Proteomic Analysis of Alfalfa (Medicago sativa L.) Roots in Response to Rhizobium Nodulation and Salt Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Rhizobia Inoculation
2.3. Salt Treatments
2.4. Physiological Measurements
2.5. Proteomic Analysis
2.6. Net K+ and H+ Flux Measurements
3. Results
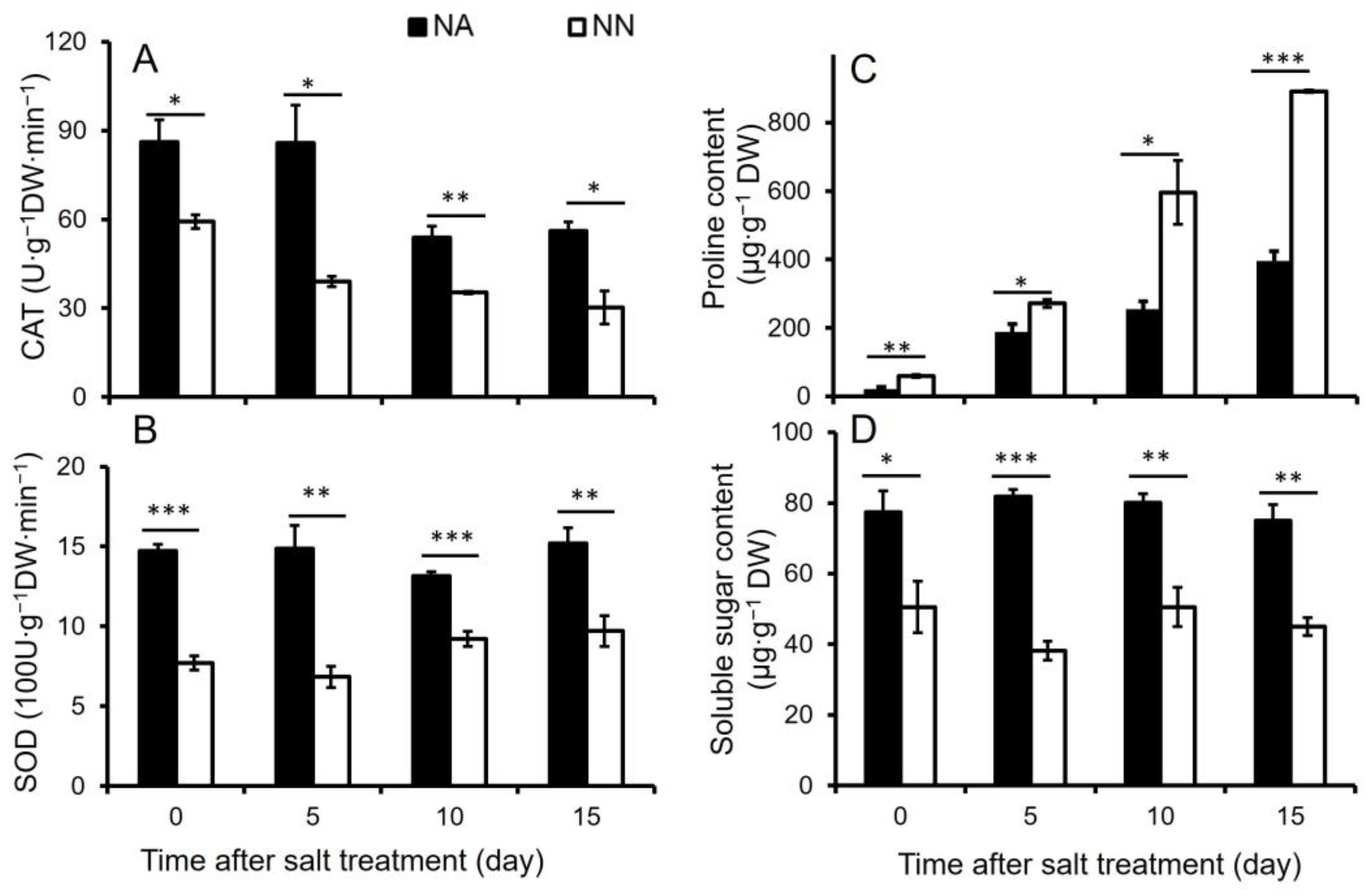
3.1. Physiological Analysis
3.2. Summary of Protein Identification
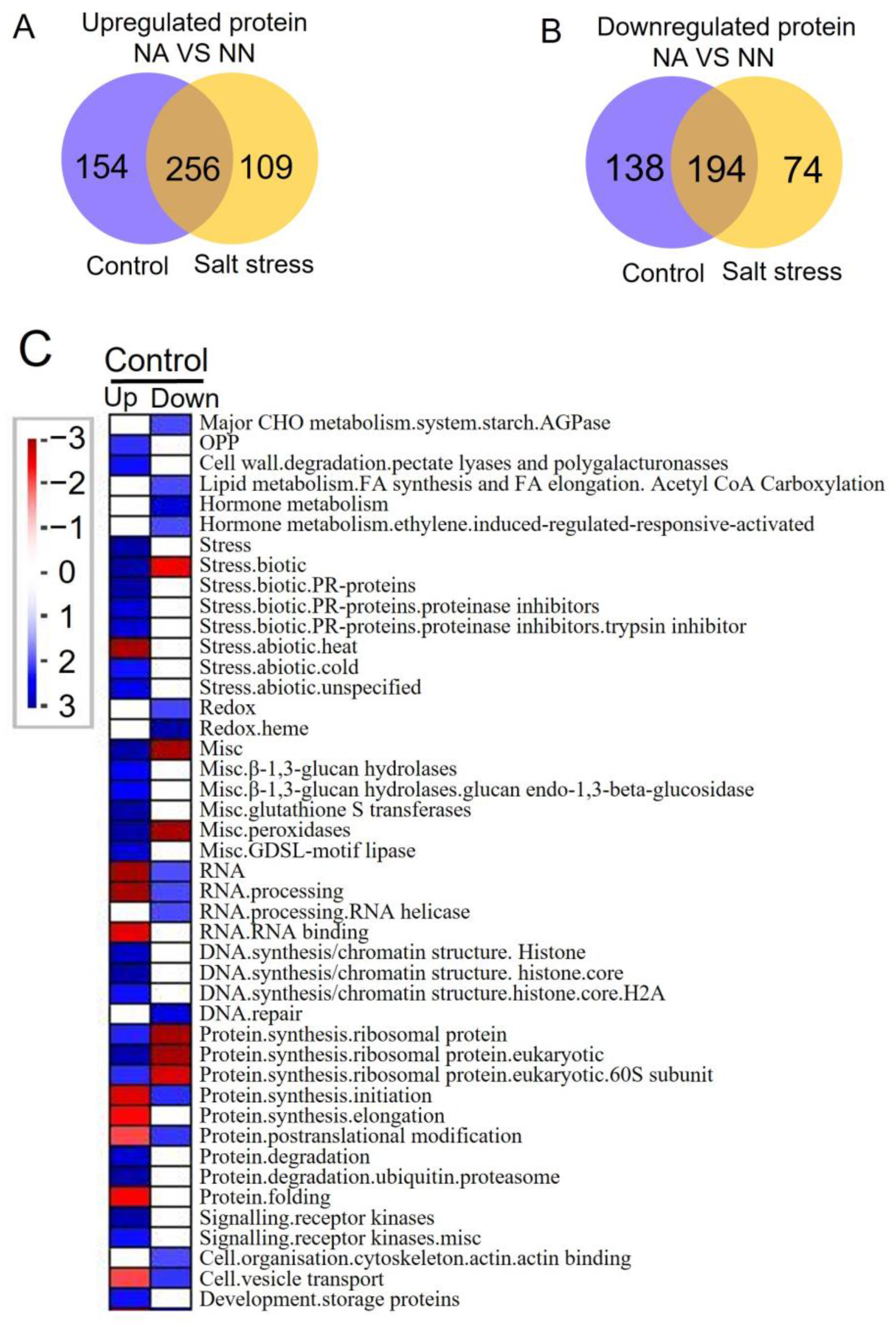
3.3. Differentially Expressed Proteins and Pathway Analysis of Alfalfa with or without Nodules
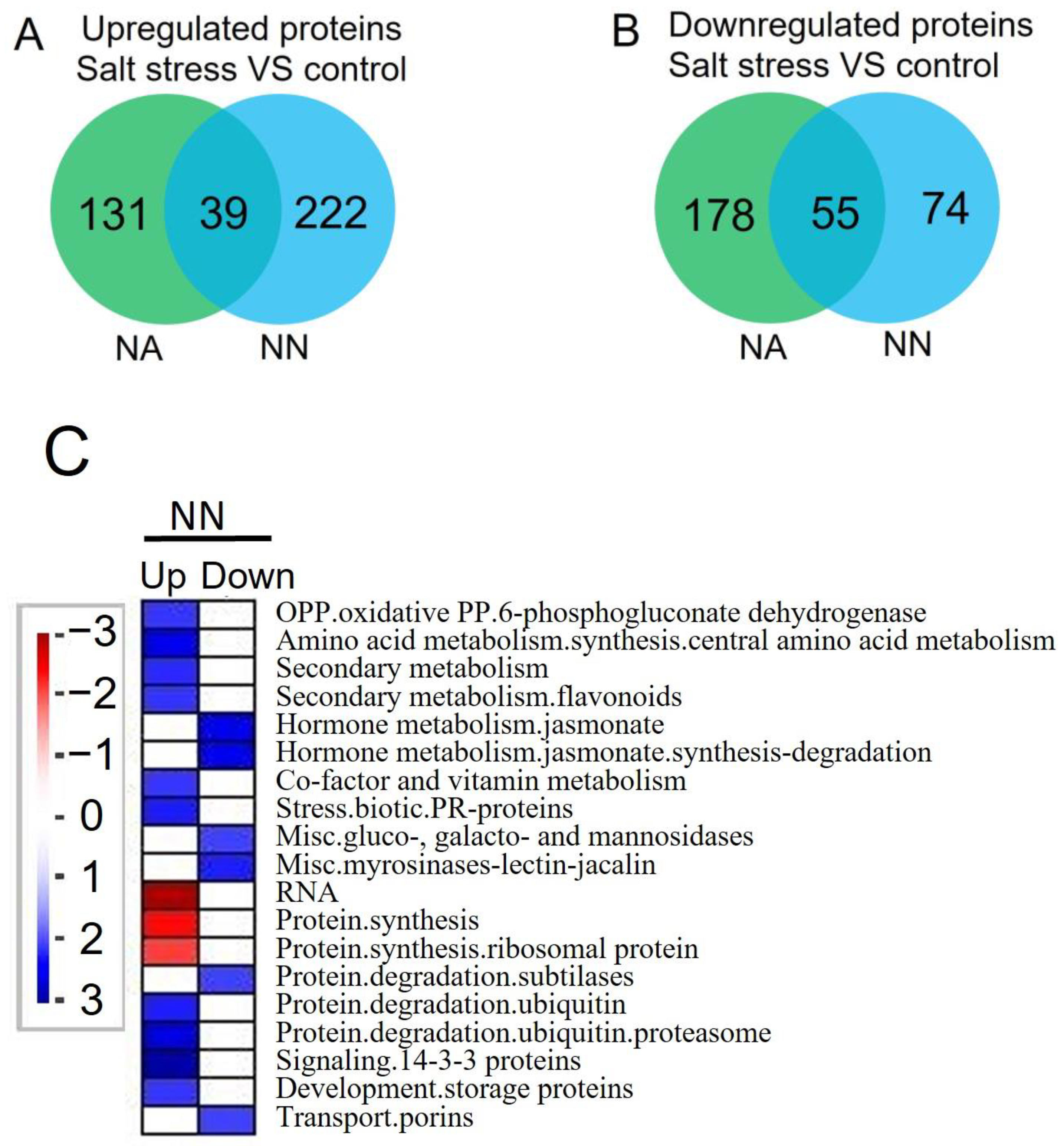
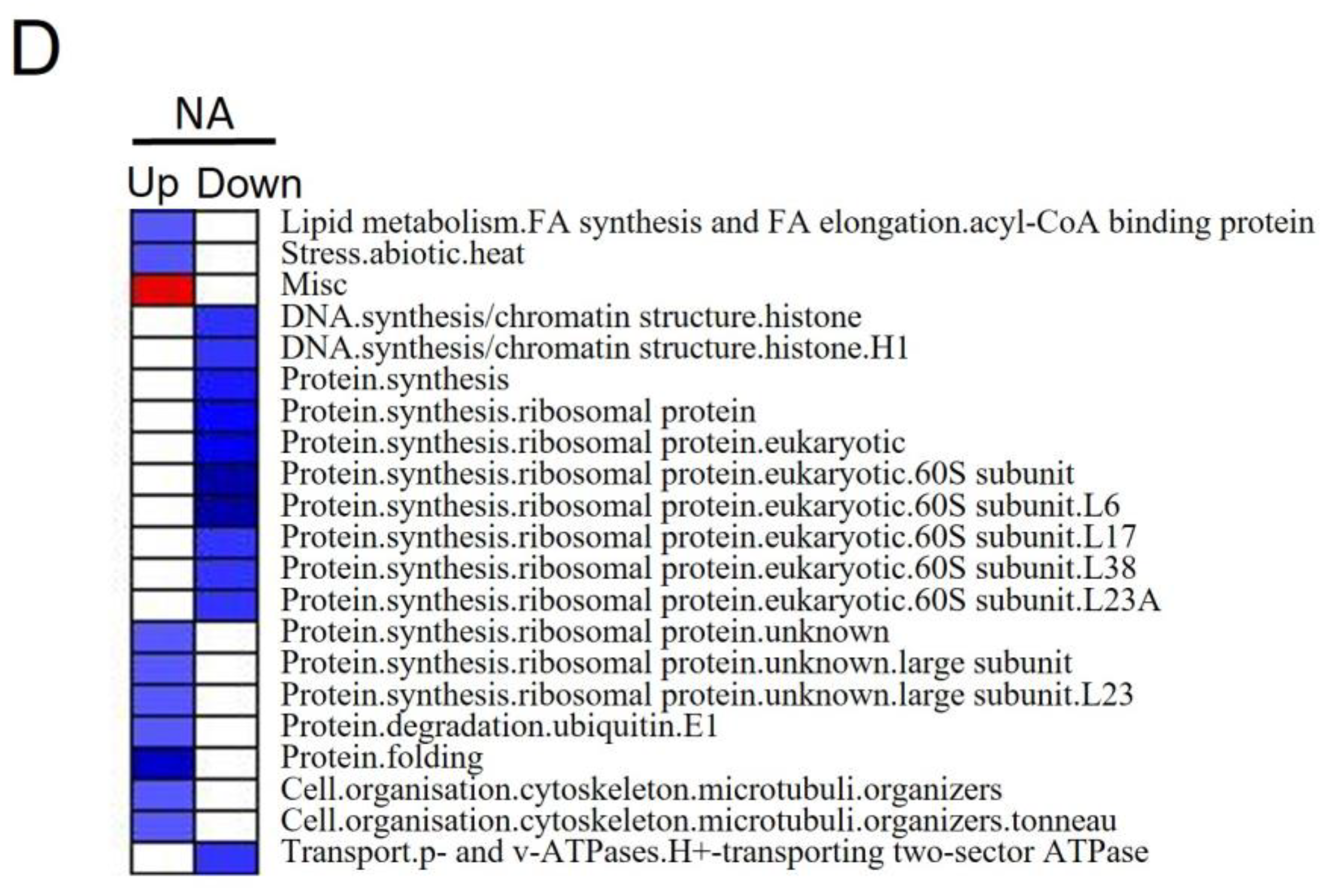
3.4. Differentially Expressed Proteins and Pathways Analysis of Alfalfa under Salt Stress
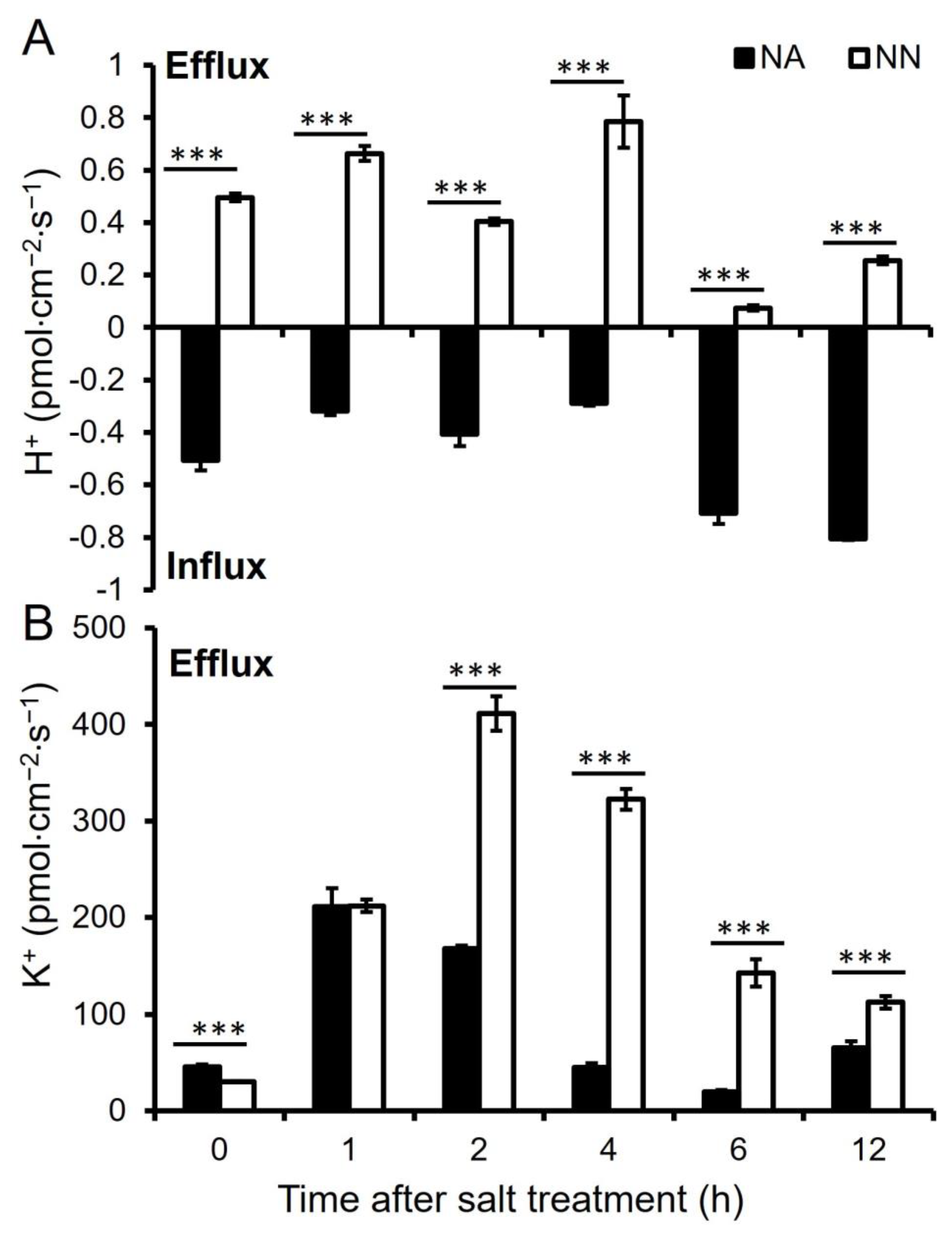
3.5. K+ and H+ Ion Flux
4. Discussion
4.1. Nodulation Triggered Pathways of the Stress Response, Antioxidant Enzymes, Energy-Saving, and Signaling Processes
4.1.1. Stress Response
4.1.2. Antioxidant Enzymes
4.1.3. Energy Consumption
4.1.4. Signaling
4.1.5. Cell Cytoskeleton
4.1.6. Protein Synthesis and Degradation
4.2. Nodulation Triggered Pathways That Conferred Salt Stress in Alfalfa Roots
4.2.1. Common Pathways for NA Roots under Control Conditions and Salt Stress
4.2.2. Unique Pathways under Salt Stress for NA Roots
4.3. Alfalfa with or without Nodules Differently Responded to Salt Stress
4.3.1. Pathways That Participated in Salt Stress in NN Roots
4.3.2. Pathways That Participated in Salt Stress in NA Roots
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tejerizo, G.T.; Papa, M.F.D.; Soria-Diaz, M.E.; Draghi, W.; Lozano, M.; Giusti, M.d.l.Á.; Manyani, H.; Megías, M.; Serrano, A.G.; Pühler, A.; et al. The nodulation of alfalfa by the acid-tolerant Rhizobium sp. strain LPU83 does not require sulfated forms of lipochitooligosaccharide nodulation signals. J. Bacteriol. 2011, 193, 30–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzaki, T.; Yoro, E.; Kawaguchi, M. Leguminous plants: Inventors of root nodules to accommodate symbiotic bacteria. Int. Rev. Cell Mol. Biol. 2015, 316, 111–158. [Google Scholar] [CrossRef] [PubMed]
