Genome-Wide Association Study of Exercise-Induced Fat Loss Efficiency
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Participants
2.2.1. Polish Women
2.2.2. Russian Athletes
2.3. Physical Exercise Training Protocol
2.4. Body Composition Measurements
2.5. Genetic Approach
2.6. Statistical Analyses
3. Results
3.1. Changes in Anthropometric Measurements
3.2. Genome-Wide Significant Markers Associated with Fat Loss Efficiency in the Polish Cohort
3.3. Study Involving Russian Athletes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boutari, C.; Mantzoros, C.S. A 2022 update on the epidemiology of obesity and a call to action: As its twin COVID-19 pandemic appears to be receding, the obesity and dysmetabolism pandemic continues to rage on. Metabolism 2022, 133, 155217. [Google Scholar] [CrossRef]
- Hinney, A.; Vogel, C.I.G.; Hebebrand, J. From monogenic to polygenic obesity: Recent advances. Eur. Child Adolesc. Psychiatry 2010, 19, 297–310. [Google Scholar] [CrossRef] [Green Version]
- Loos, R.J.F.; Yeo, G.S.H. The genetics of obesity: From discovery to biology. Nat. Rev. Genet. 2022, 23, 120–133. [Google Scholar] [CrossRef]
- McCaffery, J.M.; Papandonatos, G.D.; Bond, D.S.; Lyons, M.J.; Wing, R.R. Gene × environment interaction of vigorous exercise and body mass index among male Vietnam-era twins. Am. J. Clin. Nutr. 2009, 89, 1011–1018. [Google Scholar] [CrossRef] [Green Version]
- Min, J.; Chiu, D.T.; Wang, Y. Variation in the heritability of body mass index based on diverse twin studies: A systematic review. Obes. Rev. 2013, 14, 871–882. [Google Scholar] [CrossRef]
- Silventoinen, K.; Jelenkovic, A.; Sund, R.; Yokoyama, Y.; Hur, Y.M.; Cozen, W.; Hwang, A.E.; Mack, T.M.; Honda, C.; Inui, F.; et al. Differences in genetic and environmental variation in adult BMI by sex, age, time period, and region: An individual-based pooled analysis of 40 twin cohorts. Am. J. Clin. Nutr. 2017, 106, 457–466. [Google Scholar] [CrossRef] [Green Version]
- Locke, A.; Kahali, B.; Berndt, S.; Justice, A.; Pers, T.; Day, F. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Silventoinen, K.; Huppertz, C.; van Beijsterveldt, C.E.M.; Bartels, M.; Willemsen, G.; Boomsma, D.I. The genetic architecture of body mass index from infancy to adulthood modified by parental education. Obesity 2016, 24, 2004–2011. [Google Scholar] [CrossRef] [Green Version]
- Rose, K.M.; Newman, B.; Mayer-Davis, E.J.; Selby, J.V. Genetic and behavioral determinants of waist-hip ratio and waist circumference in women twins. Obes. Res. 1998, 6, 383–392. [Google Scholar] [CrossRef]
- Nelson, T.L.; Vogler, G.P.; Pedersen, N.L.; Miles, T.P. Genetic and Environmental Influences on Waist-to-Hip Ratio and Waist Circumference in an Older Swedish Twin Population. Int. J. Obes. Relat. Metab. Disord. 1999, 23, 449–455. [Google Scholar] [CrossRef]
