Integrated Analysis of the Fecal Metagenome and Metabolome in Bladder Cancer in a Chinese Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects and Sample Collection
2.2. Experimental Procedures of Metagenomic Sequencing
2.2.1. DNA Extraction and Sample Testing
2.2.2. Library Construction
2.2.3. Sequencing
2.3. Data Analysis of Metagenomic Project
2.3.1. Data Pretreatment and Quality Control
2.3.2. Metagenome Assembly
2.4. Gene Prediction and Abundance Analysis
2.5. Taxonomy Prediction
2.6. Functional Database Annotations
2.7. Experimental Procedures of Targeted Metabolomics
2.7.1. Metabolite Extractions
2.7.2. LC–MS/MS Analysis
2.7.3. Data Processing
2.7.4. Statistical Analysis
2.8. Integrated Analysis of Metagenomics and Metabolomics
3. Results
3.1. Patient Characterization
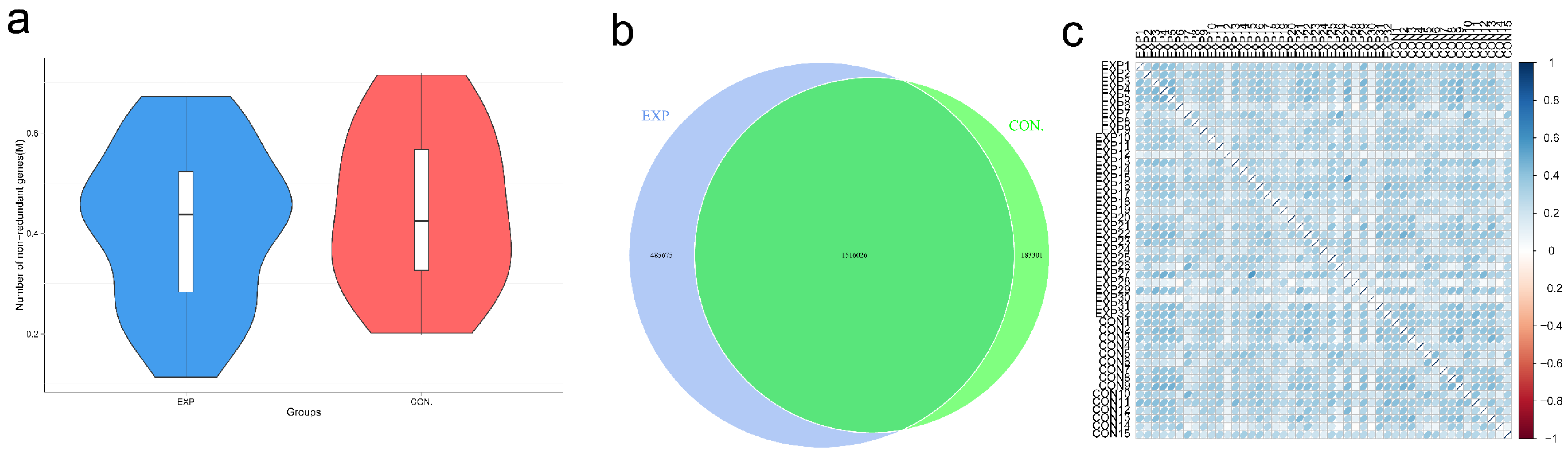
3.2. Gene Number Differences and Correlation Analysis
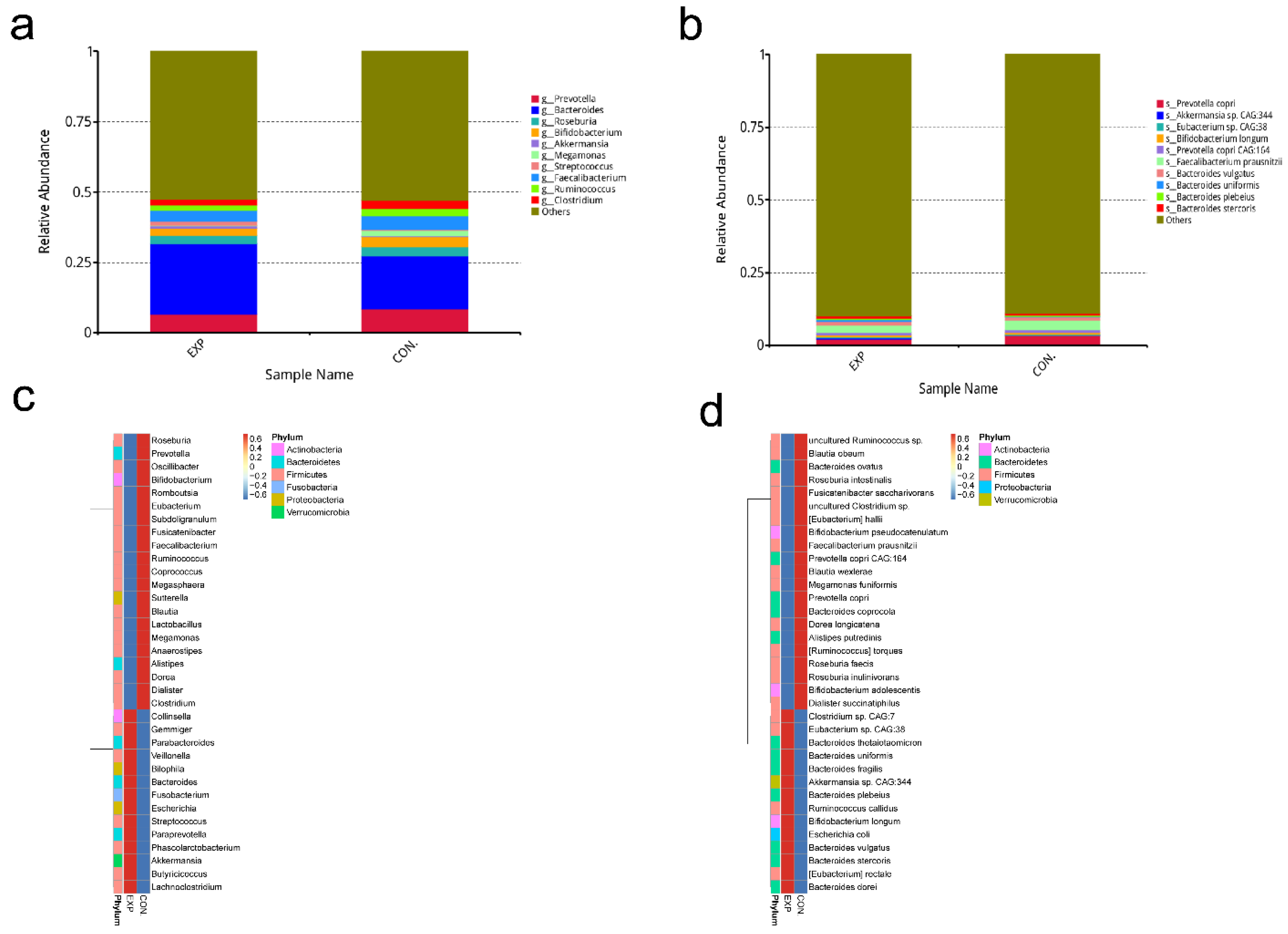
3.3. Relative Abundance Analysis of Gut Microbiota
3.4. Dimension-Reducing Analysis Based on Species Abundance
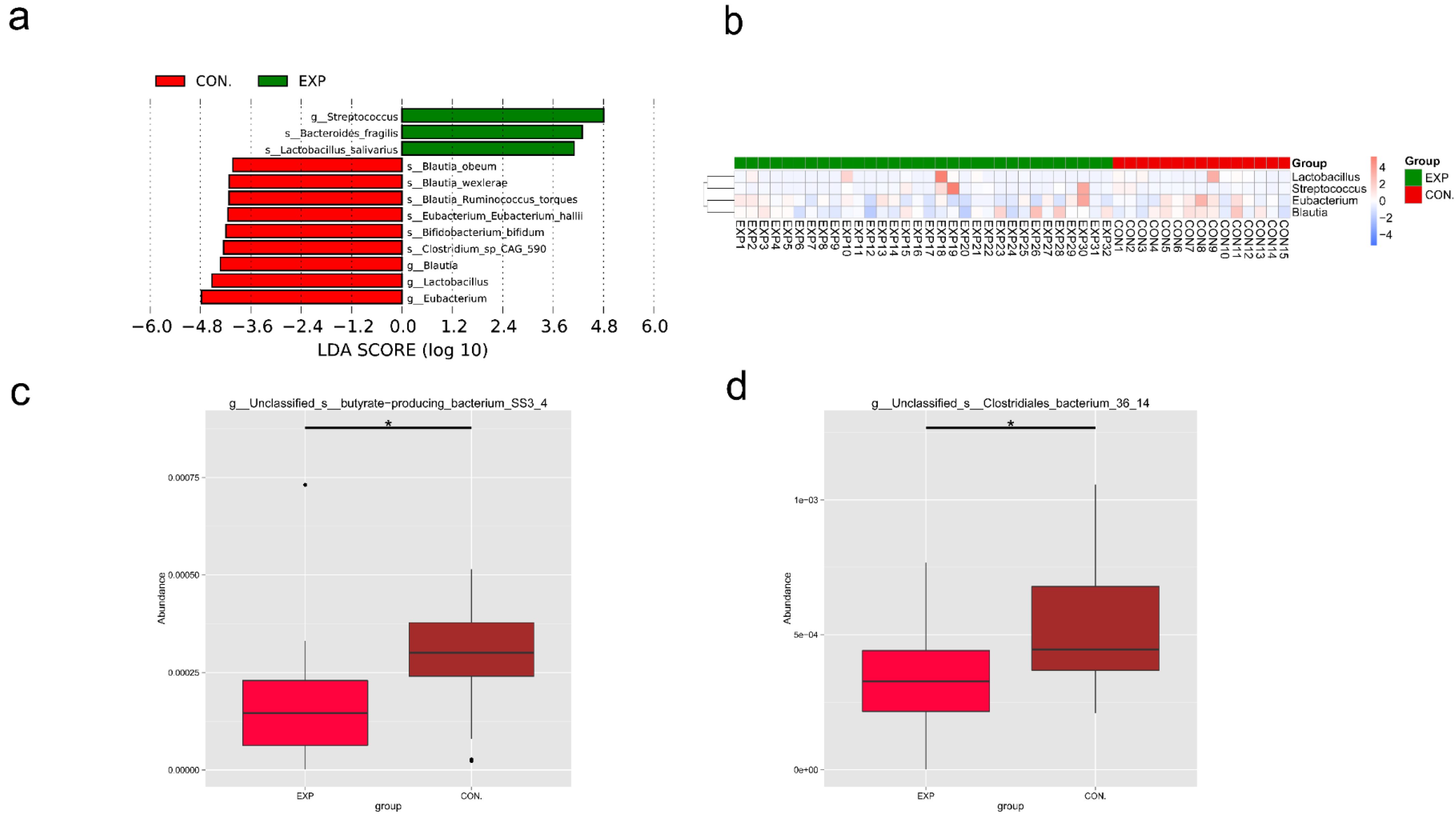
3.5. LEfSe and Metastat Analysis of Species Differing between Groups
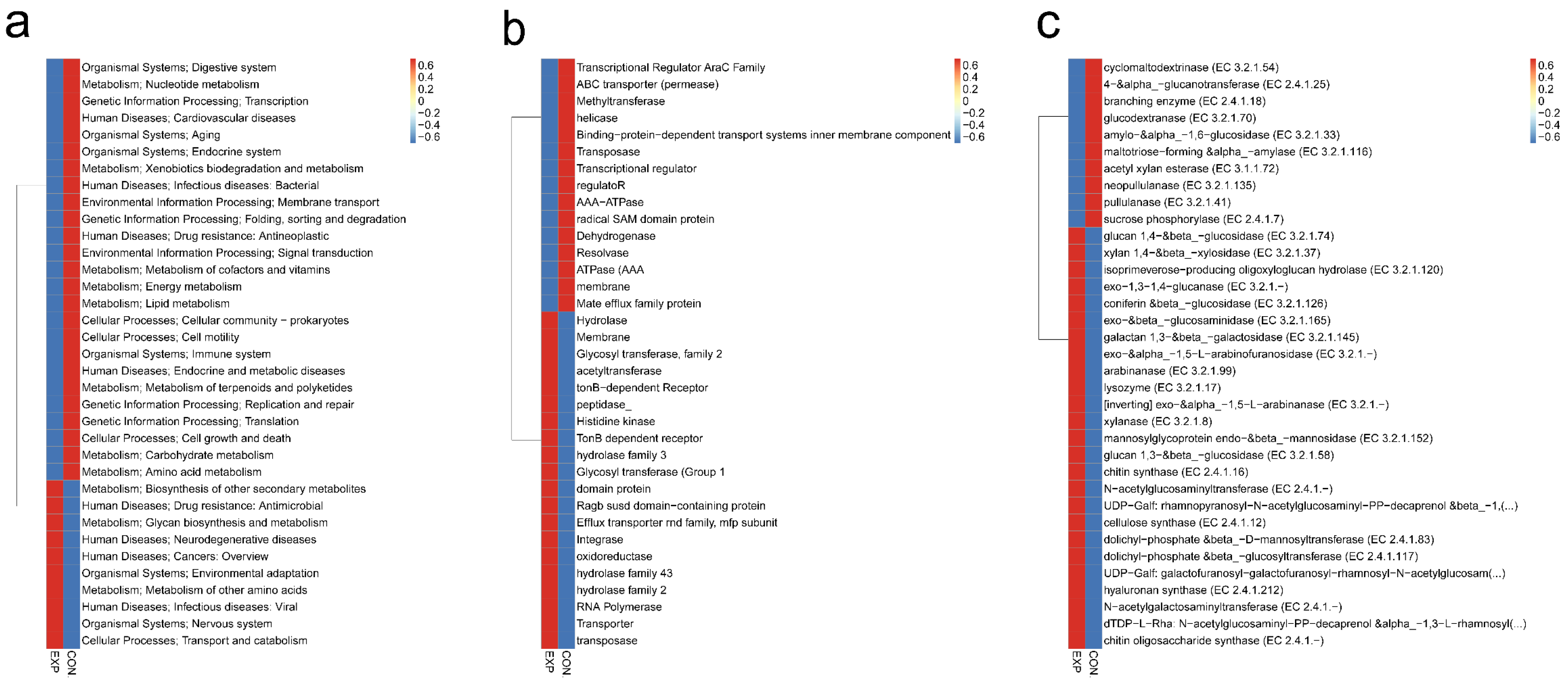
3.6. Functional Characterization of Gut Microbiota
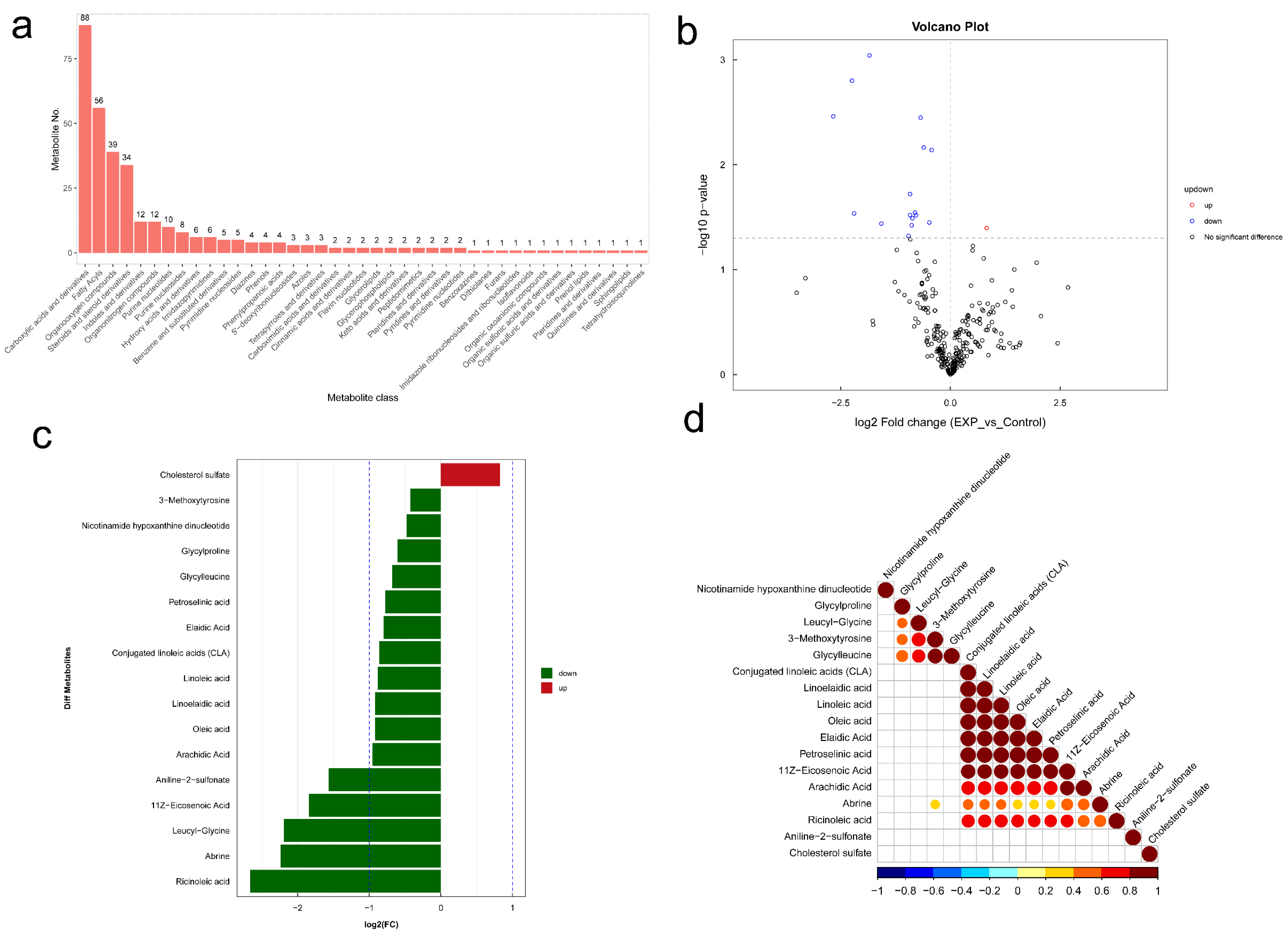
