Small Non-Coding RNAs in Human Cancer
Abstract
1. Introduction
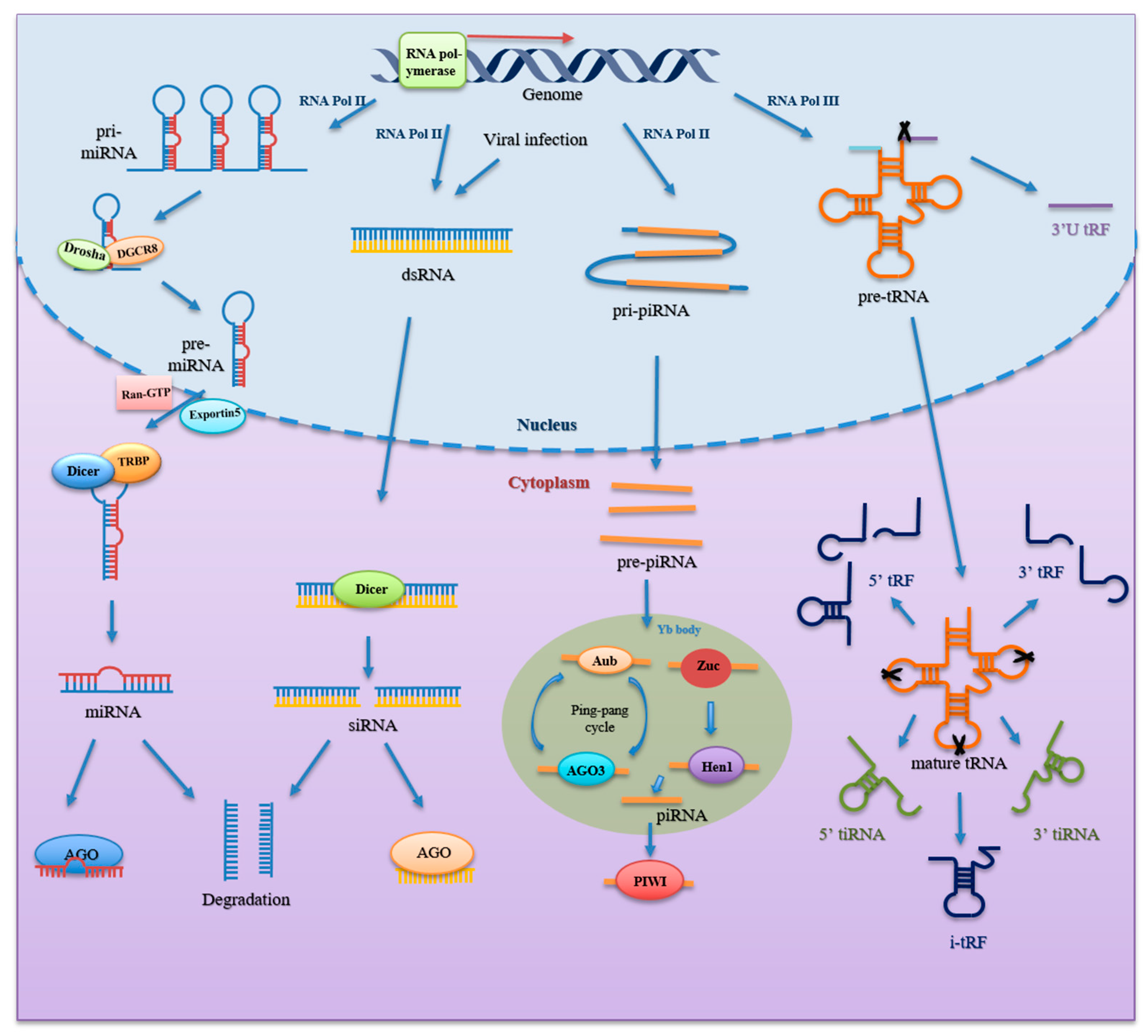
2. Biogenesis and Classification
2.1. miRNA
2.2. piRNA
2.3. tsRNA
2.4. siRNA
2.5. Other sncRNAs
3. Functions of sncRNAs
3.1. Transcriptional Regulation
3.2. Post-Transcriptional Regulation
3.3. Translation and Post-Translation Regulation
3.4. Reverse Transcription Regulation
4. Functions of Small Non-Coding RNAs in Cancer
4.1. Unlocking Phenotypic Plasticity
4.2. Non-Mutational Epigenetic Reprogramming
4.3. Polymorphic Microbiomes
4.4. Senescent Cells
5. Clinical Significances of Small Non-Coding RNAs in Cancer
5.1. Diagnostic and Prognostic Biomarker
5.2. Therapy
6. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frith, M.C.; Pheasant, M.; Mattick, J.S. The amazing complexity of the human transcriptome. Eur. J. Hum. Genet 2005, 13, 894–897. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, T.; Zhang, G.; He, Q.Y. Understanding the proteome encoded by “non-coding RNAs”: New insights into human genome. Sci. China Life Sci. 2020, 63, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Elgar, G.; Vavouri, T. Tuning in to the signals: Noncoding sequence conservation in vertebrate genomes. Trends. Genet 2008, 24, 344–352. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Grosshans, H.; Filipowicz, W. Molecular biology: The expanding world of small RNAs. Nature 2008, 451, 414–416. [Google Scholar] [CrossRef]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef]
- Babski, J.; Maier, L.K.; Heyer, R.; Jaschinski, K.; Prasse, D.; Jager, D.; Randau, L.; Schmitz, R.A.; Marchfelder, A.; Soppa, J. Small regulatory RNAs in Archaea. RNA Biol. 2014, 11, 484–493. [Google Scholar] [CrossRef]
- Shi, J.; Zhou, T.; Chen, Q. Exploring the expanding universe of small RNAs. Nat. Cell Biol. 2022, 24, 415–423. [Google Scholar] [CrossRef]
- Xiao, L.; Wang, J.; Ju, S.; Cui, M.; Jing, R. Disorders and roles of tsRNA, snoRNA, snRNA and piRNA in cancer. J. Med. Genet 2022, 59, 623–631. [Google Scholar] [CrossRef]
- Xu, D.; Qiao, D.; Lei, Y.; Zhang, C.; Bu, Y.; Zhang, Y. Transfer RNA-derived small RNAs (tsRNAs): Versatile regulators in cancer. Cancer Lett. 2022, 546, 215842. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Kim, V.N.; Nam, J.W. Genomics of microRNA. Trends. Genet 2006, 22, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Jo, M.H.; Choi, Y.G.; Park, J.; Kwon, S.C.; Hohng, S.; Kim, V.N.; Woo, J.S. Functional Anatomy of the Human Microprocessor. Cell 2015, 161, 1374–1387. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef]
- Park, J.E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5’ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.C.; Tambe, A.; Kidwell, M.A.; Noland, C.L.; Schneider, C.P.; Doudna, J.A. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell 2015, 57, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Kwak, P.B.; Tomari, Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 2012, 19, 145–151. [Google Scholar] [CrossRef]
- Gu, S.; Jin, L.; Zhang, F.; Huang, Y.; Grimm, D.; Rossi, J.J.; Kay, M.A. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc. Natl. Acad. Sci. USA 2011, 108, 9208–9213. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef]
- Grivna, S.T.; Beyret, E.; Wang, Z.; Lin, H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006, 20, 1709–1714. [Google Scholar] [CrossRef]
- Li, X.Z.; Roy, C.K.; Dong, X.; Bolcun-Filas, E.; Wang, J.; Han, B.W.; Xu, J.; Moore, M.J.; Schimenti, J.C.; Weng, Z.; et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell 2013, 50, 67–81. [Google Scholar] [CrossRef]
- Ray, R.; Pandey, P. piRNA analysis framework from small RNA-Seq data by a novel cluster prediction tool—PILFER. Genomics 2018, 110, 355–365. [Google Scholar] [CrossRef]
- Huang, Y.; Bai, J.Y.; Ren, H.T. PiRNAs biogenesis and its functions. Bioorg. Khim. 2014, 40, 320–326. [Google Scholar] [PubMed]
- Olivieri, D.; Sykora, M.M.; Sachidanandam, R.; Mechtler, K.; Brennecke, J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010, 29, 3301–3317. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Ishizu, H.; Saito, K.; Fukuhara, S.; Kamatani, M.K.; Bonnefond, L.; Matsumoto, N.; Nishizawa, T.; Nakanaga, K.; Aoki, J.; et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 2012, 491, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Han, B.W.; Wang, W.; Li, C.; Weng, Z.; Zamore, P.D. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 2015, 348, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, Y.W.; Siomi, M.C.; Siomi, H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 2015, 84, 405–433. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Hannon, G.J. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends. Biochem. Sci. 2016, 41, 324–337. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Li, Y.; Luo, J.; Zhou, H.; Liao, J.Y.; Ma, L.M.; Chen, Y.Q.; Qu, L.H. Stress-induced tRNA-derived RNAs: A novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008, 36, 6048–6055. [Google Scholar] [CrossRef]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, X.; Shi, J.; Yan, M.; Zhou, T. Origins and evolving functionalities of tRNA-derived small RNAs. Trends. Biochem. Sci. 2021, 46, 790–804. [Google Scholar] [CrossRef]
- Yamasaki, S.; Ivanov, P.; Hu, G.F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009, 583, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Collins, K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005, 280, 42744–42749. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Lu, C.; Green, P.J.; Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008, 14, 2095–2103. [Google Scholar] [CrossRef]
- Jochl, C.; Rederstorff, M.; Hertel, J.; Stadler, P.F.; Hofacker, I.L.; Schrettl, M.; Haas, H.; Huttenhofer, A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008, 36, 2677–2689. [Google Scholar] [CrossRef]