- Leońska-Duniec, A.; Jastrzębski, Z.; Jażdżewska, A.; Moska, W.; Lulińska-Kuklik, E.; Sawczuk, M.; Gubaydullina, S.I.; Shakirova, A.T.; Cięszczyk, P.; Maszczyk, A.; et al. Individual responsiveness to exercise-induced fat loss and improvement of metabolic profile in young women is associated with polymorphisms of adrenergic receptor genes. J. Sports Sci. Med. 2018, 17, 134–144. [Google Scholar] [PubMed]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Katherine, S.; Lango, H.; Rayner, N.W.; Shields, B.; et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buniello, A.; Macarthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.; Vivar, J.C.; Sarzynski, M.A.; Sung, Y.J.; Timmons, J.A.; Bouchard, C.; Rankinen, T. Integrative pathway analysis of a genome-wide association study of VO2max response to exercise training. J. Appl. Physiol. 2013, 115, 1343–1359. [Google Scholar] [CrossRef] [Green Version]
- Pickering, C.; Suraci, B.; Semenova, E.A.; Boulygina, E.A.; Kostryukova, E.S.; Kulemin, N.A.; Borisov, O.V.; Khabibova, S.A.; Larin, A.K.; Pavlenko, A.V.; et al. A Genome-Wide Association Study of Sprint Performance in Elite Youth Football Players. J. Strength Cond. Res. 2019, 33, 2344–2351. [Google Scholar] [CrossRef]
- Al-Khelaifi, F.; Yousri, N.A.; Diboun, I.; Semenova, E.A.; Kostryukova, E.S.; Kulemin, N.A.; Borisov, O.V.; Andryushchenko, L.B.; Larin, A.K.; Generozov, E.V.; et al. Genome-Wide Association Study Reveals a Novel Association Between MYBPC3 Gene Polymorphism, Endurance Athlete Status, Aerobic Capacity and Steroid Metabolism. Front. Genet. 2020, 11, 595. [Google Scholar] [CrossRef]
- Boulygina, E.A.; Borisov, O.V.; Valeeva, E.V.; Semenova, E.A.; Kostryukova, E.S.; Kulemin, N.A.; Larin, A.K.; Nabiullina, R.M.; Mavliev, F.A.; Akhatov, A.M.; et al. Whole genome sequencing of elite athletes. Biol. Sport. 2020, 37, 295–304. [Google Scholar] [CrossRef]
- Lee, I.; Shiroma, E.; Lobelo, F.; Puska, P.; Blair, S.; Katzmarzyk, P.; Lancet Physical Activity Series Working Group. Effect of physical inactivity on major non-communicable diseases worldwide: An analysis of burden of disease and life expectancy. Lancet 2012, 380, 219–229. [Google Scholar] [CrossRef] [Green Version]
- Leońska-Duniec, A.; Ahmetov, I.I.; Zmijewski, P. Genetic variants influencing effectiveness of exercise training programmes in obesity - an overview of human studies. Biol. Sport. 2016, 33, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Chung, H.C.; Keiller, D.R.; Roberts, J.D.; Gordon, D.A. Do exercise-associated genes explain phenotypic variance in the three components of fitness? A systematic review & meta-analysis. PLoS ONE 2021, 16, e0249501. [Google Scholar] [CrossRef]
- Cagnin, S.; Chemello, F.; Ahmetov, I.I. Genes and response to aerobic training. In Sports, Exercise, and Nutritional Genomics; Academic Press: Cambridge, MA, USA, 2019; pp. 169–188. [Google Scholar] [CrossRef]
- Leonska-Duniec, A.; Cieszczyk, P.; Ahmetov, I.I. Genes and individual responsiveness to exercise-induced fat loss. In Sports, Exercise, and Nutritional Genomics; Academic Press: Cambridge, MA, USA, 2019; pp. 231–247. [Google Scholar] [CrossRef]
- Leońska-Duniec, A.; Świtała, K.; Ahmetov, I.I.; Pickering, C.; Massidda, M.; Buryta, M.; Mastalerz, A.; Maculewicz, E. FABP2 Ala54Thr Polymorphism and Post-Training Changes of Body Composition and Biochemical Parameters in Caucasian Women. Genes 2021, 12, 954. [Google Scholar] [CrossRef]
