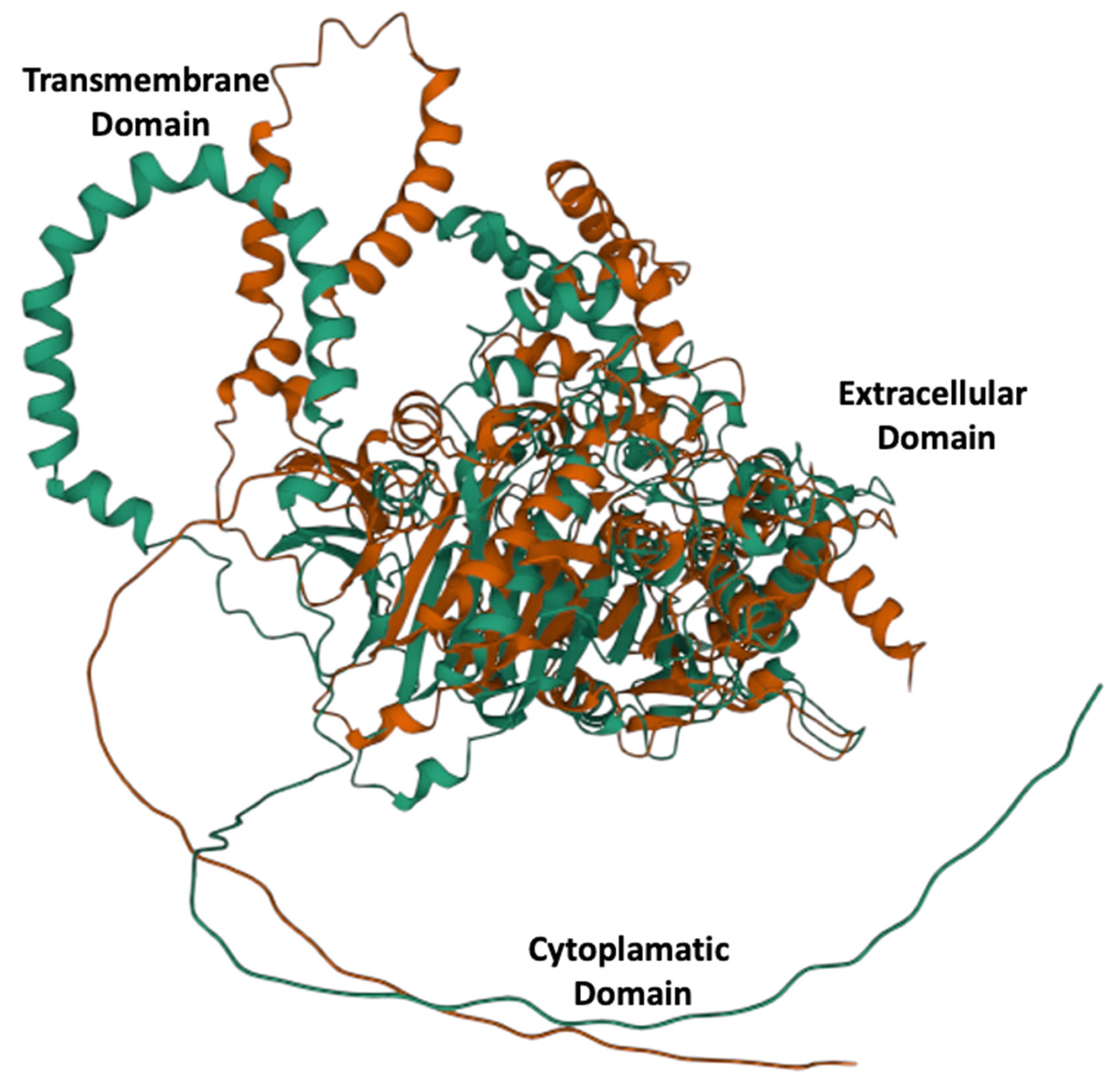
3.1. Protein Sequence Properties
3.2. Protein 3D Modeling
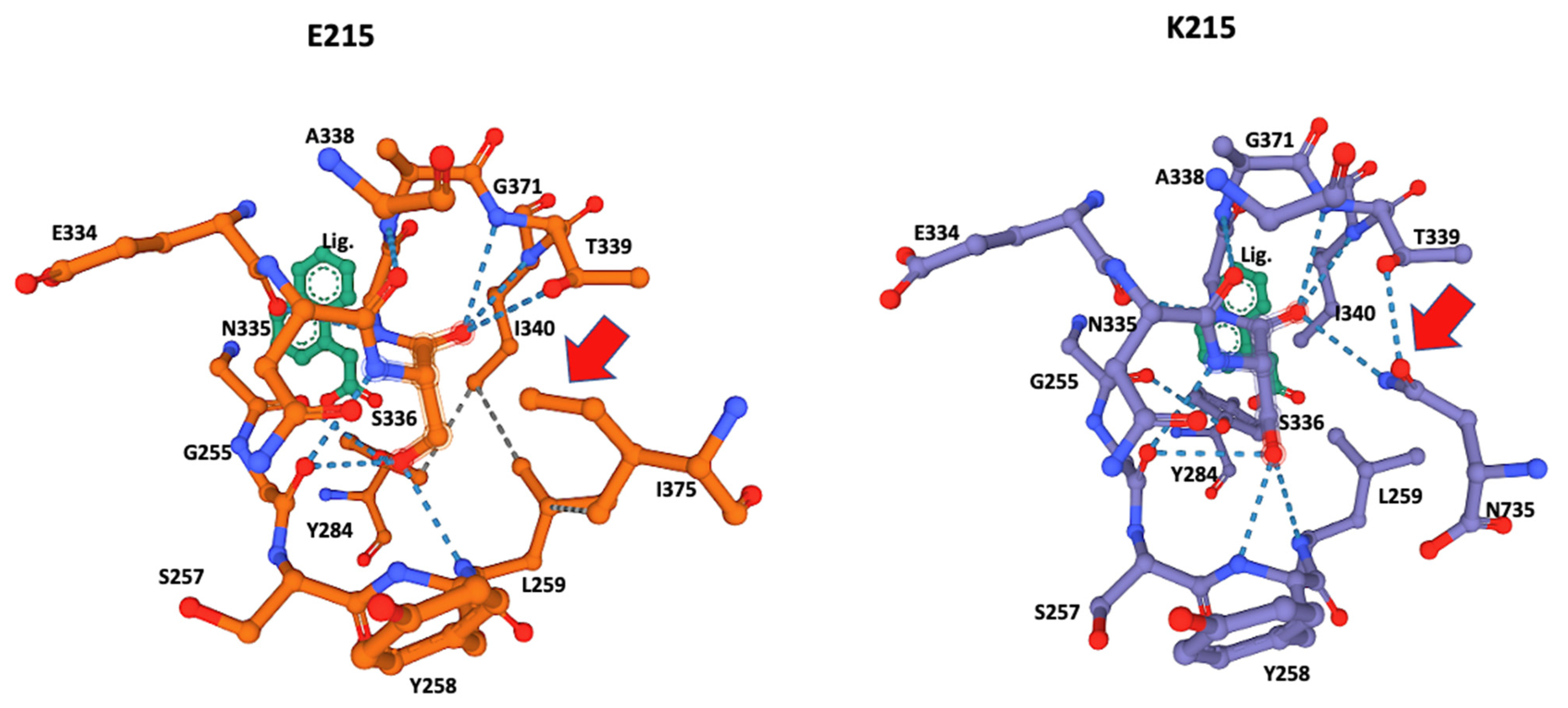
3.3. Ligand-Binding Modeling
3.4. Ligand Docking Visualization
3.5. PCR Amplicon Sequencing
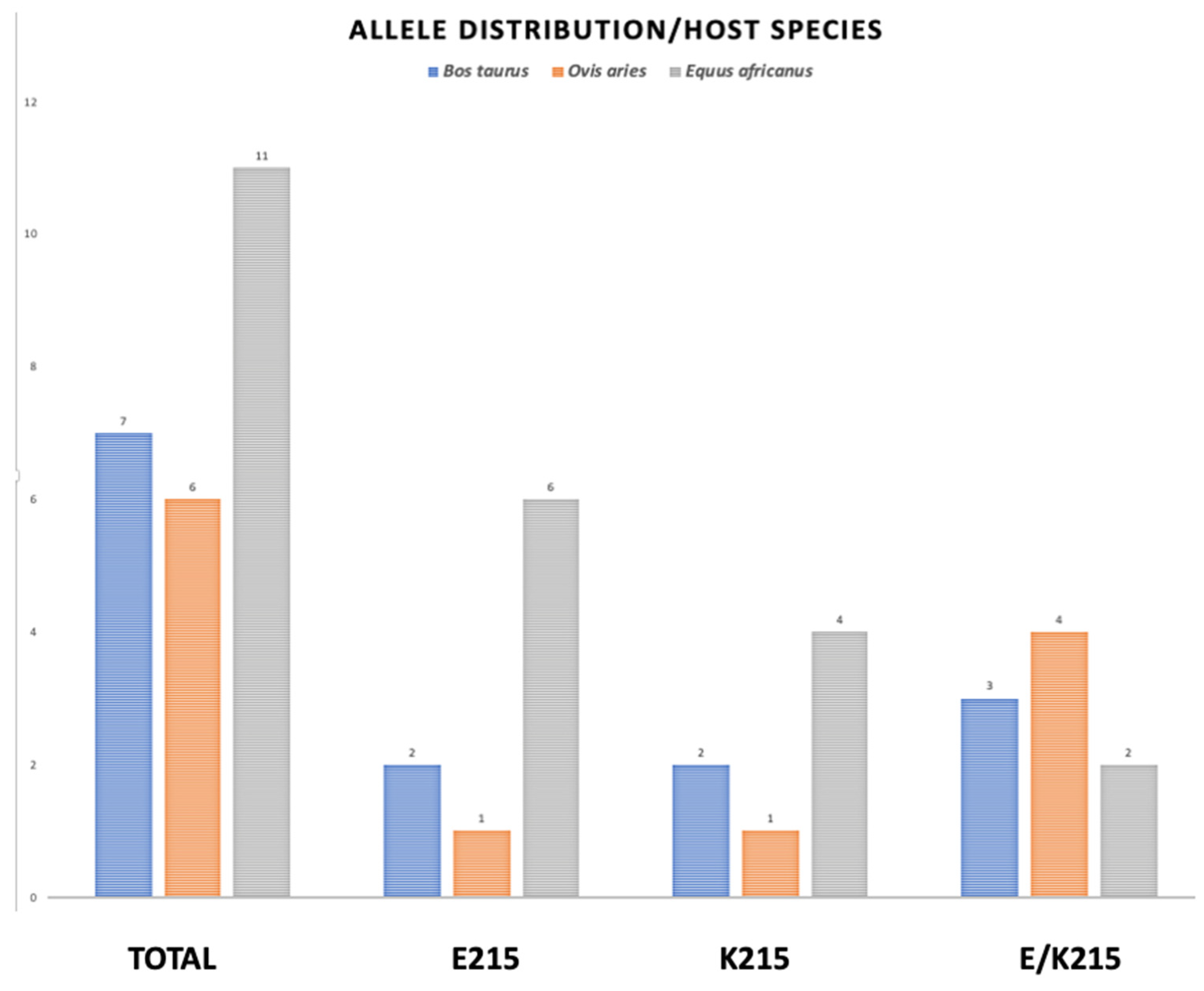
3.6. Allele Distribution by Species
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Miranda-Miranda, E.; Cossio-Bayugar, R.; Aguilar-Díaz, H.; Narváez-Padilla, V.; Sachman-Ruíz, B.; Reynaud, E. Transcriptome Assembly Dataset of Anthelmintic Response in Fasciola Hepatica. Data Brief. 2021, 35, 106808. [Google Scholar] [CrossRef] [PubMed]
- Scarcella, S.; Miranda-Miranda, E.; Cossío-Bayúgar, R.; Ceballos, L.; Fernandez, V.; Solana, H. Increase of Carboxylesterase Activity in Fasciola Hepatica Recovered from Triclabendazole Treated Sheep. Mol. Biochem. Parasitol. 2012, 185, 151–153. [Google Scholar] [CrossRef] [PubMed]
- Pedroza-Gómez, Y.J.; Cossio-Bayugar, R.; Aguilar-Díaz, H.; Scarcella, S.; Reynaud, E.; Sanchez-Carbente, M.D.R.; Narváez-Padilla, V.; Miranda-Miranda, E. Transcriptome-Based Identification of a Functional Fasciola Hepatica Carboxylesterase B. Pathogens 2021, 10, 1454. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The Selection and Use of Essential Medicines: Report of the WHO Expert Committee, 2013 (Including the 18th WHO Model List of Essential Medicines and the 4th WHO Model List of Essential Medicines for Children); WHO Technical Report Series: No. 985; World Health Organization: Geneva, Switzerland, 2014; ISBN 978-92-4-120985-4. [Google Scholar]
- Fissiha, W.; Kinde, M.Z. Anthelmintic Resistance and Its Mechanism: A Review. IDR 2021, 14, 5403–5410. [Google Scholar] [CrossRef] [PubMed]
- Matoušková, P.; Vokřál, I.; Lamka, J.; Skálová, L. The Role of Xenobiotic-Metabolizing Enzymes in Anthelmintic Deactivation and Resistance in Helminths. Trends Parasitol. 2016, 32, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, A. Biotranformation of Xenobiotics. In Cassarett and Doulls Toxicology: The Basic Science of Poisons; McGraw-Hill Medical: New York, NY, USA, 1996; pp. 113–196. [Google Scholar]