3.7. Changes of Gut-Microbiome-Associated Metabolites in BLCA Patients
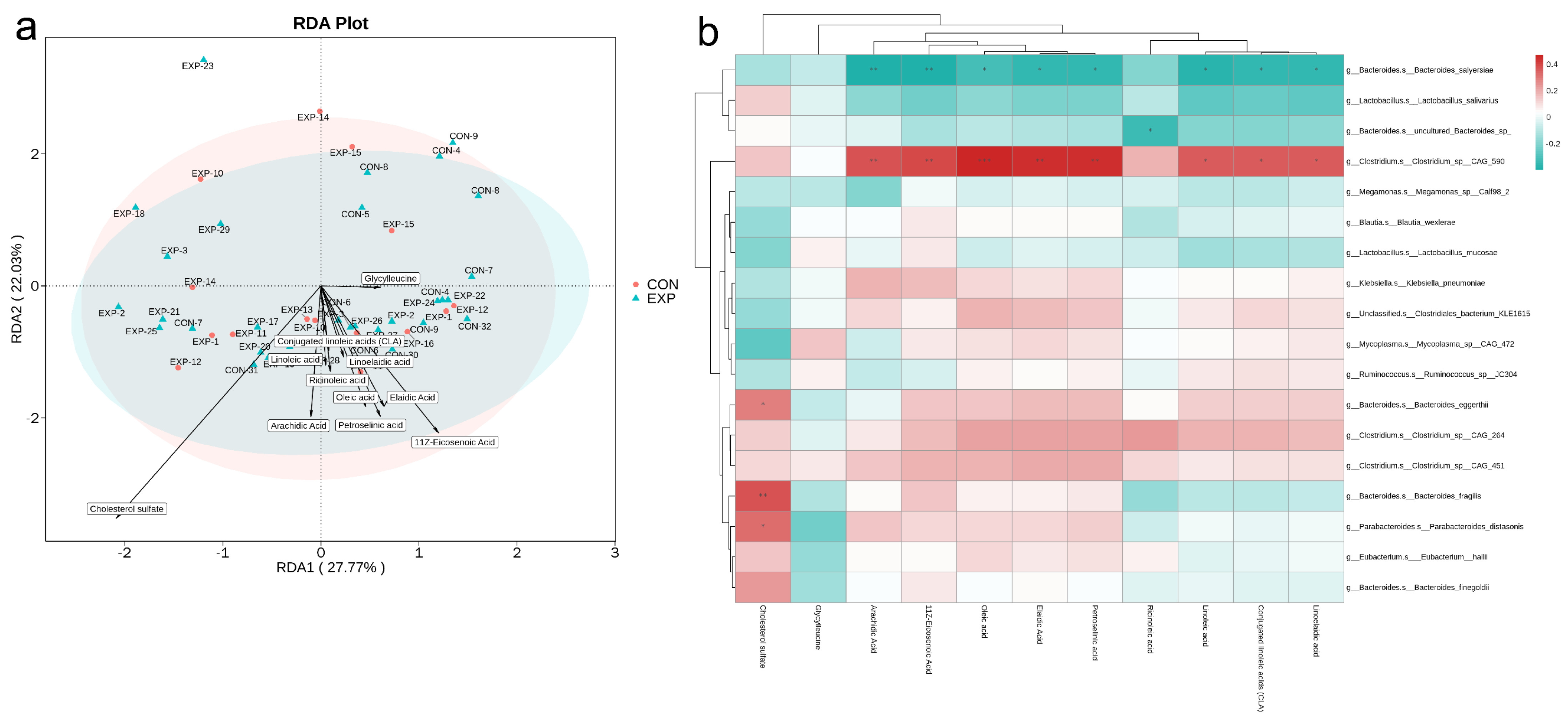
3.8. Integrative Analysis of Metagenomics and Metabolomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef]
- Prasad, S.M.; Decastro, G.J.; Steinberg, G.D. Urothelial carcinoma of the bladder: Definition, treatment and future efforts. Nat. Rev. Urol. 2011, 8, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Serag Eldien, M.M.; Abdou, A.G.; Elghrabawy, G.R.A.; Alhanafy, A.M.; Mahmoud, S.F. Stratification of urothelial bladder carcinoma depending on immunohistochemical expression of GATA3 and CK5/6. J. Immunoass. Immunochem. 2021, 42, 662–678. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between drugs and the gut microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef]
- Chen, X.; Devaraj, S. Gut Microbiome in Obesity, Metabolic Syndrome, and Diabetes. Curr. Diabetes Rep. 2018, 18, 129. [Google Scholar] [CrossRef]
- Lee, C.B.; Chae, S.U.; Jo, S.J.; Jerng, U.M.; Bae, S.K. The Relationship between the Gut Microbiome and Metformin as a Key for Treating Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2021, 22, 3566. [Google Scholar] [CrossRef]