- Mazur, I.I.; Drozdovska, S.; Andrieieva, O.; Vinnichuk, Y.; Polishchuk, A.; Dosenko, V.; Andreev, I.; Pickering, C.; Ahmetov, I.I. PPARGC1A gene polymorphism is associated with exercise-induced fat loss. Mol. Biol. Rep. 2020, 47, 7451–7457. [Google Scholar] [CrossRef]
- Little, J.; Higgins, J.P.T.; Ioannidis, J.P.A.; Moher, D.; Gagnon, F.; von Elm, E.; Khoury, M.J.; Cohen, B.; Davey-Smith, G.; Grimshaw, J.; et al. Strengthening the reporting of genetic association studies (STREGA): An extension of the STROBE Statement. Hum. Genet. 2009, 125, 131–151. [Google Scholar] [CrossRef] [Green Version]
- Zarębska, A.; Jastrzębski, Z.; Moska, W.; Leońska-Duniec, A.; Kaczmarczyk, M.; Sawczuk, M.; Maciejewska-Skrendo, A.; Żmijewski, P.; Ficek, K.; Trybek, G.; et al. The AGT gene M235T polymorphism and response of power-related variables to aerobic training. J. Sports Sci. Med. 2016, 15, 616–624. [Google Scholar]
- Kikuchi, N.; Moreland, E.; Homma, H.; Semenova, E.A.; Saito, M.; Larin, A.K.; Kobatake, N.; Yusupov, R.A.; Okamoto, T.; Nakazato, K.; et al. Genes and weightlifting performance. Genes 2022, 13, 25. [Google Scholar] [CrossRef]
- Swift, D.L.; Johannsen, N.M.; Lavie, C.J.; Earnest, C.P.; Church, T.S. The role of exercise and physical activity in weight loss and maintenance. Prog. Cardiovasc. Dis. 2014, 56, 441–447. [Google Scholar] [CrossRef] [Green Version]
- Clark, J.E. Diet, exercise or diet with exercise: Comparing the effectiveness of treatment options for weight-loss and changes in fitness for adults (18–65 years old) who are overfat, or obese; systematic review and meta-analysis. J. Diabetes Metab. Disord. 2015, 14, 31. [Google Scholar] [CrossRef] [Green Version]
- Scotto di Palumbo, A.; Guerra, E.; Orlandi, C.; Bazzucchi, I.; Sacchetti, M. Effect of combined resistance and endurance exercise training on regional fat loss. J. Sports Med. Phys. Fitness 2017, 57, 794–801. [Google Scholar] [CrossRef]
- Steger, F.L.; Donnelly, J.E.; Hull, H.R.; Li, X.; Hu, J.; Sullivan, D.K. Intermittent and continuous energy restriction result in similar weight loss, weight loss maintenance, and body composition changes in a 6 month randomized pilot study. Clin. Obes. 2021, 11, e12430. [Google Scholar] [CrossRef]
- NIDDK. Prescription Medications to Treat Overweight and Obesity. Available online: https://www.niddk.nih.gov/health-information/weight-management/prescription-medications-treat-overweight-obesity#replace (accessed on 30 September 2022).
- Holzapfel, C.; Sag, S.; Graf-Schindler, J.; Fischer, M.; Drabsch, T.; Illig, T.; Grallert, H.; Stecher, L.; Strack, C.; Caterson, I.D.; et al. Association between single nucleotide polymorphisms and weight reduction in behavioural interventions—a pooled analysis. Nutrients 2021, 13, 819. [Google Scholar] [CrossRef]
- Yoon, Y.; Park, B.L.; Cha, M.H.; Kim, K.S.; Cheong, H.S.; Choi, Y.H.; Shin, H.D. Effects of genetic polymorphisms of UCP2 and UCP3 on very low calorie diet-induced body fat reduction in Korean female subjects. Biochem. Biophys. Res. Commun. 2007, 359, 451–456. [Google Scholar] [CrossRef]
- Zhang, X.; Qi, Q.; Zhang, C.; Hu, F.B.; Sacks, F.M.; Qi, L. FTO genotype and 2-year change in body composition and fat distribution in response to weight-loss diets: The POUNDS LOST trial. Diabetes 2012, 61, 3005–3011. [Google Scholar] [CrossRef] [Green Version]
