Molecular Characterization, Purification, and Mode of Action of Enterocin KAE01 from Lactic Acid Bacteria and Its In Silico Analysis against MDR/ESBL Pseudomonas aeruginosa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Identification of Enterococcus spp.
2.3. Genomic DNA Extraction
2.4. Amplification and Sequencing of 16S rRNA Gene Using PCR
2.5. Phylogenetic Analysis
2.6. PCR Screening for Enterocin Structural Genes
2.7. Extraction and Purification of Enterocin KAE01
2.8. Molecular Mass Determination by Tricine SDS–PAGE
2.9. Antimicrobial Activity Assay
2.10. Influence of pH, Heat, and Proteolytic Enzymes on Enterocin KAE01
2.10.1. Temperature
2.10.2. pH
2.10.3. Enzymes
2.11. Statistical Analysis
2.12. Scanning Electron Microscopy (SEM)
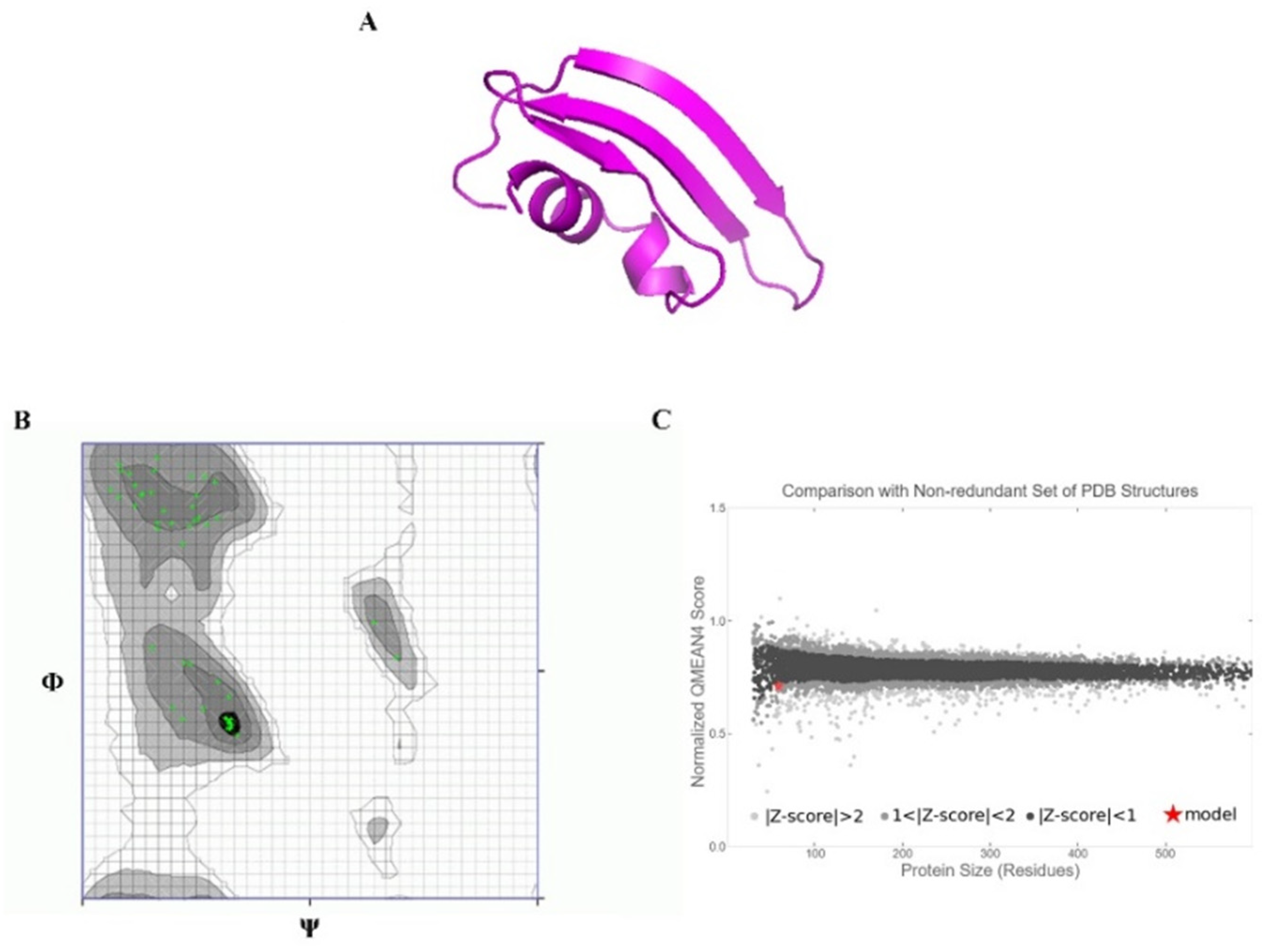
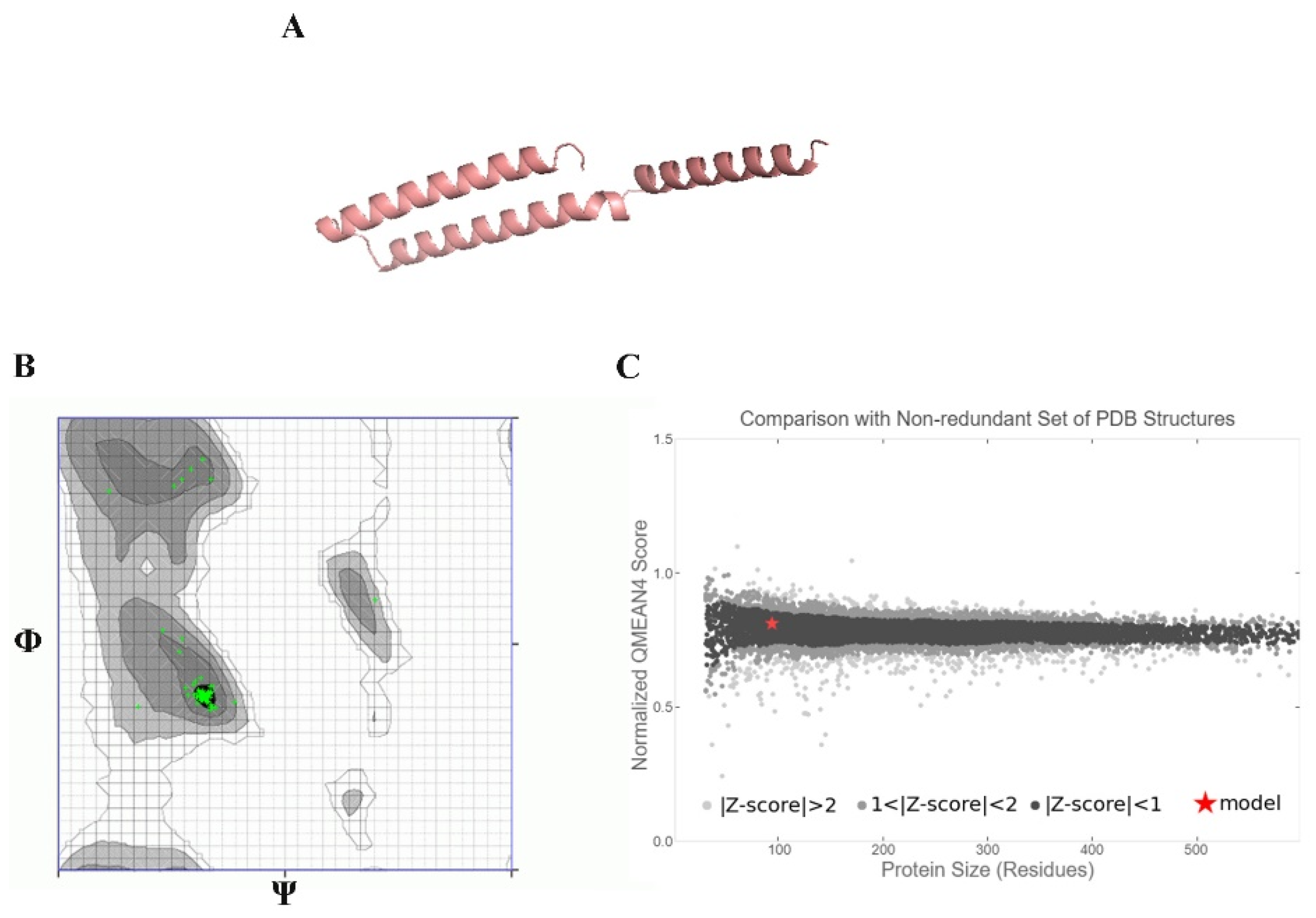
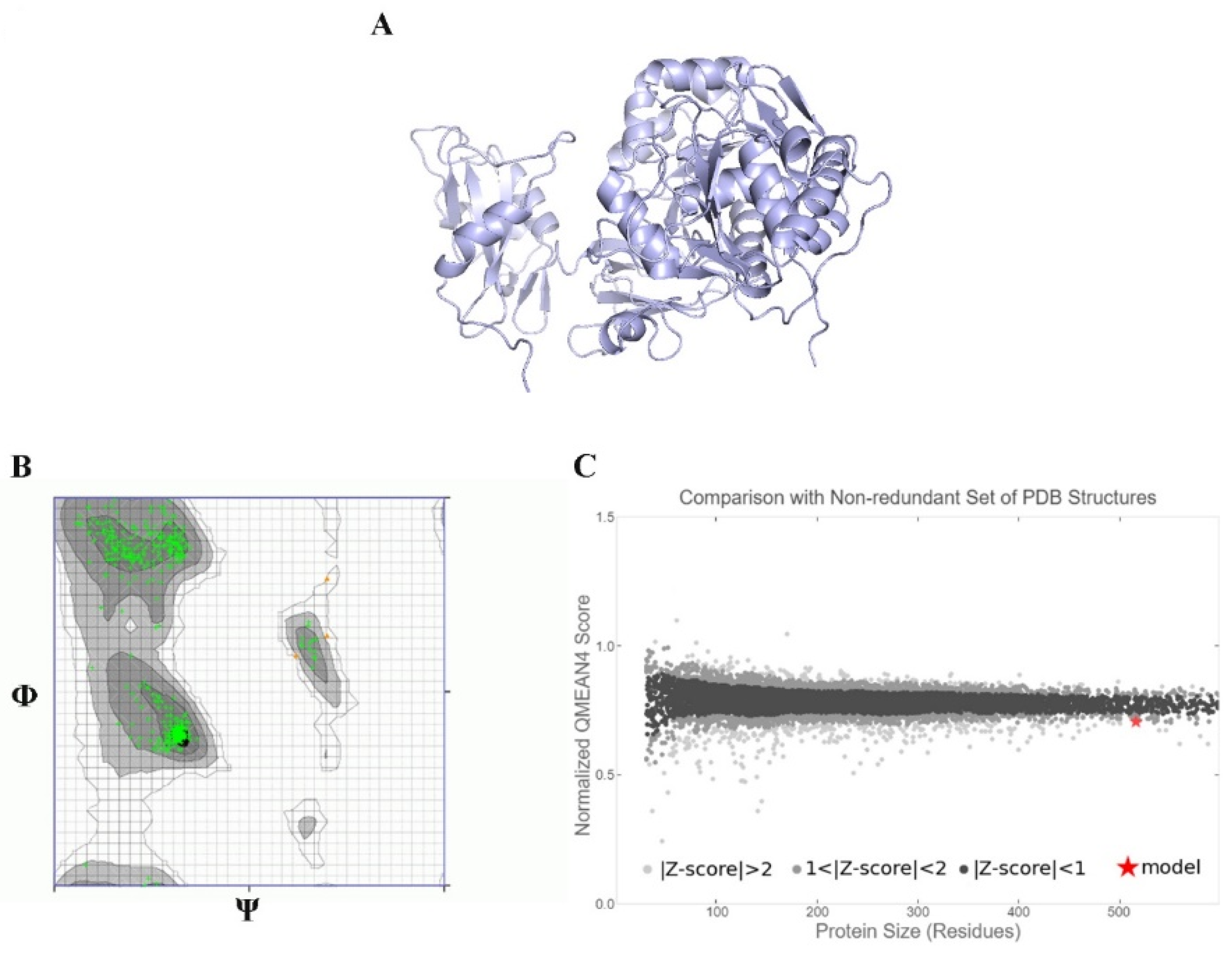
2.13. Ab Initio Modeling of KAE01
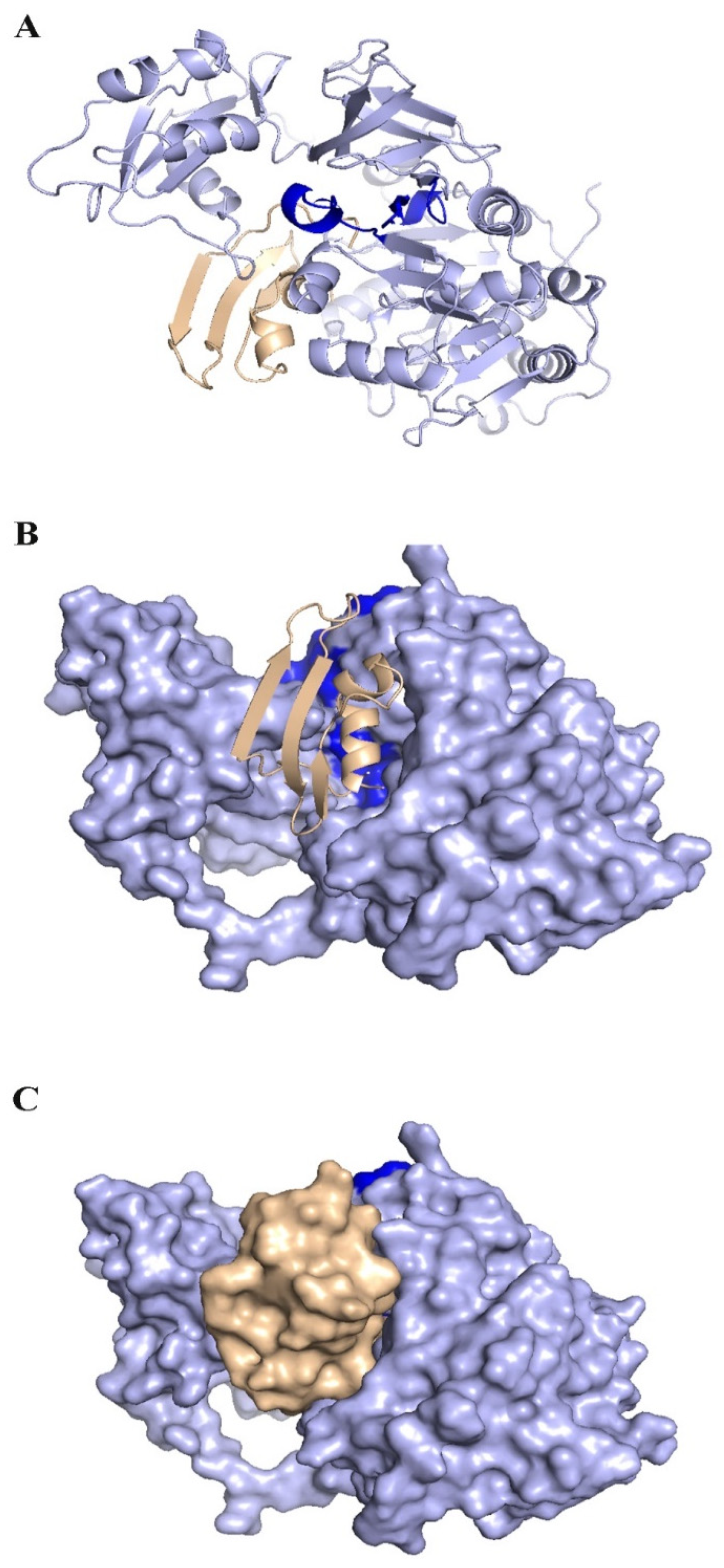
2.14. Peptide–Protein Docking
2.14.1. Protein Preparation
2.14.2. Peptide–Protein Docking
3. Results
3.1. Isolation, Identification, and Phylogenetic Analysis of Enterococcus Faecium KAE01
3.2. Analysis of Enterocin Structural Genes
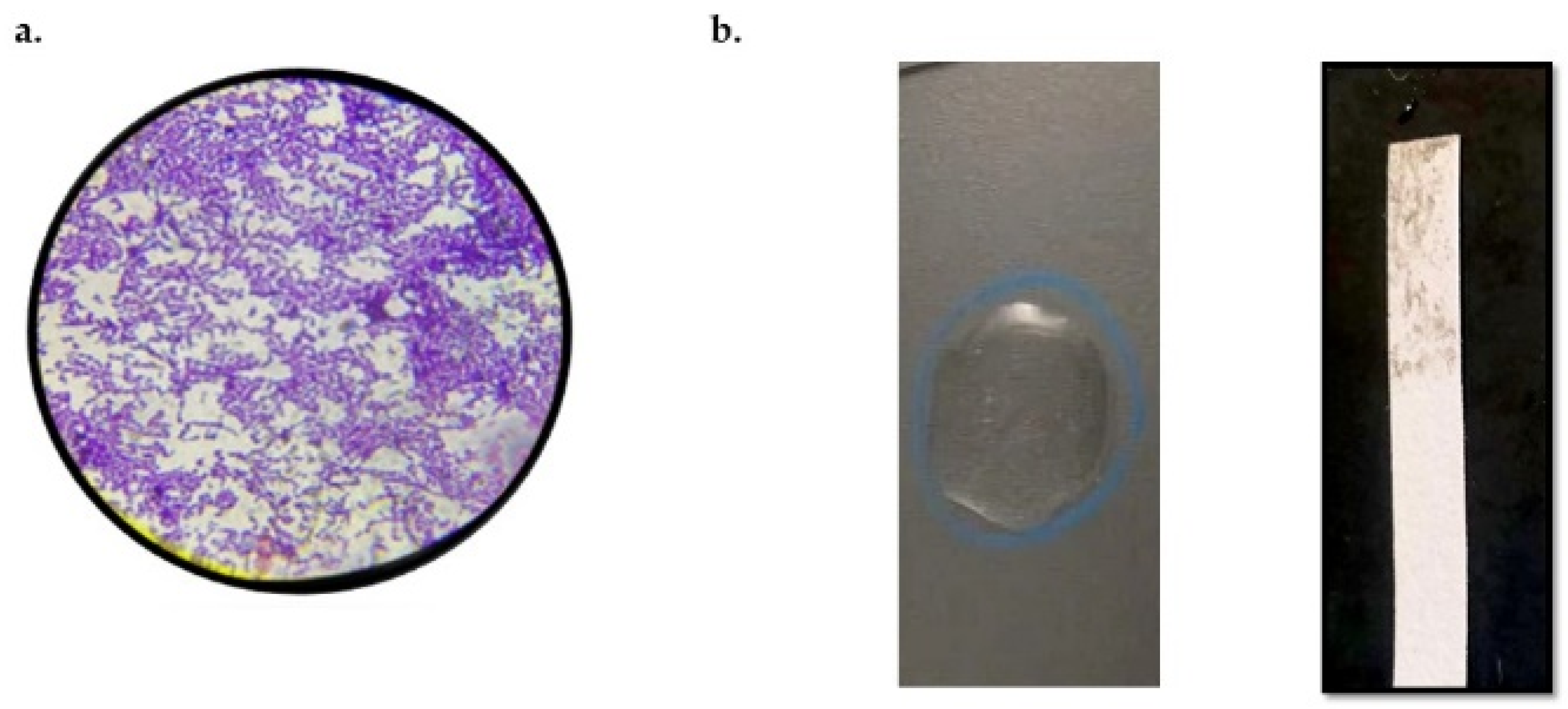
3.3. Purification of Enterocin KAE01
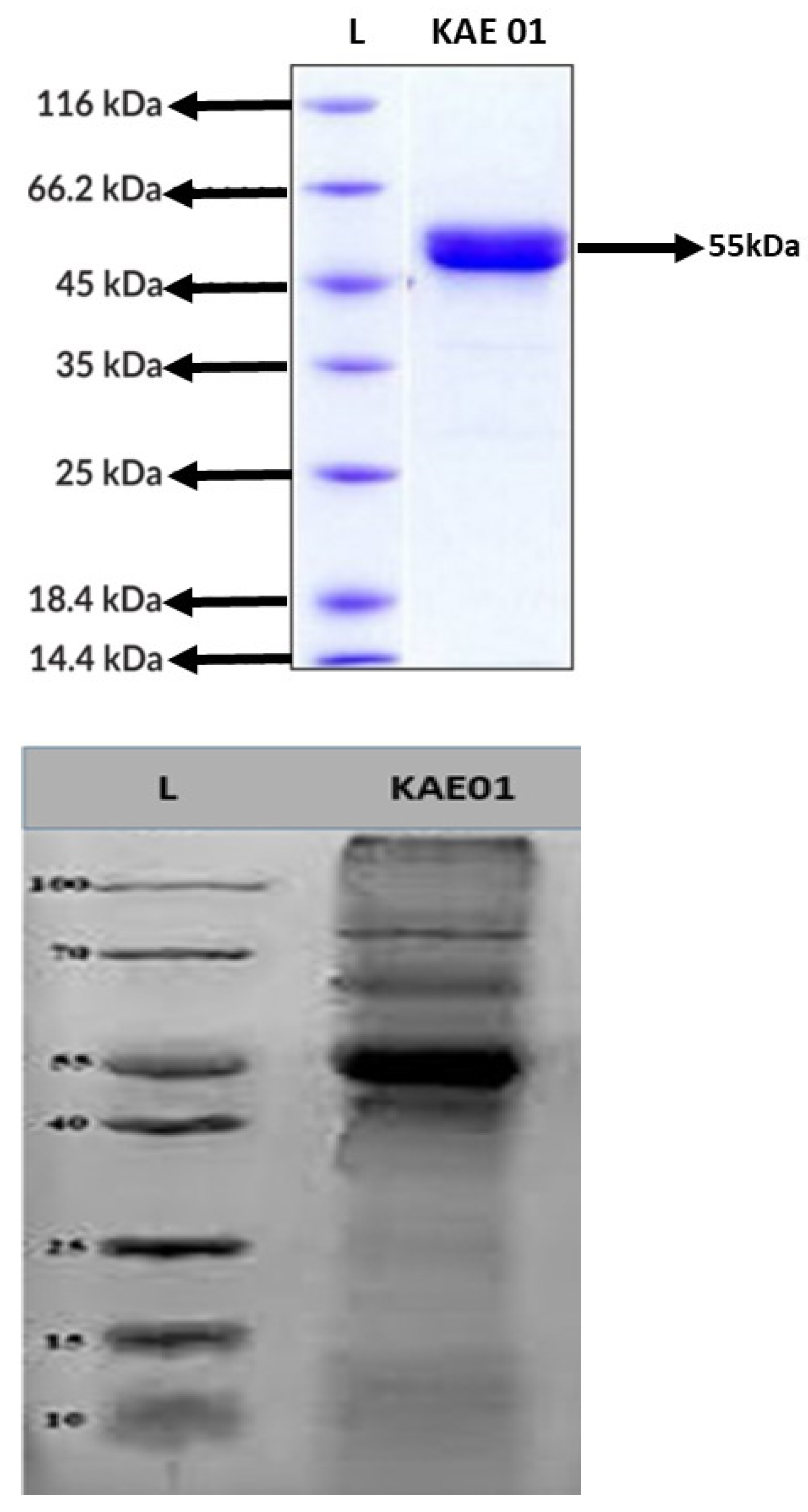
3.4. Molecular Mass Determination by Tricine SDS–PAGE
3.5. Antimicrobial Activity of Purified Enterocin KAE01
3.6. Influence of pH, Heat, and Proteolytic Enzymes on Enterocin KAE01
3.7. Scanning Electron Microscopy (SEM) Analysis
3.8. Peptide–Protein Docking
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, A.S.; De Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Prim. 2018, 4, 18033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGuinness, W.A.; Malachowa, N.; DeLeo, F.R. Focus: Infectious diseases: Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 2017, 90, 269. [Google Scholar] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, J.; Montville, T.J.; Nes, I.F.; Chikindas, M.L. Bacteriocins: Safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 2001, 71, 1–20. [Google Scholar] [CrossRef]
- Ołdak, A.; Zielińska, D. Bacteriocins from lactic acid bacteria as an alternative to antibiotics. Postep. Hig. I Med. Dosw. (Online) 2017, 71, 328–338. [Google Scholar] [CrossRef]