- Donovan, J.; Rath, S.; Kolet-Mandrikov, D.; Korennykh, A. Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 2017, 23, 1660–1671. [Google Scholar] [CrossRef]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef]
- Su, Z.; Wilson, B.; Kumar, P.; Dutta, A. Noncanonical Roles of tRNAs: tRNA Fragments and Beyond. Annu. Rev. Genet 2020, 54, 47–69. [Google Scholar] [CrossRef]
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef]
- Czech, B.; Hannon, G.J. Small RNA sorting: Matchmaking for Argonautes. Nat. Rev. Genet 2011, 12, 19–31. [Google Scholar] [CrossRef]
- Tam, O.H.; Aravin, A.A.; Stein, P.; Girard, A.; Murchison, E.P.; Cheloufi, S.; Hodges, E.; Anger, M.; Sachidanandam, R.; Schultz, R.M.; et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 2008, 453, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Shi, J.; Liu, H.; Zhang, X.; Zhou, J.J.; Li, M.; Zhou, D.; Li, R.; Lv, J.; Wen, G.; et al. Peripheral blood non-canonical small non-coding RNAs as novel biomarkers in lung cancer. Mol. Cancer 2020, 19, 159. [Google Scholar] [CrossRef] [PubMed]
- Cambier, L.; de Couto, G.; Ibrahim, A.; Echavez, A.K.; Valle, J.; Liu, W.; Kreke, M.; Smith, R.R.; Marban, L.; Marban, E. Y RNA fragment in extracellular vesicles confers cardioprotection via modulation of IL-10 expression and secretion. EMBO Mol. Med. 2017, 9, 337–352. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Heard, E. Small RNAs derived from structural non-coding RNAs. Methods 2013, 63, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Huang, R.; Li, L.; Zhu, J.; Li, Z.; Peng, C.; Zhuang, X.; Lin, H.; Shi, S.; Huang, P. CPA-seq reveals small ncRNAs with methylated nucleosides and diverse termini. Cell Discov. 2021, 7, 25. [Google Scholar] [CrossRef]
- Ender, C.; Krek, A.; Friedlander, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A human snoRNA with microRNA-like functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef]
- Hussain, S.; Sajini, A.A.; Blanco, S.; Dietmann, S.; Lombard, P.; Sugimoto, Y.; Paramor, M.; Gleeson, J.G.; Odom, D.T.; Ule, J.; et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013, 4, 255–261. [Google Scholar] [CrossRef]
- Huang, X.A.; Yin, H.; Sweeney, S.; Raha, D.; Snyder, M.; Lin, H. A major epigenetic programming mechanism guided by piRNAs. Dev. Cell 2013, 24, 502–516. [Google Scholar] [CrossRef]
- Yu, Y.; Gu, J.; Jin, Y.; Luo, Y.; Preall, J.B.; Ma, J.; Czech, B.; Hannon, G.J. Panoramix enforces piRNA-dependent cotranscriptional silencing. Science 2015, 350, 339–342. [Google Scholar] [CrossRef]
- Zhang, X.; He, X.; Liu, C.; Liu, J.; Hu, Q.; Pan, T.; Duan, X.; Liu, B.; Zhang, Y.; Chen, J.; et al. IL-4 Inhibits the Biogenesis of an Epigenetically Suppressive PIWI-Interacting RNA To Upregulate CD1a Molecules on Monocytes/Dendritic Cells. J. Immunol 2016, 196, 1591–1603. [Google Scholar] [CrossRef]
- Bai, Y.; Pan, B.; Zhan, X.; Silver, H.; Li, J. MicroRNA 195-5p Targets Foxo3 Promoter Region to Regulate Its Expression in Granulosa Cells. Int. J. Mol. Sci. 2021, 22, 6721. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.; Tao, Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer 2018, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Li, J.; Li, W.; Wang, Y.; Wu, F.; Xi, Y.; Zhang, L.; Ding, C.; Luo, H.; Li, Y.; et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017, 14, 1326–1334. [Google Scholar] [CrossRef] [PubMed]
- Elkayam, E.; Faehnle, C.R.; Morales, M.; Sun, J.; Li, H.; Joshua-Tor, L. Multivalent Recruitment of Human Argonaute by GW182. Mol. Cell 2017, 67, 646–658.e3. [Google Scholar] [CrossRef]
- Fabian, M.R.; Cieplak, M.K.; Frank, F.; Morita, M.; Green, J.; Srikumar, T.; Nagar, B.; Yamamoto, T.; Raught, B.; Duchaine, T.F.; et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 2011, 18, 1211–1217. [Google Scholar] [CrossRef]
- Braun, J.E.; Truffault, V.; Boland, A.; Huntzinger, E.; Chang, C.T.; Haas, G.; Weichenrieder, O.; Coles, M.; Izaurralde, E. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5’ exonucleolytic degradation. Nat. Struct. Mol. Biol. 2012, 19, 1324–1331. [Google Scholar] [CrossRef]
- Goh, W.S.; Falciatori, I.; Tam, O.H.; Burgess, R.; Meikar, O.; Kotaja, N.; Hammell, M.; Hannon, G.J. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 2015, 29, 1032–1044. [Google Scholar] [CrossRef]
- Peng, L.; Song, L.; Liu, C.; Lv, X.; Li, X.; Jie, J.; Zhao, D.; Li, D. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumour. Biol. 2016, 37, 2749–2756. [Google Scholar] [CrossRef]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef]
- Balatti, V.; Rizzotto, L.; Miller, C.; Palamarchuk, A.; Fadda, P.; Pandolfo, R.; Rassenti, L.Z.; Hertlein, E.; Ruppert, A.S.; Lozanski, A.; et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2015, 112, 2169–2174. [Google Scholar] [CrossRef]
- Zhong, F.; Zhou, N.; Wu, K.; Guo, Y.; Tan, W.; Zhang, H.; Zhang, X.; Geng, G.; Pan, T.; Luo, H.; et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015, 43, 10474–10491. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.; Yim, D.G.; Lakshmanan, V.; Tirumalai, V.; Koh, J.L.; Park, J.E.; Cheong, J.K.; Low, J.L.; Lim, M.J.; Sze, S.K.; et al. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 2019, 20, e47789. [Google Scholar] [CrossRef]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Rudland, P.S.; Dube, S.K. Specific interaction of an initiator tRNA fragment with 30 s ribosomal subunits. J. Mol. Biol. 1969, 43, 273–280. [Google Scholar] [CrossRef]
- Gebetsberger, J.; Wyss, L.; Mleczko, A.M.; Reuther, J.; Polacek, N. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 2017, 14, 1364–1373. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 2011, 43, 613–623. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, Y.; Zhou, T.; Chen, Q. tsRNAs: The Swiss Army Knife for Translational Regulation. Trends. Biochem. Sci. 2019, 44, 185–189. [Google Scholar] [CrossRef]