- Clúa, J.; Roda, C.; Zanetti, M.E.; Blanco, F.A. Compatibility between legumes and rhizobia for the establishment of a successful nitrogen-fixing symbiosis. Genes 2018, 9, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iantcheva, A.; Mysore, K.S.; Ratet, P. Transformation of leguminous plants to study symbiotic interactions. Int. J. Dev. Biol. 2013, 57, 577–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaiswal, S.K.; Mohammed, M.; Ibny, F.Y.I.; Dakora, F.D. Rhizobia as a source of plant growth-promoting molecules: Potential applications and possible operational mechanisms. Front. Sustain. Food Syst. 2021, 4, 619676. [Google Scholar] [CrossRef]
- Hawkins, J.P.; Oresnik, I.J. The rhizobium-legume symbiosis: Co-opting successful stress management. Front. Plant Sci. 2021, 12, 796045. [Google Scholar] [CrossRef]
- Liang, Y.; Cao, Y.; Tanaka, K.; Thibivilliers, S.; Wan, J.; Choi, J.; Kang, C.; Qiu, J.; Stacey, G. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 2013, 341, 1384–1387. [Google Scholar] [CrossRef]
- El Yahyaoui, F.; Kuster, H.; Ben Amor, B.; Hohnjec, N.; Puhler, A.; Becker, A.; Gouzy, J.; Vernie, T.; Gough, C.; Niebel, A.; et al. Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 2004, 136, 3159–3176. [Google Scholar] [CrossRef] [Green Version]
- Silva, L.R.; Bento, C.; Gonçalves, A.C.; Flores-Félix, J.D.; Ramírez-Bahena, M.H.; Peix, A.; Velázquez, E. Legume bioactive compounds: Influence of rhizobial inoculation. AIMS Microbiol. 2017, 3, 267–278. [Google Scholar] [CrossRef] [Green Version]
- Dakora, F.D.; Matiru, V.N.; Kanu, A. Rhizosphere ecology of lumichrome and riboflavin, two bacterial signal molecules eliciting developmental changes in plants. Front. Plant Sci. 2015, 6, 700. [Google Scholar] [CrossRef]