- Perez, R.H.; Zendo, T.; Sonomoto, K. Multiple bacteriocin production in lactic acid bacteria. J. Biosci. Bioeng. 2022, 134, 277–287. [Google Scholar] [CrossRef]
- Piazentin, A.C.M.; Mendonça, C.M.N.; Vallejo, M.; Mussatto, S.I.; de Souza Oliveira, R.P. Bacteriocin-like inhibitory substances production by Enterococcus faecium 135 in co-culture with Ligilactobacillus salivarius and Limosilactobacillus reuteri. Braz. J. Microbiol. 2022, 53, 131–141. [Google Scholar] [CrossRef]
- Franz, C.M.; Van Belkum, M.J.; Holzapfel, W.H.; Abriouel, H.; Gálvez, A. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol. Rev. 2007, 31, 293–310. [Google Scholar] [CrossRef]
- Rehaiem, A.; Guerra, N.P.; Belgacem, Z.B.; Bernárdez, P.F.; Castro, L.P.; Manai, M. Enhancement of enterocin A production by Enterococcus faecium MMRA and determination of its stability to temperature and pH. Biochem. Eng. J. 2011, 56, 94–106. [Google Scholar] [CrossRef]
- Herranz, C.; Chen, Y.; Chung, H.-J.; Cintas, L.; Hernandez, P.; Montville, T.; Chikindas, M. Enterocin P selectively dissipates the membrane potential of Enterococcus faecium T136. Appl. Environ. Microbiol. 2001, 67, 1689–1692. [Google Scholar] [CrossRef]
- De Vuyst, L.; Vandamme, E.J. Bacteriocins of Lactic Acid Bacteria: Microbiology, Genetics and Applications; Springer: Cham, Switzerland, 2012. [Google Scholar]
- London, N.; Raveh, B.; Cohen, E.; Fathi, G.; Schueler-Furman, O. Rosetta FlexPepDock web server—High resolution modeling of peptide–protein interactions. Nucleic Acids Res. 2011, 39, W249–W253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antes, I. DynaDock: A new molecular dynamics-based algorithm for protein-peptide docking including receptor flexibility. Proteins Struct. Funct. Bioinform. 2010, 78, 1084–1104. [Google Scholar] [CrossRef] [PubMed]
- Donsky, E.; Wolfson, H.J. PepCrawler: A fast RRT-based algorithm for high-resolution refinement and binding affinity estimation of peptide inhibitors. Bioinformatics 2011, 27, 2836–2842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trellet, M.; Melquiond, A.S.; Bonvin, A.M. A unified conformational selection and induced fit approach to protein-peptide docking. PLoS ONE 2013, 8, e58769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Zundert, G.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.; Karaca, E.; Melquiond, A.; van Dijk, M.; De Vries, S.; Bonvin, A. The HADDOCK2. 2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [Green Version]