- Kim, H.K.; Fuchs, G.; Wang, S.; Wei, W.; Zhang, Y.; Park, H.; Roy-Chaudhuri, B.; Li, P.; Xu, J.; Chu, K.; et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 2017, 552, 57–62. [Google Scholar] [CrossRef]
- Kim, H.K.; Xu, J.; Chu, K.; Park, H.; Jang, H.; Li, P.; Valdmanis, P.N.; Zhang, Q.C.; Kay, M.A. A tRNA-Derived Small RNA Regulates Ribosomal Protein S28 Protein Levels after Translation Initiation in Humans and Mice. Cell Rep. 2019, 29, 3816–3824.e3814. [Google Scholar] [CrossRef]
- Fricker, R.; Brogli, R.; Luidalepp, H.; Wyss, L.; Fasnacht, M.; Joss, O.; Zywicki, M.; Helm, M.; Schneider, A.; Cristodero, M.; et al. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat. Commun. 2019, 10, 118. [Google Scholar] [CrossRef]
- Pan, L.; Huang, X.; Liu, Z.X.; Ye, Y.; Li, R.; Zhang, J.; Wu, G.; Bai, R.; Zhuang, L.; Wei, L.; et al. Inflammatory cytokine-regulated tRNA-derived fragment tRF-21 suppresses pancreatic ductal adenocarcinoma progression. J. Clin. Investig. 2021, 131, e148130. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Gou, L.T.; Zhang, M.; Zu, L.D.; Hua, M.M.; Hua, Y.; Shi, H.J.; Li, Y.; Li, J.; Li, D.; et al. piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev. Cell 2013, 24, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Jiang, X.Y.; Qi, W.; Ji, C.G.; Xie, X.L.; Zhang, D.X.; Cui, Z.J.; Wang, C.K.; Bai, Y.; Wang, J.; et al. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci. 2017, 108, 1746–1756. [Google Scholar] [CrossRef]
- Mai, D.; Ding, P.; Tan, L.; Zhang, J.; Pan, Z.; Bai, R.; Li, C.; Li, M.; Zhou, Y.; Tan, W.; et al. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics 2018, 8, 5213–5230. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef] [PubMed]
- Nunes, A.; Ribeiro, D.R.; Marques, M.; Santos, M.A.S.; Ribeiro, D.; Soares, A.R. Emerging Roles of tRNAs in RNA Virus Infections. Trends. Biochem. Sci. 2020, 45, 794–805. [Google Scholar] [CrossRef]
- Ruggero, K.; Guffanti, A.; Corradin, A.; Sharma, V.K.; De Bellis, G.; Corti, G.; Grassi, A.; Zanovello, P.; Bronte, V.; Ciminale, V.; et al. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: A role for a tRNA fragment as a primer for reverse transcriptase. J. Virol. 2014, 88, 3612–3622. [Google Scholar] [CrossRef]
- Schorn, A.J.; Gutbrod, M.J.; LeBlanc, C.; Martienssen, R. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell 2017, 170, 61–71.e11. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ji, F.; Zhao, X.; Yang, X.; He, J.; Huang, L.; Zhang, Y. MicroRNA-371a-3p promotes progression of gastric cancer by targeting TOB1. Cancer Lett. 2019, 443, 179–188. [Google Scholar] [CrossRef]
- Cheng, J.; Deng, H.; Xiao, B.; Zhou, H.; Zhou, F.; Shen, Z.; Guo, J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012, 315, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, Z.; Yu, X.; Ruan, Y.; Shen, Y.; Shao, Y.; Zhang, X.; Ye, G.; Guo, J. The tRNA-derived fragment 5026a inhibits the proliferation of gastric cancer cells by regulating the PTEN/PI3K/AKT signaling pathway. Stem. Cell Res. Ther. 2021, 12, 418. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Zhao, Y.; Zhang, W.J.; Jiang, Y.J.; Fu, H.; Huang, F.; Li, D.J.; Shen, F.M. microRNA-802 accelerates hepatocellular carcinoma growth by targeting RUNX3. J. Cell Physiol. 2020, 235, 7128–7135. [Google Scholar] [CrossRef]
- Law, P.T.; Qin, H.; Ching, A.K.; Lai, K.P.; Co, N.N.; He, M.; Lung, R.W.; Chan, A.W.; Chan, T.F.; Wong, N. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J. Hepatol. 2013, 58, 1165–1173. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, J.; Liu, L.; Yan, M.; Zhang, Q.; Song, X.; Lin, Y.; Zhu, D.; Wei, Y.; Fu, Z.; et al. Gly-tRF enhances LCSC-like properties and promotes HCC cells migration by targeting NDFIP2. Cancer Cell Int. 2021, 21, 502. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, Y.; Li, Z.; Zhao, X.; Xi, Z.; Chen, H.; Shi, H.; Xin, T.; Shen, R.; Wang, T. MiR-4319 suppresses colorectal cancer progression by targeting ABTB1. United Eur. Gastroenterol. J. 2019, 7, 517–528. [Google Scholar] [CrossRef]
- Wang, S.; Jiang, X.; Xie, X.; Yin, J.; Zhang, J.; Liu, T.; Chen, S.; Wang, Y.; Zhou, X.; Wang, Y.; et al. piR-823 inhibits cell apoptosis via modulating mitophagy by binding to PINK1 in colorectal cancer. Cell Death Dis. 2022, 13, 465. [Google Scholar] [CrossRef]
- Tao, E.W.; Wang, H.L.; Cheng, W.Y.; Liu, Q.Q.; Chen, Y.X.; Gao, Q.Y. A specific tRNA half, 5’tiRNA-His-GTG, responds to hypoxia via the HIF1alpha/ANG axis and promotes colorectal cancer progression by regulating LATS2. J. Exp. Clin. Cancer Res. 2021, 40, 67. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tan, Z.Q.; Hu, H.; Liu, H.Z.; Wu, T.W.; Zheng, C.; Wang, X.L.; Luo, Z.Z.; Wang, J.; Liu, S.Y.; et al. microRNA-21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer 2019, 19, 738. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Mai, D.; Zhang, B.; Jiang, X.; Zhang, J.; Bai, R.; Ye, Y.; Li, M.; Pan, L.; Su, J.; et al. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol. Cancer 2019, 18, 9. [Google Scholar] [CrossRef]
- Mo, D.; Jiang, P.; Yang, Y.; Mao, X.; Tan, X.; Tang, X.; Wei, D.; Li, B.; Wang, X.; Tang, L.; et al. A tRNA fragment, 5′-tiRNA(Val), suppresses the Wnt/β-catenin signaling pathway by targeting FZD3 in breast cancer. Cancer Lett. 2019, 457, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, T.; Shang, D.; Sun, Z. miR-1254 promotes lung cancer cell proliferation by targeting SFRP1. BioMed Pharmacother. 2017, 92, 913–918. [Google Scholar] [CrossRef]
- Hu, F.; Niu, Y.J.; Mao, X.W.; Cui, J.T.; Wu, X.T.; Ii, C.B.S.; Kang, H.S.; Qin, W.X.; Jiang, L.Y. tsRNA-5001a promotes proliferation of lung adenocarcinoma cells and is associated with postoperative recurrence in lung adenocarcinoma patients. Transl. Lung Cancer Res. 2021, 10, 3957. [Google Scholar] [CrossRef]