- Bal, H.B.; Das, S.; Dangar, T.K.; Adhya, T.K. ACC deaminase and IAA producing growth promoting bacteria from the rhizosphere soil of tropical rice plants. J. Basic Microb. 2013, 53, 972–984. [Google Scholar] [CrossRef]
- Matiru, V.N.; Dakora, F.D. Xylem transport and shoot accumulation of lumichrome, a newly recognized rhizobial signal, alters root respiration, stomatal conductance, leaf transpiration and photosynthetic rates in legumes and cereals. New Phytol. 2005, 165, 847–855. [Google Scholar] [CrossRef]
- Lodeiro, A.R.; Gonzalez, P.; Hernandez, A.; Balague, L.J.; Favelukes, G. Comparison of drought tolerance in nitrogen-fixing and inorganic nitrogen-grown common beans. Plant Sci. 2000, 154, 31–41. [Google Scholar] [CrossRef]
- El-Akhal, M.R.; Rincon, A.; Coba de la Pena, T.; Lucas, M.M.; El Mourabit, N.; Barrijal, S.; Pueyo, J.J. Effects of salt stress and rhizobial inoculation on growth and nitrogen fixation of three peanut cultivars. Plant Biol. 2013, 15, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Frechilla, S.; Gonzalez, E.M.; Royuela, M.; Minchin, F.R.; Aparicio-Tejo, P.M.; Arrese-Igor, C. Source of nitrogen nutrition (nitrogen fixation or nitrate assimilation) is a major factor involved in pea response to moderate water stress. J. Plant Physiol. 2000, 157, 609–617. [Google Scholar] [CrossRef]
- Bhattarai, S.; Biswas, D.; Fu, Y.-B.; Biligetu, B. Morphological, physiological, and genetic responses to salt stress in alfalfa: A review. Agronomy 2020, 10, 577. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.F.; Zhang, Z.Q.; Zhang, P.; Cao, Y.M.; Hu, T.M.; Yang, P.Z. Rhizobium symbiosis contribution to short-term salt stress tolerance in alfalfa (Medicago sativa L.). Plant Soil 2016, 402, 247–261. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, P.; Zhou, Y.; Hu, T.; Zhang, P.; Wu, Y. A proteomic approach to understand the impact of nodulation on salinity stress response in alfalfa (Medicago sativa L.). Plant Biol. 2021, 24, 323–332. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Dreywood, R. Qualitative test for carbohydrate material. Ind. Eng. Chem. Anal. Ed. 1946, 18, 499. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutases: I. Occurrence in higher plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Raza, S.; Athar, H.; Ashraf, M.; Hameed, A. Glycinebetaine-induced modulation of antioxidant enzymes activities and ion accumulation in two wheat cultivars differing in salt tolerance. Environ. Exp. Bot. 2007, 60, 368–376. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Lansing, H.; Doering, L.; Fischer, K.; Baune, M.C.; von Schaewen, A. Analysis of potential redundancy among Arabidopsis 6-phosphogluconolactonase isoforms in peroxisomes. J. Exp. Bot. 2020, 71, 823–836. [Google Scholar] [CrossRef]
- Xiong, Y.Q.; DeFraia, C.; Williams, D.; Zhang, X.D.; Mou, Z.L. Characterization of Arabidopsis 6-phosphogluconolactonase T-DNA insertion mutants reveals an essential role for the oxidative section of the plastidic pentose phosphate pathway in plant growth and development. Plant Cell Physiol. 2009, 50, 1277–1291. [Google Scholar] [CrossRef]
- Jones, A.M.E.; Thomas, V.; Bennett, M.H.; Mansfield, J.; Grant, M. Modifications to the arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 2006, 142, 1603–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabsztunowicz, M.; Rantala, M.; Ivanauskaite, A.; Blomster, T.; Koskela, M.M.; Vuorinen, K.; Tyystjarvi, E.; Burow, M.; Overmyer, K.; Mahonen, A.P.; et al. Root-type ferredoxin-NADP+ oxidoreductase isoforms in Arabidopsis thaliana: Expression patterns, location and stress responses. Plant Cell Environ. 2021, 44, 548–558. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.N.; Singh, R.K.; Zhao, P.; Martins, V.; Aguilar, E.; Canto, T.; Tenllado, F.; Franklin, G.; Dias, A.C.P. Overexpression of polygalacturonase-inhibiting protein (PGIP) gene from Hypericum perforatum alters expression of multiple defense-related genes and modulates recalcitrance to Agrobacterium tumefaciens in tobacco. J. Plant Physiol. 2020, 253, 153268. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.; Khurana, J.P. Role of pathogenesis-related (PR) proteins in plant defense mechanism. In Molecular Aspects of Plant-Pathogen Interaction; Singh, A., Singh, I.K., Eds.; Springer: Singapore, 2018; pp. 265–281. [Google Scholar]
- Kasprzewska, A. Plant chitinases-regulation and function. Cell Mol. Biol. Lett. 2003, 8, 809–824. [Google Scholar]
- Sels, J.; Mathys, J.; De Coninck, B.M.A.; Cammue, B.P.A.; De Bolle, M.F.C. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef]
- Wojtasik, W.; Kulma, A.; Dyminska, L.; Hanuza, J.; Zebrowski, J.; Szopa, J. Fibres from flax overproducing beta-1,3-glucanase show increased accumulation of pectin and phenolics and thus higher antioxidant capacity. BMC Biotechnol. 2013, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Lim, G.H.; Kim, E.S.; Ko, C.B.; Yang, K.Y.; Jeong, J.A.; Lee, M.C.; Kim, C.S. Abiotic and biotic stress tolerance in Arabidopsis overexpressing the Multiprotein bridging factor 1a (MBF1a) transcriptional coactivator gene. Biochem. Biophys. Res. Commun. 2007, 354, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Vaish, S.; Gupta, D.; Mehrotra, R.; Mehrotra, S.; Basantani, M.K. Glutathione S-transferase: A versatile protein family. 3 Biotech 2020, 10, 321. [Google Scholar] [CrossRef] [PubMed]
- Shigeto, J.; Tsutsumi, Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2016, 209, 1395–1402. [Google Scholar] [CrossRef] [Green Version]