- Rentzsch, R.; Renard, B.Y. Docking small peptides remains a great challenge: An assessment using AutoDock Vina. Brief. Bioinform. 2015, 16, 1045–1056. [Google Scholar] [CrossRef] [Green Version]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein–ligand docking using GOLD. Proteins Struct. Funct. Bioinform. 2003, 52, 609–623. [Google Scholar] [CrossRef]
- Jain, A.N. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003, 46, 499–511. [Google Scholar] [CrossRef]
- Alam, N.; Goldstein, O.; Xia, B.; Porter, K.A.; Kozakov, D.; Schueler-Furman, O. High-resolution global peptide-protein docking using fragments-based PIPER-FlexPepDock. PLoS Comput. Biol. 2017, 13, e1005905. [Google Scholar] [CrossRef] [Green Version]
- Ansari, A.; Zohra, R.R.; Tarar, O.M.; Qader, S.A.U.; Aman, A. Screening, purification and characterization of thermostable, protease resistant Bacteriocin active against methicillin resistant Staphylococcus aureus (MRSA). BMC Microbiol. 2018, 18, 192. [Google Scholar] [CrossRef]
- Peng, J.; Long, H.; Liu, W.; Wu, Z.; Wang, T.; Zeng, Z.; Guo, G.; Wu, J. Antibacterial mechanism of peptide Cec4 against Acinetobacter baumannii. Infect. Drug Resist. 2019, 12, 2417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirubakaran, R.; ArulJothi, K.; Revathi, S.; Shameem, N.; Parray, J.A. Emerging priorities for microbial metagenome research. Bioresour. Technol. Rep. 2020, 11, 100485. [Google Scholar] [CrossRef] [PubMed]
- Perumal, V.; Venkatesan, A. Antimicrobial, cytotoxic effect and purification of bacteriocin from vancomycin susceptible Enterococcus faecalis and its safety evaluation for probiotization. LWT 2017, 78, 303–310. [Google Scholar] [CrossRef]
- Benítez-Chao, D.F.; León-Buitimea, A.; Lerma-Escalera, J.A.; Morones-Ramírez, J.R. Bacteriocins: An overview of antimicrobial, toxicity, and biosafety assessment by in vivo models. Front. Microbiol. 2021, 12, 630695. [Google Scholar] [CrossRef]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef]
- Mirkovic, N.; Radulovic, Z.; Uzelac, G.; Lozo, J.; Obradovic, D.; Topisirovic, L.; Kojic, M. Isolation and characterisation of bacteriocin and aggregation-promoting factor production in Lactococcus lactis ssp. lactis BGBM50 strain. Food Technol. 2015, 53, 237–242. [Google Scholar]