- Wu, H.; Liu, C.; Yang, Q.; Xin, C.; Du, J.; Sun, F.; Zhou, L. MIR145-3p promotes autophagy and enhances bortezomib sensitivity in multiple myeloma by targeting HDAC4. Autophagy 2020, 16, 683–697. [Google Scholar] [CrossRef]
- Yan, H.; Wu, Q.L.; Sun, C.Y.; Ai, L.S.; Deng, J.; Zhang, L.; Chen, L.; Chu, Z.B.; Tang, B.; Wang, K.; et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 2015, 29, 196–206. [Google Scholar] [CrossRef]
- Hu, T.; Chong, Y.; Lu, S.; Wang, R.; Qin, H.; Silva, J.; Kitamura, E.; Chang, C.S.; Hawthorn, L.; Cowell, J.K. miR-339 Promotes Development of Stem Cell Leukemia/Lymphoma Syndrome via Downregulation of the BCL2L11 and BAX Proapoptotic Genes. Cancer Res. 2018, 78, 3522–3531. [Google Scholar] [CrossRef]
- Han, H.Y.; Fan, G.; Song, S.; Jiang, Y.X.; Qian, C.A.; Zhang, W.M.; Su, Q.; Xue, X.F.; Zhuang, W.Z.; Li, B.Z. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood 2021, 137, 1603–1614. [Google Scholar] [CrossRef]
- Chen, S.; Xu, M.; Zhao, J.; Shen, J.Q.; Li, J.H.; Liu, Y.; Cao, G.; Ma, J.C.; He, W.Z.; Chen, X.; et al. MicroRNA-4516 suppresses pancreatic cancer development via negatively regulating orthodenticle homeobox 1. Int. J. Biol. Sci. 2020, 16, 2159–2169. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Xing, S.; Shen, B.Y.; Chen, H.T.; Sun, B.; Wang, Z.T.; Wang, J.W.; Lu, X.X. PIWIL1 interacting RNA piR-017061 inhibits pancreatic cancer growth via regulating EFNA5. Hum. Cell 2021, 34, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Yang, L.; Wang, W.; Yuan, N.; Zhan, S.; Yang, P.; Chen, X.; Ma, T.; Wang, Y. A novel class of tsRNA signatures as biomarkers for diagnosis and prognosis of pancreatic cancer. Mol. Cancer 2021, 20, 95. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Norgard, R.J.; Stanger, B.Z. Cellular Plasticity in Cancer. Cancer Discov. 2019, 9, 837–851. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, C.; Campbell, J.D.; Gerrein, J.; Tellez, C.S.; Garrison, C.B.; Walser, T.C.; Drizik, E.; Si, H.; Gower, A.C.; Vick, J.; et al. MicroRNA 4423 is a primate-specific regulator of airway epithelial cell differentiation and lung carcinogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 18946–18951. [Google Scholar] [CrossRef]
- Fuziwara, C.S.; Saito, K.C.; Kimura, E.T. Thyroid Follicular Cell Loss of Differentiation Induced by MicroRNA miR-17-92 Cluster Is Attenuated by CRISPR/Cas9n Gene Silencing in Anaplastic Thyroid Cancer. Thyroid 2020, 30, 81–94. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, Y.; Xie, C.; Zhang, Y. miR-34a exerts as a key regulator in the dedifferentiation of osteosarcoma via PAI-1-Sox2 axis. Cell Death Dis. 2018, 9, 777. [Google Scholar] [CrossRef]
- Jones, M.F.; Hara, T.; Francis, P.; Li, X.L.; Bilke, S.; Zhu, Y.; Pineda, M.; Subramanian, M.; Bodmer, W.F.; Lal, A. The CDX1-microRNA-215 axis regulates colorectal cancer stem cell differentiation. Proc. Natl. Acad. Sci. USA 2015, 112, E1550–E1558. [Google Scholar] [CrossRef]
- Ma, Y.S.; Lv, Z.W.; Yu, F.; Chang, Z.Y.; Cong, X.L.; Zhong, X.M.; Lu, G.X.; Zhu, J.; Fu, D. MicroRNA-302a/d inhibits the self-renewal capability and cell cycle entry of liver cancer stem cells by targeting the E2F7/AKT axis. J. Exp. Clin. Cancer Res. 2018, 37, 252. [Google Scholar] [CrossRef]
- Huang, S. Tumor progression: Chance and necessity in Darwinian and Lamarckian somatic (mutationless) evolution. Prog. Biophys. Mol. Biol. 2012, 110, 69–86. [Google Scholar] [CrossRef]
- Darwiche, N. Epigenetic mechanisms and the hallmarks of cancer: An intimate affair. Am. J. Cancer Res. 2020, 10, 1954–1978. [Google Scholar] [PubMed]
- Feng, Y.; Liu, X.; Pauklin, S. 3D chromatin architecture and epigenetic regulation in cancer stem cells. Protein Cell 2021, 12, 440–454. [Google Scholar] [CrossRef] [PubMed]
- Nam, A.S.; Chaligne, R.; Landau, D.A. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat. Rev. Genet 2021, 22, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.D.; Jiang, H.; Zhang, Y.F.; Zhang, Y.; Qian, L.L.; Zhang, Y.F. The regulatory function of piRNA/PIWI complex in cancer and other human diseases: The role of DNA methylation. Int. J. Biol. Sci. 2022, 18, 3358–3373. [Google Scholar] [CrossRef]
- Chen, Q.; Yan, M.; Cao, Z.; Li, X.; Zhang, Y.; Shi, J.; Feng, G.H.; Peng, H.; Zhang, X.; Zhang, Y.; et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016, 351, 397–400. [Google Scholar] [CrossRef]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef]
- Dzutsev, A.; Badger, J.H.; Perez-Chanona, E.; Roy, S.; Salcedo, R.; Smith, C.K.; Trinchieri, G. Microbes and Cancer. Annu. Rev. Immunol. 2017, 35, 199–228. [Google Scholar] [CrossRef]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Guan, L.L. Noncoding RNAs: Regulatory Molecules of Host-Microbiome Crosstalk. Trends. Microbiol. 2021, 29, 713–724. [Google Scholar] [CrossRef]
- Dong, J.; Tai, J.W.; Lu, L.F. miRNA-Microbiota Interaction in Gut Homeostasis and Colorectal Cancer. Trends. Cancer 2019, 5, 666–669. [Google Scholar] [CrossRef]
- Burns, M.B.; Montassier, E.; Abrahante, J.; Priya, S.; Niccum, D.E.; Khoruts, A.; Starr, T.K.; Knights, D.; Blekhman, R. Colorectal cancer mutational profiles correlate with defined microbial communities in the tumor microenvironment. PLoS Genet 2018, 14, e1007376. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Liao, Y.; Zhang, H.; Zhang, W.; Zhang, Z.; Zhang, J.; Wang, D.; Tang, D. Impacts of MicroRNAs Induced by the Gut Microbiome on Regulating the Development of Colorectal Cancer. Front. Cell Infect. Microbiol. 2022, 12, 804689. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 2017, 152, 851–866.e824. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Kim, N.; Park, J.H.; Nam, R.H.; Choi, Y.J.; Lee, H.S.; Yoon, H.; Shin, C.M.; Park, Y.S.; Kim, J.M.; et al. Different MicroRNA Expression Levels in Gastric Cancer Depending on Helicobacter pylori Infection. Gut Liver 2015, 9, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Rahbar Saadat, Y.; Pourseif, M.M.; Zununi Vahed, S.; Barzegari, A.; Omidi, Y.; Barar, J. Modulatory Role of Vaginal-Isolated Lactococcus lactis on the Expression of miR-21, miR-200b, and TLR-4 in CAOV-4 Cells and In Silico Revalidation. Probiot. Antimicrob. Proteins 2020, 12, 1083–1096. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.H.; Yang, X.S.; Wang, F.L.; Cui, Z.M.; Huang, Y. MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int. J. Mol. Med. 2010, 26, 819–827. [Google Scholar] [CrossRef]