- Valeeva, F.V.; Medvedeva, M.S.; Khasanova, K.B.; Valeeva, E.V.; Kiseleva, T.A.; Egorova, E.S.; Pickering, C.; Ahmetov, I.I. Association of gene polymorphisms with body weight changes in prediabetic patients. Mol. Biol. Rep. 2022, 49, 4217–4224. [Google Scholar] [CrossRef]
- Mitchell, J.A.; Church, T.S.; Rankinen, T.; Earnest, C.P.; Sui, X.; Blair, S.N. FTO genotype and the weight loss benefits of moderate intensity exercise. Obesity 2010, 18, 641–643. [Google Scholar] [CrossRef] [Green Version]
- Zarebska, A.; Jastrzebski, Z.; Cieszczyk, P.; Leonska-Duniec, A.; Kotarska, K.; Kaczmarczyk, M.; Sawczuk, M.; Maciejewska-Karlowska, A. The Pro12Ala polymorphism of the peroxisome proliferator-activated receptor gamma gene modifies the association of physical activity and body mass changes in Polish women. PPAR Res. 2014, 2014, 373782. [Google Scholar] [CrossRef] [Green Version]
- Mashek, D.G.; Bornfeldt, K.E.; Coleman, R.A.; Berger, J.; Bernlohr, D.A.; Black, P.; DiRusso, C.C.; Farber, S.A.; Guo, W.; Hashimoto, N.; et al. Revised nomenclature for the mammalian long-chain acyl-CoA synthetase gene family. J. Lipid Res. 2004, 45, 1958–1961. [Google Scholar] [CrossRef] [Green Version]
- Zhan, T.; Poppelreuther, M.; Ehehalt, R.; Füllekrug, J. Overexpressed FATP1, ACSVL4/FATP4 and ACSL1 Increase the Cellular Fatty Acid Uptake of 3T3-L1 Adipocytes but Are Localized on Intracellular Membranes. PLoS ONE 2012, 7, e45087. [Google Scholar] [CrossRef]
- Suzuki, H.; Kawarabayasi, Y.; Kondo, J.; Abe, T.; Nishikawa, K.; Kimura, S.; Hashimoto, T.; Yamamoto, T. Structure and regulation of rat long-chain acyl-CoA synthetase. J. Biol. Chem. 1990, 265, 8681–8685. [Google Scholar] [CrossRef]
- Li, L.O.; Ellis, J.M.; Paich, H.A.; Wang, S.; Gong, N.; Altshuller, G.; Thresher, R.J.; Koves, T.R.; Watkins, S.M.; Muoio, D.M.; et al. Liver-specific loss of long chain Acyl-CoA synthetase-1 decreases triacylglycerol synthesis and β-oxidation and alters phospholipid fatty acid composition. J. Biol. Chem. 2009, 284, 27816–27826. [Google Scholar] [CrossRef]
- Grevengoed, T.J.; Klett, E.L.; Coleman, R.A. Acyl-CoA Metabolism and Partitioning. Annu. Rev. Nutr. 2014, 34, 1–30. [Google Scholar] [CrossRef] [Green Version]
- Klett, E.L.; Chen, S.; Yechoor, A.; Lih, F.B.; Coleman, R.A. Long-chain acyl-CoA synthetase isoforms differ in preferences for eicosanoid species and long-chain fatty acids. J. Lipid Res. 2014, 58, 884–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durgan, D.J.; Smith, J.K.; Hotze, M.A.; Egbejimi, O.; Cuthbert, K.D.; Zaha, V.G.; Dyck, J.R.B.; Abel, E.D.; Young, M.E. Distinct transcriptional regulation of long-chain acyl-CoA synthetase isoforms and cytosolic thioesterase 1 in the rodent heart by fatty acids and insulin. Am. J. Physiol. Heart Circ. Physiol. 2006, 290, 2480–2497. [Google Scholar] [CrossRef] [PubMed]