- Saripalli, G.; Gupta, P.K. AGPase: Its role in crop productivity with emphasis on heat tolerance in cereals. Theor. Appl. Genet. 2015, 128, 1893–1916. [Google Scholar] [CrossRef]
- Ohlrogge, J.B.; Jaworski, J.G. Regulation of fatty acid synthesis. Annu. Rev. Plant Physiol. 1997, 48, 109–136. [Google Scholar] [CrossRef] [Green Version]
- Matos, A.R.; Gigon, A.; Laffray, D.; Petres, S.; Zuily-Fodil, Y.; Pham-Thi, A.T. Effects of progressive drought stress on the expression of patatin-like lipid acyl hydrolase genes in Arabidopsis leaves. Physiol. Plant 2008, 134, 110–120. [Google Scholar] [CrossRef]
- Lin, F.; Li, S.; Wang, K.; Tian, H.; Gao, J.; Zhao, Q.; Du, C. A leucine-rich repeat receptor-like kinase, OsSTLK, modulates salt tolerance in rice. Plant Sci. 2020, 296, 110465. [Google Scholar] [CrossRef]
- Janke, C.; Magiera, M.M. The tubulin code and its role in controlling microtubule properties and functions. Nat Rev. Mol. Cell Biol. 2020, 21, 307–326. [Google Scholar] [CrossRef]
- Mao, T.; Jin, L.; Li, H.; Liu, B.; Yuan, M. Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiol. 2005, 138, 654–662. [Google Scholar] [CrossRef] [Green Version]
- Ostrowska, Z.; Moraczewska, J. Cofilin—A protein controlling dynamics of actin filaments. Postepy Hig. Med. Dosw. Online 2017, 71, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Kusters, R.; Caorsi, V.; Allard, A.; Abou-Ghali, M.; Manzi, J.; Di Cicco, A.; Lévy, D.; Lenz, M.; Joanny, J.-F.; et al. Actin dynamics drive cell-like membrane deformation. Nat. Phys. 2019, 15, 602–609. [Google Scholar] [CrossRef]
- Ramakrishnan, V. The eukaryotic ribosome. Science 2011, 331, 681–682. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.K.; Shabala, S. Mechanisms of plant responses and adaptation to soil salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, S.A.; Kini, K.R.; Raj, S.N.; Moerschbacher, B.M.; Shetty, H.S. Polygalacturonase-inhibitor proteins in pearl millet: Possible involvement in resistance against downy mildew. Acta Biochim. Biophys. Sin. 2012, 44, 415–423. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Cosgrove, D.J. Adaptation of roots to low water potentials by changes in cell wall extensibility and cell wall proteins. J. Exp. Bot. 2000, 51, 1543–1553. [Google Scholar] [CrossRef] [Green Version]
- Ishida, K.; Yokoyama, R. Reconsidering the function of the xyloglucan endotransglucosylase/hydrolase family. J. Plant Res. 2022, 135, 145–156. [Google Scholar] [CrossRef]
- Bellande, K.; Bono, J.J.; Savelli, B.; Jamet, E.; Canut, H. Plant lectins and lectin receptor-like kinases: How do they sense the outside? Int. J. Mol. Sci. 2017, 18, 1164. [Google Scholar] [CrossRef]
- Katoch, R.; Tripathi, A. Research advances and prospects of legume lectins. J. Biosci. 2021, 46, 104. [Google Scholar] [CrossRef]
- Sun, Y.; Wu, Z.; Wang, Y.; Yang, J.; Wei, G.; Chou, M. Identification of phytocyanin gene family in legume plants and their involvement in nodulation of Medicago truncatula. Plant Cell Physiol. 2019, 60, 900–915. [Google Scholar] [CrossRef]
- Kong, Y.; Li, X.; Wang, B.; Li, W.; Du, H.; Zhang, C. The soybean purple acid phosphatase GmPAP14 predominantly enhances external phytate utilization in plants. Front. Plant Sci. 2018, 9, 292. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Tateda, C. The functions of voltage-dependent anion channels in plants. Apoptosis 2013, 18, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Duan, X.; Wang, Z.; Yin, H.; Zang, J.; Zhu, K.; Wang, Y.; Zhang, P. Overexpression of a voltage-dependent anion-selective channel (VDAC) protein-encoding gene, MsVDAC, from Medicago sativa confers cold and drought tolerance to transgenic tobacco. Genes 2021, 12, 1706. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, C.; Xue, Y.; Liu, X.; Chen, S.; Song, C.; Yang, Y.; Guo, Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 2019, 10, 1199. [Google Scholar] [CrossRef] [Green Version]