- Di Costanzo, M.; Carucci, L.; Berni Canani, R.; Biasucci, G. Gut Microbiome Modulation for Preventing and Treating Pediatric Food Allergies. Int. J. Mol. Sci. 2020, 21, 5275. [Google Scholar] [CrossRef]
- Zhang, J.; Xia, Y.; Sun, J. Breast and gut microbiome in health and cancer. Genes Dis. 2021, 8, 581–589. [Google Scholar] [CrossRef]
- Fujita, K.; Matsushita, M.; Banno, E.; De Velasco, M.A.; Hatano, K.; Nonomura, N.; Uemura, H. Gut microbiome and prostate cancer. Int. J. Urol. 2022, 29, 793–798. [Google Scholar] [CrossRef]
- Fong, W.; Li, Q.; Yu, J. Gut microbiota modulation: A novel strategy for prevention and treatment of colorectal cancer. Oncogene 2020, 39, 4925–4943. [Google Scholar] [CrossRef]
- Matsushita, M.; Fujita, K.; Hayashi, T.; Kayama, H.; Motooka, D.; Hase, H.; Jingushi, K.; Yamamichi, G.; Yumiba, S.; Tomiyama, E.; et al. Gut Microbiota–Derived Short-Chain Fatty Acids Promote Prostate Cancer Growth via IGF1 Signaling. Cancer Res. 2021, 81, 4014–4026. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Wolf, P.G.; Gaskins, H.R. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes 2016, 7, 201–215. [Google Scholar] [CrossRef]
- Holbert, C.E.; Cullen, M.T.; Casero, R.A.; Stewart, T.M. Polyamines in cancer: Integrating organismal metabolism and antitumour immunity. Nat. Rev. Cancer 2022, 22, 467–480. [Google Scholar] [CrossRef]