- Hosokawa, M. Structure and Catalytic Properties of Carboxylesterase Isozymes Involved in Metabolic Activation of Prodrugs. Molecules 2008, 13, 412–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Istvan, E.S.; Mallari, J.P.; Corey, V.C.; Dharia, N.V.; Marshall, G.R.; Winzeler, E.A.; Goldberg, D.E. Esterase Mutation Is a Mechanism of Resistance to Antimalarial Compounds. Nat. Commun. 2017, 8, 14240. [Google Scholar] [CrossRef] [Green Version]
- Butler, J.H.; Baptista, R.P.; Valenciano, A.L.; Zhou, B.; Kissinger, J.C.; Tumwebaze, P.K.; Rosenthal, P.J.; Cooper, R.A.; Yue, J.-M.; Cassera, M.B. Resistance to Some But Not Other Dimeric Lindenane Sesquiterpenoid Esters Is Mediated by Mutations in a Plasmodium Falciparum Esterase. ACS Infect. Dis. 2020, 6, 2994–3003. [Google Scholar] [CrossRef]
- Dahan-Moss, Y.L.; Koekemoer, L.L. Analysis of Esterase Enzyme Activity in Adults of the Major Malaria Vector Anopheles Funestus. Parasites Vectors 2016, 9, 110. [Google Scholar] [CrossRef] [Green Version]
- Shin, D.; Smartt, C.T. Assessment of Esterase Gene Expression as a Risk Marker for Insecticide Resistance in Florida Culex Nigripalpus (Diptera: Culicidae). J. Vector Ecol. 2016, 41, 63–71. [Google Scholar] [CrossRef]
- Schama, R.; Pedrini, N.; Juárez, M.P.; Nelson, D.R.; Torres, A.Q.; Valle, D.; Mesquita, R.D. Rhodnius Prolixus Supergene Families of Enzymes Potentially Associated with Insecticide Resistance. Insect Biochem. Mol. Biol. 2016, 69, 91–104. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern Web App for 3D Visualization and Analysis of Large Biomolecular Structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef]