- Meng, W.; Su, Y.C.F.; Saunders, R.M.K.; Chye, M.L. The rice acyl-CoA-binding protein gene family: Phylogeny, expression and functional analysis. New Phytol. 2011, 189, 1170–1184. [Google Scholar] [CrossRef]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Akhmanova, A. Microtubule-organizing centers. Annu. Rev. Cell Dev. Biol. 2017, 33, 51–75. [Google Scholar] [CrossRef]
- Zhang, X.H.; Li, B.; Hu, Y.G.; Chen, L.; Min, D.H. The wheat E subunit of V-type H+-ATPase is involved in the plant response to osmotic stress. Int. J. Mol. Sci. 2014, 15, 16196–16210. [Google Scholar] [CrossRef] [Green Version]
- Assaha, D.V.M.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The role of Na+ and K+ transporters in salt stress adaptation in Glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhang, P.; Li, L.; Li, D.; Liang, Z.; Cao, Y.; Hu, T.; Yang, P. Proteomic Analysis of Alfalfa (Medicago sativa L.) Roots in Response to Rhizobium Nodulation and Salt Stress. Genes 2022, 13, 2004. https://doi.org/10.3390/genes13112004
Wang Y, Zhang P, Li L, Li D, Liang Z, Cao Y, Hu T, Yang P. Proteomic Analysis of Alfalfa (Medicago sativa L.) Roots in Response to Rhizobium Nodulation and Salt Stress. Genes. 2022; 13(11):2004. https://doi.org/10.3390/genes13112004
Chicago/Turabian StyleWang, Yafang, Pan Zhang, Le Li, Danning Li, Zheng Liang, Yuman Cao, Tianming Hu, and Peizhi Yang. 2022. "Proteomic Analysis of Alfalfa (Medicago sativa L.) Roots in Response to Rhizobium Nodulation and Salt Stress" Genes 13, no. 11: 2004. https://doi.org/10.3390/genes13112004
APA StyleWang, Y., Zhang, P., Li, L., Li, D., Liang, Z., Cao, Y., Hu, T., & Yang, P. (2022). Proteomic Analysis of Alfalfa (Medicago sativa L.) Roots in Response to Rhizobium Nodulation and Salt Stress. Genes, 13(11), 2004. https://doi.org/10.3390/genes13112004