- Koutsaki, M.; Spandidos, D.A.; Zaravinos, A. Epithelial-mesenchymal transition-associated miRNAs in ovarian carcinoma, with highlight on the miR-200 family: Prognostic value and prospective role in ovarian cancer therapeutics. Cancer Lett. 2014, 351, 173–181. [Google Scholar] [CrossRef]
- Lee, S.; Schmitt, C.A. The dynamic nature of senescence in cancer. Nat. Cell Biol. 2019, 21, 94–101. [Google Scholar] [CrossRef]
- Ramalho-Carvalho, J.; Graca, I.; Gomez, A.; Oliveira, J.; Henrique, R.; Esteller, M.; Jeronimo, C. Downregulation of miR-130b similar to 301b cluster is mediated by aberrant promoter methylation and impairs cellular senescence in prostate cancer. J. Hematol. Oncol. 2017, 10, 281–297. [Google Scholar] [CrossRef]
- Su, W.; Hong, L.; Xu, X.; Huang, S.; Herpai, D.; Li, L.; Xu, Y.; Truong, L.; Hu, W.Y.; Wu, X.; et al. miR-30 disrupts senescence and promotes cancer by targeting both p16(INK4A) and DNA damage pathways. Oncogene 2018, 37, 5618–5632. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, X.; Qin, Z.L.; Wei, L.Y.; Lu, Y.J.; Peng, Q.; Gao, Y.X.; Zhang, X.M.; Zhang, X.Y.; Li, Z.S.; et al. Epstein-Barr virus miR-BART3-3p promotes tumorigenesis by regulating the senescence pathway in gastric cancer. J. Biol. Chem. 2019, 294, 4854–4866. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.Q.; Chen, H.B.; Zhang, T.Y.; Chen, Z.; Tian, L.; Gu, D.N. MicroRNA-7 modulates cellular senescence to relieve gemcitabine resistance by targeting PARP1/NF-kappaB signaling in pancreatic cancer cells. Oncol. Lett. 2021, 21, 139. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Kohli, J.; Demaria, M. Senescent Cells in Cancer Therapy: Friends or Foes? Trends. Cancer 2020, 6, 838–857. [Google Scholar] [CrossRef]
- Kowald, A.; Passos, J.F.; Kirkwood, T.B.L. On the evolution of cellular senescence. Aging Cell 2020, 19, e13270. [Google Scholar] [CrossRef] [PubMed]
- Birch, J.; Gil, J. Senescence and the SASP: Many therapeutic avenues. Genes Dev. 2020, 34, 1565–1576. [Google Scholar] [CrossRef]
- Faget, D.V.; Ren, Q.; Stewart, S.A. Unmasking senescence: Context-dependent effects of SASP in cancer. Nat. Rev. Cancer 2019, 19, 439–453. [Google Scholar] [CrossRef]
- He, S.; Sharpless, N.E. Senescence in Health and Disease. Cell 2017, 169, 1000–1011. [Google Scholar] [CrossRef]
- Kabir, T.D.; Leigh, R.J.; Tasena, H.; Mellone, M.; Coletta, R.D.; Parkinson, E.K.; Prime, S.S.; Thomas, G.J.; Paterson, I.C.; Zhou, D.; et al. A miR-335/COX-2/PTEN axis regulates the secretory phenotype of senescent cancer-associated fibroblasts. Aging 2016, 8, 1608–1635. [Google Scholar] [CrossRef]
- Imaoka, H.; Toiyama, Y.; Fujikawa, H.; Hiro, J.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Mori, T.; Kato, T.; et al. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann. Oncol. 2016, 27, 1879–1886. [Google Scholar] [CrossRef]
- Nakamura, K.; Sawada, K.; Yoshimura, A.; Kinose, Y.; Nakatsuka, E.; Kimura, T. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol. Cancer 2016, 15, 48. [Google Scholar] [CrossRef]
- Nassar, F.J.; Nasr, R.; Talhouk, R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol. Ther. 2017, 172, 34–49. [Google Scholar] [CrossRef] [PubMed]
- McDonald, A.C.; Vira, M.; Shen, J.; Sanda, M.; Raman, J.D.; Liao, J.; Patil, D.; Taioli, E. Circulating microRNAs in plasma as potential biomarkers for the early detection of prostate cancer. Prostate 2018, 78, 411–418. [Google Scholar] [CrossRef]
- Zhu, L.; Li, J.; Gong, Y.; Wu, Q.; Tan, S.; Sun, D.; Xu, X.; Zuo, Y.; Zhao, Y.; Wei, Y.Q.; et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer 2019, 18, 74. [Google Scholar] [CrossRef] [PubMed]
- Preethi, K.A.; Selvakumar, S.C.; Ross, K.; Jayaraman, S.; Tusubira, D.; Sekar, D. Liquid biopsy: Exosomal microRNAs as novel diagnostic and prognostic biomarkers in cancer. Mol. Cancer 2022, 21, 54. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Lin, Y.; Luo, Y.; Xiong, X.; Wang, L.; Durante, K.; Li, J.; Zhou, F.; Guo, Y.; Chen, S.; et al. A signature of saliva-derived exosomal small RNAs as predicting biomarker for esophageal carcinoma: A multicenter prospective study. Mol. Cancer 2022, 21, 21. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, W.; Wang, F.; Yang, S.; Hu, J.; Lu, B.; Pan, Z.; Ma, Y.; Zheng, M.; Zhou, L.; et al. Plasma-derived exosomal miR-15a-5p as a promising diagnostic biomarker for early detection of endometrial carcinoma. Mol. Cancer 2021, 20, 57. [Google Scholar] [CrossRef]
- Zhong, S.; Golpon, H.; Zardo, P.; Borlak, J. miRNAs in lung cancer. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl. Res. 2021, 230, 164–196. [Google Scholar] [CrossRef]
- Gao, Z.; Jijiwa, M.; Nasu, M.; Borgard, H.; Gong, T.; Xu, J.; Chen, S.; Fu, Y.; Chen, Y.; Hu, X.; et al. Comprehensive landscape of tRNA-derived fragments in lung cancer. Mol. Ther. Oncolytics 2022, 26, 207–225. [Google Scholar] [CrossRef]
- Li, D.; Luo, Y.; Gao, Y.; Yang, Y.; Wang, Y.; Xu, Y.; Tan, S.; Zhang, Y.; Duan, J.; Yang, Y. piR-651 promotes tumor formation in non-small cell lung carcinoma through the upregulation of cyclin D1 and CDK4. Int. J. Mol. Med. 2016, 38, 927–936. [Google Scholar] [CrossRef]
- Tang, S.; Li, S.; Liu, T.; He, Y.; Hu, H.; Zhu, Y.; Tang, S.; Zhou, H. MicroRNAs: Emerging oncogenic and tumor-suppressive regulators, biomarkers and therapeutic targets in lung cancer. Cancer Lett. 2021, 502, 71–83. [Google Scholar] [CrossRef]