- Lakshminarayanan, B.; Guinane, C.; O’Connor, P.; Coakley, M.; Hill, C.; Stanton, C.; O’Toole, P.; Ross, R. Isolation and characterization of bacteriocin-producing bacteria from the intestinal microbiota of elderly Irish subjects. J. Appl. Microbiol. 2013, 114, 886–898. [Google Scholar] [CrossRef]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef] [Green Version]
- Abdulhussain Kareem, R.; Razavi, S.H. Plantaricin bacteriocins: As safe alternative antimicrobial peptides in food preservation—A review. J. Food Saf. 2020, 40, e12735. [Google Scholar] [CrossRef]
- Li, H.-W.; Xiang, Y.-Z.; Zhang, M.; Jiang, Y.-H.; Zhang, Y.; Liu, Y.-Y.; Lin, L.-B.; Zhang, Q.-L. A novel bacteriocin from Lactobacillus salivarius against Staphylococcus aureus: Isolation, purification, identification, antibacterial and antibiofilm activity. LWT 2021, 140, 110826. [Google Scholar] [CrossRef]
- Pei, J.; Jin, W.; Abd El-Aty, A.; Baranenko, D.A.; Gou, X.; Zhang, H.; Geng, J.; Jiang, L.; Chen, D.; Yue, T. Isolation, purification, and structural identification of a new bacteriocin made by Lactobacillus plantarum found in conventional kombucha. Food Control 2020, 110, 106923. [Google Scholar] [CrossRef]
- Zou, J.; Jiang, H.; Cheng, H.; Fang, J.; Huang, G. Strategies for screening, purification and characterization of bacteriocins. Int. J. Biol. Macromol. 2018, 117, 781–789. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Heilig, H.G.; Klaassens, E.S.; Booijink, C.C.; Kleerebezem, M.; Smidt, H.; De Vos, W.M. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat. Protoc. 2006, 1, 870–873. [Google Scholar] [CrossRef] [PubMed]
- Vivodík, M.; Balážová, Ž.; Gálová, Z. DNA analysis of ricin using RAPD technique. J. Microbiol. Biotechnol. Food Sci. 2021, 2021, 181–183. [Google Scholar]
- Hall, M.A. Correlation-Based Feature Selection for Machine Learning. Ph.D. Thesis, The University of Waikato, Hamilton, New Zealand, 1999. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Shittu, O.B.; Adelaja, O.M.; Obuotor, T.M.; Sam-Wobo, S.O.; Adenaike, A.S. PCR-Internal Transcribed Spacer (ITS) genes sequencing and phylogenetic analysis of clinical and environmental Aspergillus species associated with HIV-TB co infected patients in a hospital in Abeokuta, southwestern Nigeria. Afr. Health Sci. 2016, 16, 141–148. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Phylogenies and the comparative method. Am. Nat. 1985, 125, 1–15. [Google Scholar] [CrossRef]