- Wu, Q.; Peng, Z.; Zhang, Y.; Yang, J. COACH-D: Improved Protein–Ligand Binding Sites Prediction with Refined Ligand-Binding Poses through Molecular Docking. Nucleic Acids Res. 2018, 46, W438–W442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Roy, A.; Zhang, Y. Protein–Ligand Binding Site Recognition Using Complementary Binding-Specific Substructure Comparison and Sequence Profile Alignment. Bioinformatics 2013, 29, 2588–2595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvani, N.E.D.; Windsor, P.A.; Bush, R.D.; Šlapeta, J. Scrambled Eggs: A Highly Sensitive Molecular Diagnostic Workflow for Fasciola Species Specific Detection from Faecal Samples. PLoS Negl. Trop. Dis. 2017, 11, e0005931. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.F.; Rusell, D.W. (Eds.) Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; ISBN 978-0-87969-577-4. [Google Scholar]
- Scarcella, S.; Miranda-Miranda, E.; Solana, M.V.; Solana, H. Approach to Molecular Characterization of Different Strains of Fasciola Hepatica Using Random Amplified Polymorphic DNA Polymerase Chain Reaction. Parasitol. Res. 2015, 114, 1341–1345. [Google Scholar] [CrossRef]
- Boyko, K.M.; Kryukova, M.V.; Petrovskaya, L.E.; Nikolaeva, A.Y.; Korzhenevsky, D.A.; Novototskaya-Vlasova, K.A.; Rivkina, E.M.; Dolgikh, D.A.; Kirpichnikov, M.P.; Popov, V.O. Crystal Structure of PMGL2 Esterase from the Hormone-Sensitive Lipase Family with GCSAG Motif around the Catalytic Serine. PLoS ONE 2020, 15, e0226838. [Google Scholar] [CrossRef]
- Klaassen, C.D. (Ed.) Casarett & Doull’s Toxicology: The Basic Science of Poisons, 9th ed.; McGraw Hill Medical: New York, NY, USA, 2019; ISBN 978-1-259-86374-5. [Google Scholar]
- Abu-Qare, A.W.; Abou-Donia, M.B. Sarin: Health Effects, Metabolism, and Methods of Analysis. Food Chem. Toxicol. 2002, 40, 1327–1333. [Google Scholar] [CrossRef]
- Manchenko, G.P. Handbook of Detection of Enzymes on Electrophoretic Gels, 2nd ed.; CRC Press LLC: Boca Raton, FL, USA, 2003. [Google Scholar]