- Yang, W.; Gao, K.; Qian, Y.; Huang, Y.; Xiang, Q.; Chen, C.; Chen, Q.; Wang, Y.; Fang, F.; He, Q.; et al. A novel tRNA-derived fragment AS-tDR-007333 promotes the malignancy of NSCLC via the HSPB1/MED29 and ELK4/MED29 axes. J. Hematol. Oncol. 2022, 15, 53. [Google Scholar] [CrossRef] [PubMed]
- Giglio, S.; De Nunzio, C.; Cirombella, R.; Stoppacciaro, A.; Faruq, O.; Volinia, S.; Baldassarre, G.; Tubaro, A.; Ishii, H.; Croce, C.M.; et al. A preliminary study of micro-RNAs as minimally invasive biomarkers for the diagnosis of prostate cancer patients. J. Exp. Clin. Cancer Res. 2021, 40, 79. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.P.; Lai, H.M.; Guo, Z. Prostate cancer early diagnosis: Circulating microRNA pairs potentially beyond single microRNAs upon 1231 serum samples. Brief. Bioinform. 2021, 22, bbaa111. [Google Scholar] [CrossRef] [PubMed]
- Baumann, B.; Acosta, A.M.; Richards, Z.; Deaton, R.; Sapatynska, A.; Murphy, A.; Kajdacsy-Balla, A.; Gann, P.H.; Nonn, L. Association of High miR-182 Levels with Low-Risk Prostate Cancer. Am. J. Pathol. 2019, 189, 911–923. [Google Scholar] [CrossRef]
- Wang, J.; Ni, J.; Beretov, J.; Thompson, J.; Graham, P.; Li, Y. Exosomal microRNAs as liquid biopsy biomarkers in prostate cancer. Crit. Rev. Oncol. Hematol. 2020, 145, 102860. [Google Scholar] [CrossRef]
- So, J.B.Y.; Kapoor, R.; Zhu, F.; Koh, C.; Zhou, L.; Zou, R.; Tang, Y.C.; Goo, P.C.K.; Rha, S.Y.; Chung, H.C.; et al. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut 2021, 70, 829–837. [Google Scholar] [CrossRef]
- Ouyang, J.; Xie, Z.; Lei, X.; Tang, G.; Gan, R.; Yang, X. Clinical crosstalk between microRNAs and gastric cancer (Review). Int. J. Oncol. 2021, 58, 7. [Google Scholar] [CrossRef]
- Abe, S.; Matsuzaki, J.; Sudo, K.; Oda, I.; Katai, H.; Kato, K.; Takizawa, S.; Sakamoto, H.; Takeshita, F.; Niida, S.; et al. A novel combination of serum microRNAs for the detection of early gastric cancer. Gastric. Cancer 2021, 24, 835–843. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, X.; Qin, X.; Huang, Y.; Ju, S. Evaluation of serum tRF-23-Q99P9P9NDD as a potential biomarker for the clinical diagnosis of gastric cancer. Mol. Med. 2022, 28, 63. [Google Scholar] [CrossRef]
- Chen, B.; Xia, Z.; Deng, Y.N.; Yang, Y.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. Emerging microRNA biomarkers for colorectal cancer diagnosis and prognosis. Open Biol. 2019, 9, 180212. [Google Scholar] [CrossRef]
- Chen, H.; Xu, Z.; Cai, H.; Peng, Y.; Yang, L.; Wang, Z. Identifying Differentially Expressed tRNA-Derived Small Fragments as a Biomarker for the Progression and Metastasis of Colorectal Cancer. Dis. Markers 2022, 2022, 2646173. [Google Scholar] [CrossRef] [PubMed]
- Ahluwalia, P.; Kolhe, R.; Gahlay, G.K. The clinical relevance of gene expression based prognostic signatures in colorectal cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188513. [Google Scholar] [CrossRef] [PubMed]
- Tsiakanikas, P.; Adamopoulos, P.G.; Tsirba, D.; Artemaki, P.I.; Papadopoulos, I.N.; Kontos, C.K.; Scorilas, A. High Expression of a tRNA(Pro) Derivative Associates with Poor Survival and Independently Predicts Colorectal Cancer Recurrence. Biomedicines 2022, 10, 1120. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Bhowmik, S.; Majumdar, S.; Goswami, A.; Chakraborty, J.; Gupta, S.; Aggarwal, S.; Ray, S.; Chatterjee, R.; Bhattacharyya, S.; et al. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low α-fetoprotein. Int. J. Cancer 2020, 147, 2934–2947. [Google Scholar] [CrossRef]
- Zhan, S.; Yang, P.; Zhou, S.; Xu, Y.; Xu, R.; Liang, G.; Zhang, C.; Chen, X.; Yang, L.; Jin, F.; et al. Serum mitochondrial tsRNA serves as a novel biomarker for hepatocarcinoma diagnosis. Front. Med. 2022, 16, 216–226. [Google Scholar] [CrossRef]
- Yang, B.; Feng, X.; Liu, H.; Tong, R.; Wu, J.; Li, C.; Yu, H.; Chen, Y.; Cheng, Q.; Chen, J.; et al. High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene 2020, 39, 6529–6543. [Google Scholar] [CrossRef]
- Wei, X.; Zhao, L.; Ren, R.; Ji, F.; Xue, S.; Zhang, J.; Liu, Z.; Ma, Z.; Wang, X.W.; Wong, L.; et al. MiR-125b Loss Activated HIF1alpha/pAKT Loop, Leading to Transarterial Chemoembolization Resistance in Hepatocellular Carcinoma. Hepatology 2021, 73, 1381–1398. [Google Scholar] [CrossRef]
- Zhao, J.; Li, H.; Zhao, S.; Wang, E.; Zhu, J.; Feng, D.; Zhu, Y.; Dou, W.; Fan, Q.; Hu, J.; et al. Epigenetic silencing of miR-144/451a cluster contributes to HCC progression via paracrine HGF/MIF-mediated TAM remodeling. Mol. Cancer 2021, 20, 46. [Google Scholar] [CrossRef]
- Yokota, Y.; Noda, T.; Okumura, Y.; Kobayashi, S.; Iwagami, Y.; Yamada, D.; Tomimaru, Y.; Akita, H.; Gotoh, K.; Takeda, Y.; et al. Serum exosomal miR-638 is a prognostic marker of HCC via downregulation of VE-cadherin and ZO-1 of endothelial cells. Cancer Sci. 2021, 112, 1275–1288. [Google Scholar] [CrossRef]
- Sharma, G.G.; Okada, Y.; Von Hoff, D.; Goel, A. Non-coding RNA biomarkers in pancreatic ductal adenocarcinoma. Semin Cancer Biol. 2021, 75, 153–168. [Google Scholar] [CrossRef]
- Yang, J.; Xu, R.; Wang, C.; Qiu, J.; Ren, B.; You, L. Early screening and diagnosis strategies of pancreatic cancer: A comprehensive review. Cancer Commun. 2021, 41, 1257–1274. [Google Scholar] [CrossRef] [PubMed]
- Daoud, A.Z.; Mulholland, E.J.; Cole, G.; McCarthy, H.O. MicroRNAs in Pancreatic Cancer: Biomarkers, prognostic, and therapeutic modulators. BMC Cancer 2019, 19, 1130. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yoo, J.; Ho, J.Y.; Jung, Y.; Lee, S.; Hur, S.Y.; Choi, Y.J. Plasma-derived exosomal miR-4732-5p is a promising noninvasive diagnostic biomarker for epithelial ovarian cancer. J. Ovarian Res. 2021, 14, 59. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Chen, Z.; Wang, M.; Zhang, M.; Chen, Y.; Yang, X.; Zhou, C.; Liu, Y.; Hong, L.; Zhang, L. Detection of plasma exosomal miRNA-205 as a biomarker for early diagnosis and an adjuvant indicator of ovarian cancer staging. J. Ovarian Res. 2022, 15, 27. [Google Scholar] [CrossRef] [PubMed]