- Georgieva, R.N.; Iliev, I.N.; Chipeva, V.A.; Dimitonova, S.P.; Samelis, J.; Danova, S.T. Identification and in vitro characterisation of Lactobacillus plantarum strains from artisanal Bulgarian white brined cheeses. J. Basic Microbiol. 2008, 48, 234–244. [Google Scholar] [CrossRef]
- Schägger, H.; Aquila, H.; Von Jagow, G. Coomassie blue-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for direct visualization of polypeptides during electrophoresis. Anal. Biochem. 1988, 173, 201–205. [Google Scholar] [CrossRef]
- Hidayat, B.; Wea, A.; Andriati, N. Physicochemical, sensory attributes and protein profile by SDS-PAGE of beef sausage substituted with texturized vegetable protein. Food Res. 2018, 2, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Alekseenko, A.; Kozakov, D.; Miao, Y. Improved modeling of peptide-protein binding through global docking and accelerated molecular dynamics simulations. Front. Mol. Biosci. 2019, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Ghafary, S.; Ranjbar, S.; Larijani, B.; Amini, M.; Biglar, M.; Mahdavi, M.; Bakhshaei, M.; Khoshneviszadeh, M.; Sakhteman, A.; Khoshneviszadeh, M. Novel morpholine containing cinnamoyl amides as potent tyrosinase inhibitors. Int. J. Biol. Macromol. 2019, 135, 978–985. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [Green Version]
- Anderson, R.J.; Weng, Z.; Campbell, R.K.; Jiang, X. Main-chain conformational tendencies of amino acids. Proteins Struct. Funct. Bioinform. 2005, 60, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.R.; Heo, L.; Seok, C. Effective protein model structure refinement by loop modeling and overall relaxation. Proteins Struct. Funct. Bioinform. 2016, 84, 293–301. [Google Scholar] [CrossRef]
- Vajda, S.; Yueh, C.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Kozakov, D. New additions to the C lus P ro server motivated by CAPRI. Proteins Struct. Funct. Bioinform. 2017, 85, 435–444. [Google Scholar] [CrossRef] [Green Version]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein—Protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins Struct. Funct. Bioinform. 2013, 81, 2159–2166. [Google Scholar] [CrossRef] [Green Version]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and its limits in rigid body protein-protein docking. Structure 2020, 28, 1071–1081.e1073. [Google Scholar] [CrossRef]