- Nestor, S.L.; Bancroft, J.D. Enzyme Histochemistry and Its Diagnostic Applications. In Theory and Practice of Histological Techniques; Elsevier: Amsterdam, The Netherlands, 2008; pp. 405–432. ISBN 978-0-443-10279-0. [Google Scholar]
- Heinen, T.; Secchia, S.; Reddington, J.P.; Zhao, B.; Furlong, E.E.M.; Stegle, O. ScDALI: Modeling Allelic Heterogeneity in Single Cells Reveals Context-Specific Genetic Regulation. Genome Biol. 2022, 23, 8. [Google Scholar] [CrossRef] [PubMed]
- Hickey, D.A.; Golding, G.B. The Advantage of Recombination When Selection Is Acting at Many Genetic Loci. J. Theor. Biol. 2018, 442, 123–128. [Google Scholar] [CrossRef] [PubMed]

| Cestb genbank/Uniprot Accession Numbers | MT843326/A0A8A1L7B4 |
| Genbank/Uniprot Identity to other Proteins | A0A4E0S0J7, QSS48625, THD28967, MW655750, D915_000180 |
| KEGG | K03927//carboxylesterase 2 [EC:3.1.1.1 3.1.1.84 3.1.1.56]; K07378//neuroligin; K01050//cholinesterase [EC:3.1.1.8]; K07377//neurexin |
| GO | 0016021//integral component of membrane |
| Interpro Protein Domains | AAs 141-184, 214-281 α-β hydrolase AAs 197-681 Carboxylesterase type B AAs 1-73 Cytoplasmatic Domain AAs 93-735 Extracellular Domain AAs 172-672 Carboxylesterase AAs 73-93 Transmembranal Helix AAs 74-95 Transmembranal Peptide |
| Literature Description | [2,3] |
| AA Substitution | E215 | K215 |
|---|---|---|
| Binding Site AA Prediction | K210, N211, G214, E215, L216, V217, G256, L259, Y284, S336, T339, I340 | K210, N211, G214, K215, L216, V217, G256, Y258, L259, S336, T339, I340, N735 |
| Predicted S336 Hydrogen Bonds to Adjacent AAs | G256, L259, Y284, T339, I340 | G256, L259, Y258, T339, I340, N735 |
| Predicted Hydrophobic Bonds at Catalytic Site | Y284-I340 I340-L259 L259-I375 | N735-Y258 |