- Wang, R.; Yang, X.; Liu, J.; Zhong, F.; Zhang, C.; Chen, Y.; Sun, T.; Ji, C.; Ma, D. Gut microbiota regulates acute myeloid leukaemia via alteration of intestinal barrier function mediated by butyrate. Nat. Commun. 2022, 13, 2522. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Fåk, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Bäckhed, F.; Nielsen, J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012, 3, 1245. [Google Scholar] [CrossRef]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef]
- Nielsen, H.B.; Almeida, M.; Juncker, A.S.; Rasmussen, S.; Li, J.; Sunagawa, S.; Plichta, D.R.; Gautier, L.; Pedersen, A.G.; Le Chatelier, E.; et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 2014, 32, 822–828. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical Methods for Detecting Differentially Abundant Features in Clinical Metagenomic Samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef] [PubMed]
- Mármol, I.; Sánchez-De-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef] [PubMed]
- Clos-Garcia, M.; Garcia, K.; Alonso, C.; Iruarrizaga-Lejarreta, M.; D’Amato, M.; Crespo, A.; Iglesias, A.; Cubiella, J.; Bujanda, L.; Falcón-Pérez, J.M. Integrative Analysis of Fecal Metagenomics and Metabolomics in Colorectal Cancer. Cancers 2020, 12, 1142. [Google Scholar] [CrossRef]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial driver–passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef]
- Martin, A.; Woolbright, B.L.; Umar, S.; Ingersoll, M.A.; Taylor, J.A. Bladder cancer, inflammageing and microbiomes. Nat. Rev. Urol. 2022, 19, 495–509. [Google Scholar] [CrossRef]
- Gondo, T.; Nakashima, J.; Ohno, Y.; Choichiro, O.; Horiguchi, Y.; Namiki, K.; Yoshioka, K.; Ohori, M.; Hatano, T.; Tachibana, M. Prognostic Value of Neutrophil-to-lymphocyte Ratio and Establishment of Novel Preoperative Risk Stratification Model in Bladder Cancer Patients Treated with Radical Cystectomy. Urology 2012, 79, 1085–1091. [Google Scholar] [CrossRef]
- Kawai, K.; Yamamoto, M.; Kameyama, S.; Kawamata, H.; Rademaker, A.; Oyasu, R. Enhancement of rat urinary bladder tumorigenesis by lipopolysaccharide-induced inflammation. Cancer Res. 1993, 53, 5172–5175. [Google Scholar]
- He, C.; Li, B.; Huang, L.; Teng, C.; Bao, Y.; Ren, M.; Shan, Y. Gut microbial composition changes in bladder cancer patients: A case-control study in Harbin, China. Asia Pac. J. Clin. Nutr. 2020, 29, 395–403. [Google Scholar] [CrossRef]