- Abdel-Daim, A.; Hassouna, N.; Hafez, M.; Ashor, M.S.A.; Aboulwafa, M.M. Antagonistic activity of Lactobacillus isolates against Salmonella typhi in vitro. BioMed Res. Int. 2013, 2013, 680605. [Google Scholar] [CrossRef] [Green Version]
- Shaker, B.; Ahmad, S.; Thai, T.D.; Eyun, S.-i.; Na, D. Rational drug design for Pseudomonas aeruginosa PqsA enzyme: An in silico guided study to block biofilm formation. Front. Mol. Biosci. 2020, 7, 577316. [Google Scholar] [CrossRef] [PubMed]
- Dimitonova, S.P.; Bakalov, B.V.; Aleksandrova-Georgieva, R.N.; Danova, S.T. Phenotypic and molecular identification of lactobacilli isolated from vaginal secretions. J. Microbiol. Immunol. Infect. 2008, 41, 469–477. [Google Scholar] [PubMed]
- Kemperman, R.; Kuipers, A.; Karsens, H.; Nauta, A.; Kuipers, O.; Kok, J. Identification and characterization of two novel clostridial bacteriocins, circularin A and closticin 574. Appl. Environ. Microbiol. 2003, 69, 1589–1597. [Google Scholar] [CrossRef] [Green Version]
- Darbandi, A.; Asadi, A.; Mahdizade Ari, M.; Ohadi, E.; Talebi, M.; Halaj Zadeh, M.; Darb Emamie, A.; Ghanavati, R.; Kakanj, M. Bacteriocins: Properties and potential use as antimicrobials. J. Clin. Lab. Anal. 2022, 36, e24093. [Google Scholar] [CrossRef]
- de Almeida Ramos, E.T.; Meneses, C.H.S.G.; Vidal, M.S.; Baldani, J.I. Characterisation and action mode of Gluconacin, a bacteriocin with antimicrobial activity against Xanthomonas albilineans. Ann. Appl. Biol. 2022, 180, 163–175. [Google Scholar] [CrossRef]
- Gao, X.; Huang, Q.; Zhao, Z.; Han, Q.; Ke, X.; Qin, H.; Huang, L. Studies on the infection, colonization, and movement of Pseudomonas syringae pv. actinidiae in kiwifruit tissues using a GFPuv-labeled strain. PLoS ONE 2016, 11, e0151169. [Google Scholar] [CrossRef] [PubMed]
- Klaenhammer, T.R. Bacteriocins of lactic acid bacteria. Biochimie 1988, 70, 337–349. [Google Scholar] [CrossRef]
- Sharma, P.; Kaur, S.; Chadha, B.S.; Kaur, R.; Kaur, M.; Kaur, S. Anticancer and antimicrobial potential of enterocin 12a from Enterococcus faecium. BMC Microbiol. 2021, 21, 39. [Google Scholar] [CrossRef]