- Mashek, D.G.; Li, L.O.; Coleman, R.A. Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. J. Lipid Res. 2006, 47, 2004–2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuku, N.; He, Z.H.; Sanchis-Gomar, F.; Pareja-Galeano, H.; Tian, Y.; Arai, Y.; Abe, Y.; Murakami, H.; Miyachi, M.; Zempo, H.; et al. Exceptional longevity and muscle and fitness related genotypes: A functional in vitro analysis and case-control association replication study with SNPs THRH rs7832552, IL6 rs1800795 and ACSL1 rs6552828. Front. Aging Neurosci. 2015, 7, 59. [Google Scholar] [CrossRef]
- Bouchard, C.; Sarzynski, M.A.; Rice, T.K.; Kraus, W.E.; Church, T.S.; Sung, Y.J.; Rao, D.C.; Rankinen, T. Genomic predictors of the maximal O2 uptake response to standardized exercise training programs. J. Appl. Physiol. 2011, 110, 1160–1170. [Google Scholar] [CrossRef] [Green Version]
- Yvert, T.; He, Z.H.; Santiago, C.; Hu, Y.; Li, Y.C.; Gómez-Gallego, F.; Fiuza-Luces, C.; Verde, Z.; Muniesa, C.A.; Oliván, J.; et al. Acyl coenzyme A synthetase long-chain 1 (ACSL1) gene polymorphism (rs6552828) and elite endurance athletic status: A replication study. PLoS ONE 2012, 7, e41268. [Google Scholar] [CrossRef] [Green Version]
- Oikawa, E.; Iijima, H.; Suzuki, T.; Sasano, H.; Kamataki, A.; Nagura, H.; Kang, M.; Fujino, T.; Oikawa, E. A novel acyl-CoA synthetase, ACS5, expressed in intestinal epithelial cells and proliferating preadipocytes. J. Biochem. 1998, 124, 679–685. [Google Scholar] [CrossRef] [Green Version]
- Ellis, J.M.; Li, L.O.; Wu, P.C.; Koves, T.R.; Ilkayeva, O.; Stevens, R.D.; Watkins, S.M.; Muoio, D.M.; Coleman, R.A. Adipose Acyl-CoA synthetase-1 directs fatty acids toward β-oxidation and is required for cold thermogenesis. Cell Metab. 2010, 12, 53–64. [Google Scholar] [CrossRef] [Green Version]
- Li, L.O.; Grevengoed, T.J.; Paul, D.S.; Ilkayeva, O.; Koves, T.R.; Pascual, F.; Newgard, C.B.; Muoio, D.M.; Coleman, R.A. Compartmentalized Acyl-CoA metabolism in skeletal muscle regulates systemic glucose homeostasis. Diabetes 2015, 64, 23–35. [Google Scholar] [CrossRef]
- Bolotta, A.; Filardo, G.; Abruzzo, P.M.; Astolfi, A.; De Sanctis, P.; Di Martino, A.; Hofer, C.; Indio, V.; Kern, H.; Löfler, S.; et al. Skeletal muscle gene expression in long-term endurance and resistance trained elderly. Int. J. Mol. Sci. 2020, 21, 3988. [Google Scholar] [CrossRef]
- Woodlief, T.L.; Sweazey, M.A.; File, C.; Hickner, R.C.; Neufer, P.D.; Cortright, R.N. Effects of exercise on expression of skeletal muscle acyl-CoA synthetase isoform gene expression in obese African-American and Caucasian women. FASEB J. 2008, 22, 958. [Google Scholar] [CrossRef]
- Lefai, E.; Blanc, S.; Momken, I.; Antoun, E.; Chery, I.; Zahariev, A.; Gabert, L.; Bergouignan, A.; Simon, C. Exercise training improves fat metabolism independent of total energy expenditure in sedentary overweight men, but does not restore lean metabolic phenotype. Int. J. Obes. 2017, 41, 1728–1736. [Google Scholar] [CrossRef] [PubMed]
- Schneider, H.; Staudacher, S.; Poppelreuther, M.; Stremmel, W.; Ehehalt, R.; Füllekrug, J. Protein mediated fatty acid uptake: Synergy between CD36/FAT-facilitated transport and acyl-CoA synthetase-driven metabolism. Arch. Biochem. Biophys. 2014, 546, 8–18. [Google Scholar] [CrossRef] [PubMed]
- NCBI. Gene Functions. Available online: https://www.ncbi.nlm.nih.gov/gene (accessed on 30 September 2022).