| Ligand Affinity Prediction/Ligand description | Ethyl hydrogen propylamidophosphate/serine hydrolase inhibitor Naphthyl acetate/synthetic esterase substrate N-acetylneuraminic acid/neurone membrane component Cyclohexyl (s)-methylphosphonofluoridate/ww2 cyclosarin Ethyl hydrogen phosphonate/serine hydrolase inhibitor 1-(2-nitrophenyl)-2,2,2-trifluoroethyl]-arsenocholine/neurotransmiter analogous Choline/neurotransmitter precursor Edrophonium\/serine hydrolase inhibitor | Ethyl hydrogen propylamidophosphate/serine hydrolase inhibitor Naphthyl acetate/synthetic esterase substrate N-ethoxyphosphonoyl-n-methyl-methanamine/serine hydrolase inhibitor Sialic acid/neurone membrane component (4r)-4-hydroxy-n,n,n-trimethylpentan-1-aminium/cholinesterase inhibitor Butyric acid/fatty acid 1-(2-nitrophenyl)-2,2,2-trifluoroethyl]-arsenocholine/cholinesterase inhibitor Ethyl hydrogen diethylamidophosphate/serine hydrolase inhibitor Tacrine/cholinesterase inhibitor Methylphosphonic acid ester/cholinesterae inhibitor Huperzine/alkaloid 3-[(1s)-1-(dimethylamino)ethyl]phenol/cholinesterase inhibitor Galanthamine/cholinesterase inhibitor |
| Isolate Location/Coordinates | Host Species | Parasite Stage | SNP643 | AA215 |
|---|---|---|---|---|
| MEX. 19.3026° N 98.5500 W | Bos taurus 1 | Adult | A | K |
| MEX. 19.5032° N 98.5846° W | B. taurus 2 | Eggs | R | E/K |
| MEX. 18.5454° N 98.2723° W | B. taurus 3 | Adult | A | K |
| MEX. 20.0623° N 98.4548° W | Ovis aries 1 | Eggs | G | E |
| MEX. 20.0737° N 98.4012° W | O. aries 2 | Eggs | R | E/K |
| MEX. 20.1304° N 98.3459° W | O. aries 3 | Eggs | R | E/K |
| MEX. 19.6861° N 98.8116° W | B. taurus 4 | Adult | R | E/K |
| MEX. 19.4942° N 99.1220° W | O. aries 4 | Eggs | R | E/K |
| MEX. 20.0619° N 98.2333° W | O. aries 5 | Eggs | A | K |
| MEX. 18.5939° N 99.0322° W | B. taurus 5 | Eggs | R | E/K |
| MEX. 18.8892° N, 99.0626° W | B. taurus 6 | Eggs | G | E |
| MEX.18.8126° N, 98.9548° W | B. taurus 7 | Eggs | G | E |
| MEX. 18.8995° N, 99.1733° W | B. taurus 8 | Eggs | G | E |