- Güllüce, M.; Karadayı, M.; Barış, Ö. Bacteriocins: Promising natural antimicrobials. Local Environ. 2013, 3, 1016–1027. [Google Scholar]
- Belguesmia, Y.; Naghmouchi, K.; Chihib, N.-E.; Drider, D. Class IIa bacteriocins: Current knowledge and perspectives. In Prokaryotic Antimicrobial Peptides; Springer: New York, NY, USA, 2011; pp. 171–195. [Google Scholar]
- Nissen-Meyer, J.; Oppegård, C.; Rogne, P.; Haugen, H.S.; Kristiansen, P.E. Structure and mode-of-action of the two-peptide (class-IIb) bacteriocins. Probiotics Antimicrob. Proteins 2010, 2, 52–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Zhang, C.; Wang, Y.; Shi, J.; Zhang, L.; Ding, Z.; Qu, X.; Cui, H. Class IIa bacteriocins: Diversity and new developments. Int. J. Mol. Sci. 2012, 13, 16668–16707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanatani, K.; Tahara, T.; Yoshida, K.; Miura, H.; Sakamoto, M.; Oshimura, M. Plasmid-associated bacteriocin production by and immunity of Lactobacillus acidophilus TK8912. Biosci. Biotechnol. Biochem. 1992, 56, 648–651. [Google Scholar] [CrossRef] [PubMed]
- Acedo, J.Z.; Chiorean, S.; Vederas, J.C.; van Belkum, M.J. The expanding structural variety among bacteriocins from Gram-positive bacteria. FEMS Microbiol. Rev. 2018, 42, 805–828. [Google Scholar] [CrossRef] [PubMed]
- Deraz, S.F.; Karlsson, E.N.; Khalil, A.A.; Mattiasson, B. Mode of action of acidocin D20079, a bacteriocin produced by the potential probiotic strain, Lactobacillus acidophilus DSM 20079. J. Ind. Microbiol. Biotechnol. 2007, 34, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Chumchalova, J.; Stiles, J.; Josephsen, J.; Plockova, M. Characterization and purification of acidocin CH5, a bacteriocin produced by Lactobacillus acidophilus CH5. J. Appl. Microbiol. 2004, 96, 1082–1089. [Google Scholar] [CrossRef]







| Enterococcus | Indicator Strain | ZOI (mm) | p-Value |
|---|---|---|---|
| KAE 01 | PA 01 | 12 ± 0.1 | 0.001 * |
| PA 02 | 11 ± 0.2 | ||
| PA 03 | 12 ± 0.5 |
| pH Value | ZOI (mm) | p-Value |
|---|---|---|
| 2.0 | 17.9 ± 0.9 | 0.001 * |
| 3.0 | 18.8 ± 0.1 | |
| 4.0 | 19.5 ± 2 | |
| 5.0 | 23.2 ± 0.1 | |
| 6.0 | 22.4 ± 0.1 | |
| 7.0 | 20.8 ± 0.2 | |
| 8.0 | 17.9 ± 0.1 | |
| 9.0 | 16.2 ± 0.5 | |
| 10.0 | 15.7 ± 0.6 |