- Fadista, J.; Manning, A.K.; Florez, J.C.; Groop, L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur. J. Hum. Genet. 2016, 24, 1202–1205. [Google Scholar] [CrossRef] [Green Version]
- Kraft, P.; Zeggini, E.; Ioannidis, J.P.A. Replication in genome-wide association studies. Stat. Sci. 2009, 24, 561–573. [Google Scholar] [CrossRef] [Green Version]
- Willoughby, D.; Hewlings, S.; Kalman, D. Body composition changes in weight loss: Strategies and supplementation for maintaining lean body mass, a brief review. Nutrients 2018, 10, 1876. [Google Scholar] [CrossRef] [Green Version]
- Switala, K.; Leonska-Duniec, A. Physical activity and gene association with human obesity. Balt. J. Health. Phys. Act. 2021, 13, 99–111. [Google Scholar] [CrossRef]

| Parameters | Before Training | After Training | % Change from Baseline | p |
|---|---|---|---|---|
| Body mass, kg | 60.94 (7.09) | 60.33 (6.94) | −0.94 (2.44) | <0.0001 * |
| BMI, kg/m2 | 21.71 (2.44) | 21.50 (2.39) | −0.92 (2.41) | <0.0001 * |
| FM, kg | 15.13 (4.83) | 14.28 (4.85) | −5.53 (11.77) | <0.0001 * |
| FM% | 24.32 (5.03) | 23.13 (5.09) | −4.68 (9.53) | <0.0001 * |
| FFM, kg | 45.81 (2.95) | 46.15 (3.03) | 0.79 (2.99) | 0.0044 * |
| Gene/Near Gene | SNP | Chromo-some | Allele 1/ Allele 2 | MAF | FM Change, % | FFM Change, % | ||
|---|---|---|---|---|---|---|---|---|
| beta | p | beta | p | |||||
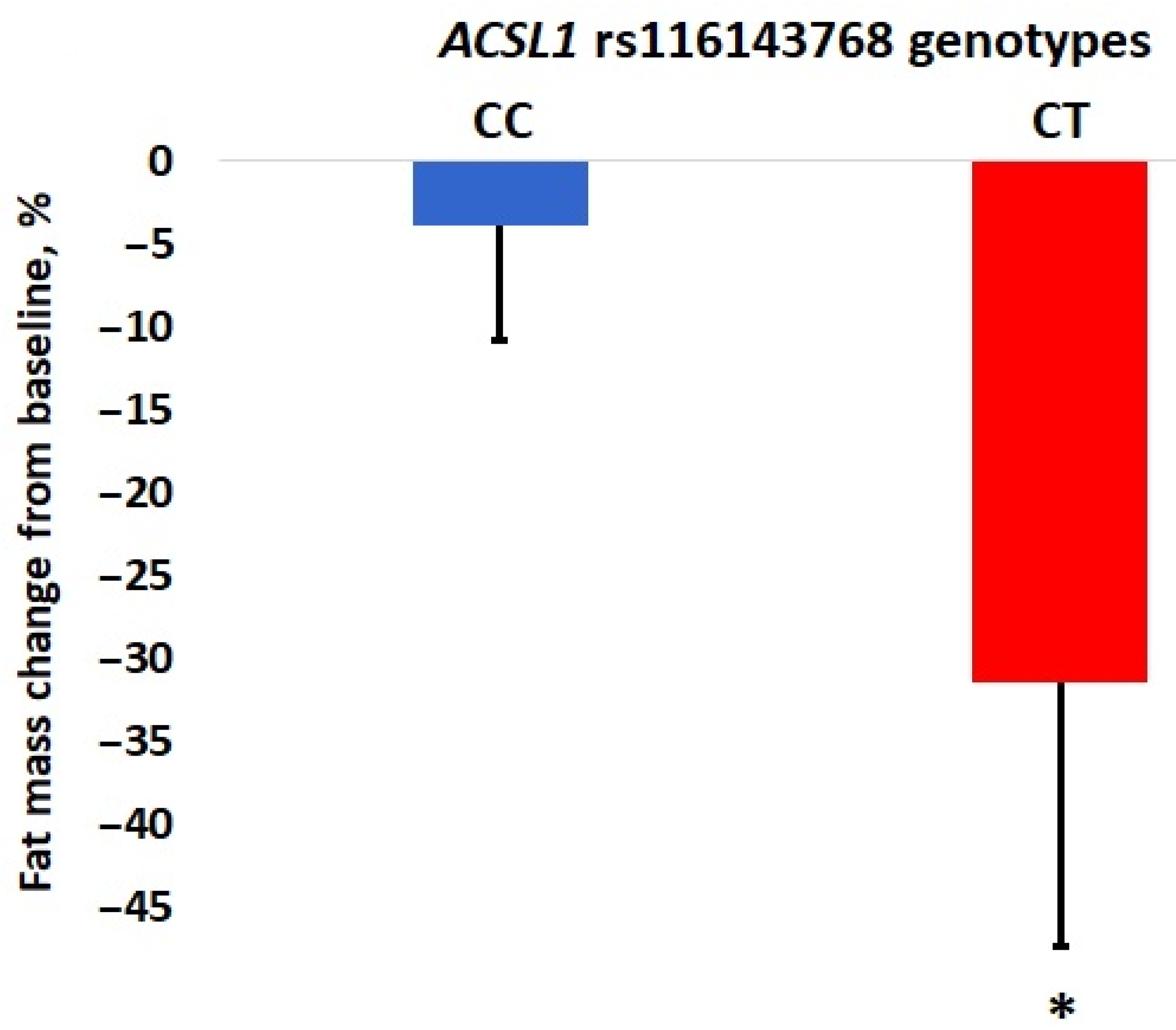
| ACSL1 | rs116143768 | 4 | T/C | 0.0158 | −27.55 | 1.18 × 10−9 * | 6.91 | 2.35 × 10−6 |
| PTPRZ1 | rs79300430 | 7 | G/T | 0.0082 | −34.91 | 6.14 × 10−8 | 7.32 | 4.19 × 10−4 |
| KANK1 | rs7867795 | 9 | G/A | 0.02 | −21.81 | 1.48 × 10−7 | 6.13 | 3.41 × 10−6 |