- Hulstaert, E.; Morlion, A.; Levanon, K.; Vandesompele, J.; Mestdagh, P. Candidate RNA biomarkers in biofluids for early diagnosis of ovarian cancer: A systematic review. Gynecol. Oncol. 2021, 160, 633–642. [Google Scholar] [CrossRef]
- Tung, C.H.; Kuo, L.W.; Huang, M.F.; Wu, Y.Y.; Tsai, Y.T.; Wu, J.E.; Hsu, K.F.; Chen, Y.L.; Hong, T.M. MicroRNA-150-5p promotes cell motility by inhibiting c-Myb-mediated Slug suppression and is a prognostic biomarker for recurrent ovarian cancer. Oncogene 2020, 39, 862–876. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, W.; Xu, L.; Yi, T.; Zhao, X.; Wei, Y.; Vermeulen, L.; Goel, A.; Zhou, S.; Wang, X. Integrative network biology analysis identifies miR-508-3p as the determinant for the mesenchymal identity and a strong prognostic biomarker of ovarian cancer. Oncogene 2019, 38, 2305–2319. [Google Scholar] [CrossRef]
- Todeschini, P.; Salviato, E.; Romani, C.; Raimondi, V.; Ciccarese, F.; Ferrari, F.; Tognon, G.; Marchini, S.; D’Incalci, M.; Zanotti, L.; et al. Comprehensive Profiling of Hypoxia-Related miRNAs Identifies miR-23a-3p Overexpression as a Marker of Platinum Resistance and Poor Prognosis in High-Grade Serous Ovarian Cancer. Cancers 2021, 13, 3358. [Google Scholar] [CrossRef]
- Panoutsopoulou, K.; Avgeris, M.; Mavridis, K.; Dreyer, T.; Dorn, J.; Obermayr, E.; Reinthaller, A.; Michaelidou, K.; Mahner, S.; Vergote, I.; et al. miR-203 is an independent molecular predictor of prognosis and treatment outcome in ovarian cancer: A multi-institutional study. Carcinogenesis 2020, 41, 442–451. [Google Scholar] [CrossRef]
- Panoutsopoulou, K.; Dreyer, T.; Dorn, J.; Obermayr, E.; Mahner, S.; Gorp, T.V.; Braicu, I.; Zeillinger, R.; Magdolen, V.; Avgeris, M.; et al. tRNA(GlyGCC)-Derived Internal Fragment (i-tRF-GlyGCC) in Ovarian Cancer Treatment Outcome and Progression. Cancers 2021, 14, 24. [Google Scholar] [CrossRef]
- Bao, Z.; Zhang, N.; Niu, W.; Mu, M.; Zhang, X.; Hu, S.; Niu, C. Exosomal miR-155-5p derived from glioma stem-like cells promotes mesenchymal transition via targeting ACOT12. Cell Death Dis. 2022, 13, 725. [Google Scholar] [CrossRef] [PubMed]
- Shao, N.; Xue, L.; Wang, R.; Luo, K.; Zhi, F.; Lan, Q. miR-454-3p Is an Exosomal Biomarker and Functions as a Tumor Suppressor in Glioma. Mol. Cancer Ther. 2019, 18, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Xiao, Z.; Li, B.; Li, H.; Yang, B.; Li, T.; Mei, Z. miRNA-21 may serve as a promising noninvasive marker of glioma with a high diagnostic performance: A pooled analysis of 997 patients. Ther. Adv. Med. Oncol. 2021, 13, 1758835920987650. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Zhao, X.; Ming, J.; Liu, X.; Liu, D.; Jiang, C. Stepwise detection and evaluation reveal miR-10b and miR-222 as a remarkable prognostic pair for glioblastoma. Oncogene 2019, 38, 6142–6157. [Google Scholar] [CrossRef]
- Lin, J.; Ding, S.; Xie, C.; Yi, R.; Wu, Z.; Luo, J.; Huang, T.; Zeng, Y.; Wang, X.; Xu, A.; et al. MicroRNA-4476 promotes glioma progression through a miR-4476/APC/β-catenin/c-Jun positive feedback loop. Cell Death Dis. 2020, 11, 269. [Google Scholar] [CrossRef]
- Kandettu, A.; Radhakrishnan, R.; Chakrabarty, S.; Sriharikrishnaa, S.; Kabekkodu, S.P. The emerging role of miRNA clusters in breast cancer progression. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188413. [Google Scholar] [CrossRef]
- Wang, J.; Ma, G.; Ge, H.; Han, X.; Mao, X.; Wang, X.; Veeramootoo, J.S.; Xia, T.; Liu, X.; Wang, S. Circulating tRNA-derived small RNAs (tsRNAs) signature for the diagnosis and prognosis of breast cancer. NPJ Breast Cancer 2021, 7, 4. [Google Scholar] [CrossRef]
- Takagawa, Y.; Gen, Y.; Muramatsu, T.; Tanimoto, K.; Inoue, J.; Harada, H.; Inazawa, J. miR-1293, a Candidate for miRNA-Based Cancer Therapeutics, Simultaneously Targets BRD4 and the DNA Repair Pathway. Mol. Ther. 2020, 28, 1494–1505. [Google Scholar] [CrossRef]
- Zeng, A.; Wei, Z.; Yan, W.; Yin, J.; Huang, X.; Zhou, X.; Li, R.; Shen, F.; Wu, W.; Wang, X.; et al. Exosomal transfer of miR-151a enhances chemosensitivity to temozolomide in drug-resistant glioblastoma. Cancer Lett. 2018, 436, 10–21. [Google Scholar] [CrossRef]
- Chaudhary, A.K.; Mondal, G.; Kumar, V.; Kattel, K.; Mahato, R.I. Chemosensitization and inhibition of pancreatic cancer stem cell proliferation by overexpression of microRNA-205. Cancer Lett. 2017, 402, 1–8. [Google Scholar] [CrossRef]
- Cortez, M.A.; Valdecanas, D.; Niknam, S.; Peltier, H.J.; Diao, L.; Giri, U.; Komaki, R.; Calin, G.A.; Gomez, D.R.; Chang, J.Y.; et al. In Vivo Delivery of miR-34a Sensitizes Lung Tumors to Radiation Through RAD51 Regulation. Mol. Ther. Nucleic. Acids 2015, 4, e270. [Google Scholar] [CrossRef]
- Ma, W.; Ma, C.N.; Zhou, N.N.; Li, X.D.; Zhang, Y.J. Up-regulation of miR-328-3p sensitizes non-small cell lung cancer to radiotherapy. Sci. Rep. 2016, 6, 31651. [Google Scholar] [CrossRef] [PubMed]
- Ohno, M.; Ohkuri, T.; Kosaka, A.; Tanahashi, K.; June, C.H.; Natsume, A.; Okada, H. Expression of miR-17-92 enhances anti-tumor activity of T-cells transduced with the anti-EGFRvIII chimeric antigen receptor in mice bearing human GBM xenografts. J. Immunother. Cancer 2013, 1, 21. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Q.; Kumar, S.; Calin, G.A.; Li, Z.; Chim, C.S. Frequent methylation of the tumour suppressor miR-1258 targeting PDL1: Implication in multiple myeloma-specific cytotoxicity and prognostification. Br. J. Haematol. 2020, 190, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Mollaei, H.; Safaralizadeh, R.; Rostami, Z. MicroRNA replacement therapy in cancer. J. Cell Physiol. 2019, 234, 12369–12384. [Google Scholar] [CrossRef]