- Kato, I.; Endo, K.; Yokokura, T. Effects of oral administration of Lactobacillus casei on antitumor responses induced by tumor resection in mice. Int. J. Immunopharmacol. 1994, 16, 29–36. [Google Scholar] [CrossRef]
- Kitagawa, K.; Tatsumi, M.; Kato, M.; Komai, S.; Doi, H.; Hashii, Y.; Katayama, T.; Fujisawa, M.; Shirakawa, T. An oral cancer vaccine using a Bifidobacterium vector suppresses tumor growth in a syngeneic mouse bladder cancer model. Mol. Ther.-Oncolytics 2021, 22, 592–603. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef]
- Tan, J.; Mckenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef]
- Zeng, H.; Umar, S.; Rust, B.; Lazarova, D.; Bordonaro, M. Secondary Bile Acids and Short Chain Fatty Acids in the Colon: A Focus on Colonic Microbiome, Cell Proliferation, Inflammation, and Cancer. Int. J. Mol. Sci. 2019, 20, 1214. [Google Scholar] [CrossRef]
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V.; et al. Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018, 360, eaan5931. [Google Scholar] [CrossRef]
- Pujo, J.; Petitfils, C.; Le Faouder, P.; Eeckhaut, V.; Payros, G.; Maurel, S.; Perez-Berezo, T.; Van Hul, M.; Barreau, F.; Blanpied, C.; et al. Bacteria-derived long chain fatty acid exhibits anti-inflammatory properties in colitis. Gut 2021, 70, 1088–1097. [Google Scholar] [CrossRef]