| ARG. 33.0500° S 68.5300° W | Equus africanus 7 | Adult | G | E |
| ARG. 33.0500° S 68.5300° W | E. africanus 8 | Adult | A | K |
| ARG. 33.0500° S 68.5300° W | E. africanus 9 | Adult | G | E |
| ARG. 33.0500° S 68.5300° W | E. africanus 10 | Adult | G | E |
| ARG. 33.0500° S 68.5300° W | E. africanus 12 | Adult | G | E |
| ARG. 33.0500° S 68.5300° W | E. africanus 13 | Adult | A | K |
| ARG. 33.0500° S 68.5300° W | E. africanus 14 | Adult | G | E |
| ARG. 33.0500° S 68.5300° W | O. aries 6 | Adult | R | E/K |
| ARG. 33.0500° S 68.5300° W | E. africanus 15 | Adult | G | E |
| ARG. 33.0500° S 68.5300° W | E. africanus 17 | Adult | R | E/K |
| ARG. 33.0500° S 68.5300° W | E. africanus 19 | Adult | R | E/K |
| ARG. 33.0500° S 68.5300° W | E. africanus 21 | Adult | A | K |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miranda-Miranda, E.; Scarcella, S.; Reynaud, E.; Narváez-Padilla, V.; Neira, G.; Mera-y-Sierra, R.; Aguilar-Díaz, H.; Cossio-Bayugar, R. A Single Nucleotide Polymorphism Translates into a Radical Amino Acid Substitution at the Ligand-Binding Site in Fasciola hepatica Carboxylesterase B. Genes 2022, 13, 1899. https://doi.org/10.3390/genes13101899
Miranda-Miranda E, Scarcella S, Reynaud E, Narváez-Padilla V, Neira G, Mera-y-Sierra R, Aguilar-Díaz H, Cossio-Bayugar R. A Single Nucleotide Polymorphism Translates into a Radical Amino Acid Substitution at the Ligand-Binding Site in Fasciola hepatica Carboxylesterase B. Genes. 2022; 13(10):1899. https://doi.org/10.3390/genes13101899
Chicago/Turabian StyleMiranda-Miranda, Estefan, Silvana Scarcella, Enrique Reynaud, Verónica Narváez-Padilla, Gisela Neira, Roberto Mera-y-Sierra, Hugo Aguilar-Díaz, and Raquel Cossio-Bayugar. 2022. "A Single Nucleotide Polymorphism Translates into a Radical Amino Acid Substitution at the Ligand-Binding Site in Fasciola hepatica Carboxylesterase B" Genes 13, no. 10: 1899. https://doi.org/10.3390/genes13101899