- Di Paolo, D.; Pastorino, F.; Brignole, C.; Corrias, M.V.; Emionite, L.; Cilli, M.; Tamma, R.; Priddy, L.; Amaro, A.; Ferrari, D.; et al. Combined Replenishment of miR-34a and let-7b by Targeted Nanoparticles Inhibits Tumor Growth in Neuroblastoma Preclinical Models. Small 2020, 16, e1906426. [Google Scholar] [CrossRef]
| Cancer Type | Small RNA (Examples) | Expression | Function | Mechanism | References |
|---|---|---|---|---|---|
| Gastric cancer | miR-371-3 | Up | Promotes cell proliferation, migration and invasion | Suppresses TOB1 | [93] |
| piR-823 | Down | Inhibits cell growth | Unknown | [94] | |
| tRF-5026a | Down | Inhibits cell proliferation, migration, and cell cycle | Regulates the PTEN/PI3K/AKT signaling pathway | [95] | |
| Liver cancer | miR-802 | Up | Promotes cell proliferation, cell cycle and migration | Suppresses RUNX3 | [96] |
| piR-Hep1 | Up | Promotes cell viability, motility, and invasiveness | Reduces phosphorylated AKT | [97] | |
| Gly-tRF | Up | Promotes liver cancer stem cell-like cell properties and cell migration | Suppresses NDFIP2 and increases phosphorylated AKT | [98] | |
| Colorectal cancer | miR-4319 | Down | Inhibits cell proliferation, migration and invasion | Suppresses ABTB1 | [99] |
| piR-823 | Down | Inhibits cell apoptosis | Interacts with PINK1 | [100] | |
| 5′tiRNA-His-GTG | Up | Promotes cell proliferation | Suppresses LATS2 | [101] | |
| Breast cancer | miR-21 | Up | Promotes cell proliferation and metastasis | Suppresses LZTFL1 | [102] |
| piRNA-36712 | Down | Inhibits cell proliferation and cell cycle | Interacts with SEPW1P RNA | [103] | |
| 5′-tiRNA-Val | Down | Inhibits cell proliferation and migration | Inhibits FZD3/Wnt/β-Catenin signaling pathway | [104] | |
| Lung cancer | miR-1254 | Up | Promotes cell proliferation | Suppresses SFRP1 | [105] |
| tsRNA-5001a | Up | Promotes cell proliferation | May target GADD45G | [106] | |
| Multiple myeloma | miR-145-3p | Down | Inhibits cell proliferation and promotes apoptosis | Suppresses HDAC4 | [107] |
| piRNA-823 | Up | Promotes cell proliferation, apoptosis and cell cycle | Regulates de novo DNA methylation and angiogenesis | [108] | |
| Lympho-ma | miR-339-5p | Up | Promotes cell proliferation, cell cycle and reduces apoptosis | Suppresses BCL2L11 and BAX | [109] |
| piRNA-30473 | Up | Promotes cell proliferation and cell cycle | Regulates m6A RNA methylation | [110] | |
| Pancreatic cancer | miR-4516 | Down | Inhibits cell proliferation, migration and invasion, while promotes cell apoptosis | Suppresses OTX1 | [111] |
| piR-017061 | Down | inhibits cell growth | Suppresses EFNA5 | [112] | |
| tRF-Pro-AGG-004 and tRF-Leu-CAG-002 | Up | Promotes cell growth and invasion | unknown | [113] |
| Cancer Type | Diagnosis | Prognosis |
|---|---|---|
| Lung cancer | miR-20a, miR-10b, miR-150, miR-223, miR-205 [157]; 5a_tRF-Ile-AAT/GAT, 5a_tRF-Asp-GTC, 3P_tRNA-Arg-TCG-1-, 3P_tRNA-Arg-TCT-4-1, 5P_tRNA-Gly-TCC-1-1, 5P_tRNA-Asn-GTT-2-3 [158]; piR-651 [159] | miR-21, miR-155, miR-200c and miR-125b, miR-21, miR-155, miR-200c and miR-125b [160]; AS-tDR-007333 [161], 5a_tRF-Ile-AAT/GAT [158]; piR-651 [159] |
| Prostate cancer | miR-26b-5p, miR-98-5p [162], miR-5100, miR-1290 [163] | miR-182 [164]; miR-1290, miR-375 [165] |
| Gastric cancer | miR-140, miR-183, miR-30e, miR-103a, miR-126, miR-93, miR-142, miR-21, miR-29c, miR-424 and miR-181a, miR-340 [166]; miR-21, miR-222, miR-99a-5p; miR-17, miR-25, miR-214 [167]; miR-4257, miR-6785-5p, miR-187-5p, and miR-5739 [168]; tRF-23-Q99P9P9NDD [169] | miR-501-5p, miR-208a, miR-718, miR-15b-3p, miR-519a, miR-153, miR-187, miR-345, miR-28-5p, miR-124-3p, miR-383-5p [167]; tRF-23-Q99P9P9NDD [169] |
| Colorectal cancer | miR-21, miR-24-2, miR-139-3p, miR-135a-5p, miR-21, miR-29, miR-92, miR-125, miR-223, miR-6803-5p, miR-21, miR-4478 [170]; tRF-phe-GAA-031, tRF-VAL-TCA-002 [171] | miR-21, miR-6826, miR-122, miR-139-5p, miR-203, miR-6803-5p, miR-17-5p [170]; miR-5091, miR-10b-3p, miR-9-5p, miR-187-3p, miR-32-5p, miR-652-3p, miR-342-3p, miR-501-3p, miR-328-3p [172]; tRF-phe-GAA-031, tRF-VAL-TCA-002 [171]; 5′-tiRNA-Pro-TGG [173] |
| Liver cancer | miR-10b-5p, miR-221-3p, miR-223-3p, miR-21-5p [174]; tRNA-ValTAC-3, tRNA-GlyTCC-5, tRNA-ValAAC-5; tRNA-GluCTC-5 [153]; tRF-Gln-TTG-006 [175] | miR92a-3p [176], miR-125b [177], miR-144, miR-451a [178]; miR-638 [179] |
| Pancreatic cancer | miR-21, miR-155, miR-196a, miR-196b, miR-16, miR-18a, miR-20a [180], let-7b-5p, miR-192- 5p, miR-19a-3p, miR-19b-3p, miR-223-3p, and miR-25-3p [181]; tRF-Pro-AGG-004, tRF-Leu-CAG-002 [113] | miR-10b, miR-21, miR-34a, miR-155, miR-200 family, miR-216, let-7 family [182]; tRF-Pro-AGG-004, tRF-Leu-CAG-002 [113] |
| Ovarian cancer | miR-4732-5p [183]; miRNA-205 [184]; miR-21, miR-200 family, miR-205, miR-10a, miR-346 [185] | miRNA-150-5p [186]; miR-508-3p [187]; miR-23a-3p [188]; miR-203 [189]; i-tRF-Gly-GCC [190] |
| Glioma | miR-155-5p [191]; miR-454-3p [192]; miRNA-21[193] | miR-10b, miR-222 [194]; miR-4476 [195]; miR-454-3p [192] |
| Breast cancer | miR-21, miR-96, miR-183, miR-182, miR-141, miR-200a, miR-429, miR-139, miR-145 [196]; RF-Arg-CCT-017, tRF-Gly-CCC-001, tiRNA-Phe-GAA-003 [197] | miR-17, miR-18a, miR-19a, miR-19b, miR-20a, miR-27b-3p, miR-92, miR-497, miR-532-5p [196]; piRNA-36712 [103]; tRF-Arg-CCT-017, tiRNA-Phe-GAA-003 [197] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, Q.; Zhang, Y.; Li, J.; Zhu, Q. Small Non-Coding RNAs in Human Cancer. Genes 2022, 13, 2072. https://doi.org/10.3390/genes13112072
Xiong Q, Zhang Y, Li J, Zhu Q. Small Non-Coding RNAs in Human Cancer. Genes. 2022; 13(11):2072. https://doi.org/10.3390/genes13112072
Chicago/Turabian StyleXiong, Qunli, Yaguang Zhang, Junjun Li, and Qing Zhu. 2022. "Small Non-Coding RNAs in Human Cancer" Genes 13, no. 11: 2072. https://doi.org/10.3390/genes13112072
APA StyleXiong, Q., Zhang, Y., Li, J., & Zhu, Q. (2022). Small Non-Coding RNAs in Human Cancer. Genes, 13(11), 2072. https://doi.org/10.3390/genes13112072