- Ilag, L. Are Long-Chain Polyunsaturated Fatty Acids the Link between the Immune System and the Microbiome towards Modulating Cancer? Medicines 2018, 5, 102. [Google Scholar] [CrossRef]
- Weng, Y.J.; Gan, H.Y.; Li, X.; Huang, Y.; Li, Z.C.; Deng, H.M.; Chen, S.Z.; Zhou, Y.; Wang, L.S.; Han, Y.P.; et al. Correlation of diet, microbiota and metabolite networks in inflammatory bowel disease. J. Dig. Dis. 2019, 20, 447–459. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, X.; Zhang, X.; Xiao, F.; Hu, H.; Li, X.; Dong, F.; Sun, M.; Xiao, Y.; Ge, T.; et al. Microbial and metabolic features associated with outcome of infliximab therapy in pediatric Crohn’s disease. Gut Microbes 2021, 13, 1865708. [Google Scholar] [CrossRef]
- Le, H.H.; Lee, M.; Besler, K.R.; Comrie, J.M.C.; Johnson, E.L. Characterization of interactions of dietary cholesterol with the murine and human gut microbiome. Nat. Microbiol. 2022, 7, 1390–1403. [Google Scholar] [CrossRef]
- Johnson, C.H.; Santidrian, A.F.; Leboeuf, S.E.; Kurczy, M.E.; Rattray, N.J.W.; Rattray, Z.; Warth, B.; Ritland, M.; Hoang, L.T.; Loriot, C.; et al. Metabolomics guided pathway analysis reveals link between cancer metastasis, cholesterol sulfate, and phospholipids. Cancer Metab. 2017, 5, 9. [Google Scholar] [CrossRef]