| Temperature (°C) | ZOI (mm) | p-Value |
|---|---|---|
| −20 | 23.8 ± 0.1 | 0.001 * |
| 4.0 | 24.0 ± 0.1 | |
| 30 | 24.5 ± 0.05 | |
| 60 | 22.8 ± 0.1 | |
| 90 | 21.1 ± 0.3 | |
| 100 | 20.8 ± 0.6 | |
| 121 * | 17.4 ± 0.5 |
| Proteolytic Enzymes | ZOI (mm) | p-value |
|---|---|---|
| Pepsin | 5.5 ± 0.1 | 0.001 * |
| Trypsin | 8.6 ± 0.6 | |
| Proteinase K | 10.5 ± 0.2 | |
| Alkaline Protease | 7.5 ± 0.2 |
| Peptides | Cluster | Members (Docked Conformations) | Representative | Weighted Score (KJ/mol) | ΔG (Kcal/mol) | Kd (M) at 25 °C |
|---|---|---|---|---|---|---|
| KAE01 EntA | 3 | 46 | Center Lowest-energy structure | −955.8 −1046.0 | −12.4 | 8 × 10−10 |
| KAE01 EntP | 2 | 49 | Center Lowest-energy structure | −732.4 −732.4 | −16.4 | 8.9 × 10−13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bashir, A.; Ali, K.; Bux, K.; Farid, N.; Khaireabadi, M.; Hassan, K.A.; Hussain, A.; Fatima, K.; Mehmood, S.; Haider, S.A.; et al. Molecular Characterization, Purification, and Mode of Action of Enterocin KAE01 from Lactic Acid Bacteria and Its In Silico Analysis against MDR/ESBL Pseudomonas aeruginosa. Genes 2022, 13, 2333. https://doi.org/10.3390/genes13122333
Bashir A, Ali K, Bux K, Farid N, Khaireabadi M, Hassan KA, Hussain A, Fatima K, Mehmood S, Haider SA, et al. Molecular Characterization, Purification, and Mode of Action of Enterocin KAE01 from Lactic Acid Bacteria and Its In Silico Analysis against MDR/ESBL Pseudomonas aeruginosa. Genes. 2022; 13(12):2333. https://doi.org/10.3390/genes13122333
Chicago/Turabian StyleBashir, Asma, Kashif Ali, Khair Bux, Neha Farid, Mitra Khaireabadi, Khwaja Ali Hassan, Abrar Hussain, Kiran Fatima, Shahab Mehmood, Syed Ali Haider, and et al. 2022. "Molecular Characterization, Purification, and Mode of Action of Enterocin KAE01 from Lactic Acid Bacteria and Its In Silico Analysis against MDR/ESBL Pseudomonas aeruginosa" Genes 13, no. 12: 2333. https://doi.org/10.3390/genes13122333
APA StyleBashir, A., Ali, K., Bux, K., Farid, N., Khaireabadi, M., Hassan, K. A., Hussain, A., Fatima, K., Mehmood, S., Haider, S. A., & Herwig, R. (2022). Molecular Characterization, Purification, and Mode of Action of Enterocin KAE01 from Lactic Acid Bacteria and Its In Silico Analysis against MDR/ESBL Pseudomonas aeruginosa. Genes, 13(12), 2333. https://doi.org/10.3390/genes13122333