| TENT5A | rs112141659 | 6 | G/A | 0.0317 | −17.01 | 3.11 × 10−7 | 3.25 | 2.59 × 10−3 |
| GALR1 | rs144060810 | 18 | T/C | 0.0119 | −27.17 | 3.27 × 10−7 | 8.33 | 6.55 × 10−7 |
| LINC00478 | rs238997 | 21 | T/C | 0.0158 | −22.96 | 7.46 × 10−7 | 5.36 | 3.26 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bojarczuk, A.; Boulygina, E.A.; Dzitkowska-Zabielska, M.; Łubkowska, B.; Leońska-Duniec, A.; Egorova, E.S.; Semenova, E.A.; Andryushchenko, L.B.; Larin, A.K.; Generozov, E.V.; et al. Genome-Wide Association Study of Exercise-Induced Fat Loss Efficiency. Genes 2022, 13, 1975. https://doi.org/10.3390/genes13111975
Bojarczuk A, Boulygina EA, Dzitkowska-Zabielska M, Łubkowska B, Leońska-Duniec A, Egorova ES, Semenova EA, Andryushchenko LB, Larin AK, Generozov EV, et al. Genome-Wide Association Study of Exercise-Induced Fat Loss Efficiency. Genes. 2022; 13(11):1975. https://doi.org/10.3390/genes13111975
Chicago/Turabian StyleBojarczuk, Aleksandra, Eugenia A. Boulygina, Magdalena Dzitkowska-Zabielska, Beata Łubkowska, Agata Leońska-Duniec, Emiliya S. Egorova, Ekaterina A. Semenova, Liliya B. Andryushchenko, Andrey K. Larin, Edward V. Generozov, and et al. 2022. "Genome-Wide Association Study of Exercise-Induced Fat Loss Efficiency" Genes 13, no. 11: 1975. https://doi.org/10.3390/genes13111975
APA StyleBojarczuk, A., Boulygina, E. A., Dzitkowska-Zabielska, M., Łubkowska, B., Leońska-Duniec, A., Egorova, E. S., Semenova, E. A., Andryushchenko, L. B., Larin, A. K., Generozov, E. V., Cięszczyk, P., & Ahmetov, I. I. (2022). Genome-Wide Association Study of Exercise-Induced Fat Loss Efficiency. Genes, 13(11), 1975. https://doi.org/10.3390/genes13111975