- Vickman, R.E.; Crist, S.A.; Kerian, K.; Eberlin, L.; Cooks, R.G.; Burcham, G.N.; Buhman, K.K.; Hu, C.; Mesecar, A.D.; Cheng, L.; et al. Cholesterol Sulfonation Enzyme, SULT2B1b, Modulates AR and Cell Growth Properties in Prostate Cancer. Mol. Cancer Res. 2016, 14, 776–786. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, C.; Chen, Z.; Cao, R.; Shi, M.; Tian, Y. Integrated Analysis of the Fecal Metagenome and Metabolome in Bladder Cancer in a Chinese Population. Genes 2022, 13, 1967. https://doi.org/10.3390/genes13111967
Qin C, Chen Z, Cao R, Shi M, Tian Y. Integrated Analysis of the Fecal Metagenome and Metabolome in Bladder Cancer in a Chinese Population. Genes. 2022; 13(11):1967. https://doi.org/10.3390/genes13111967
Chicago/Turabian StyleQin, Chuan, Zhenghao Chen, Rui Cao, Mingjun Shi, and Ye Tian. 2022. "Integrated Analysis of the Fecal Metagenome and Metabolome in Bladder Cancer in a Chinese Population" Genes 13, no. 11: 1967. https://doi.org/10.3390/genes13111967
APA StyleQin, C., Chen, Z., Cao, R., Shi, M., & Tian, Y. (2022). Integrated Analysis of the Fecal Metagenome and Metabolome in Bladder Cancer in a Chinese Population. Genes, 13(11), 1967. https://doi.org/10.3390/genes13111967




