In Silico and In Vivo Evaluation of microRNA-181c-5p’s Role in Hepatocellular Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Database Analysis of the Expression of miR-181c-5p
2.2. Materials and Cells
2.3. Recombinant pmiR-181c-5p Construct Synthesis
2.4. Polyplex Formulation of pEGP-miR-181c-5p
2.5. LA-PAMAM-pmiR-181c-5p Transfection
2.6. Cell Viability Study
2.7. Animal Experiment
2.8. Blood and Tissue Sampling
2.9. Biochemical Analysis
2.10. Histopathological Analysis
2.11. Prediction of the Targets of miR-181c-5p
2.12. Functional Enrichment Analysis
2.13. RNA Extraction and Quantitative Real Time PCR (qRT-PCR)
2.14. Statistical Analysis
3. Results
3.1. MiR-181c-5p Expression Data in HCC
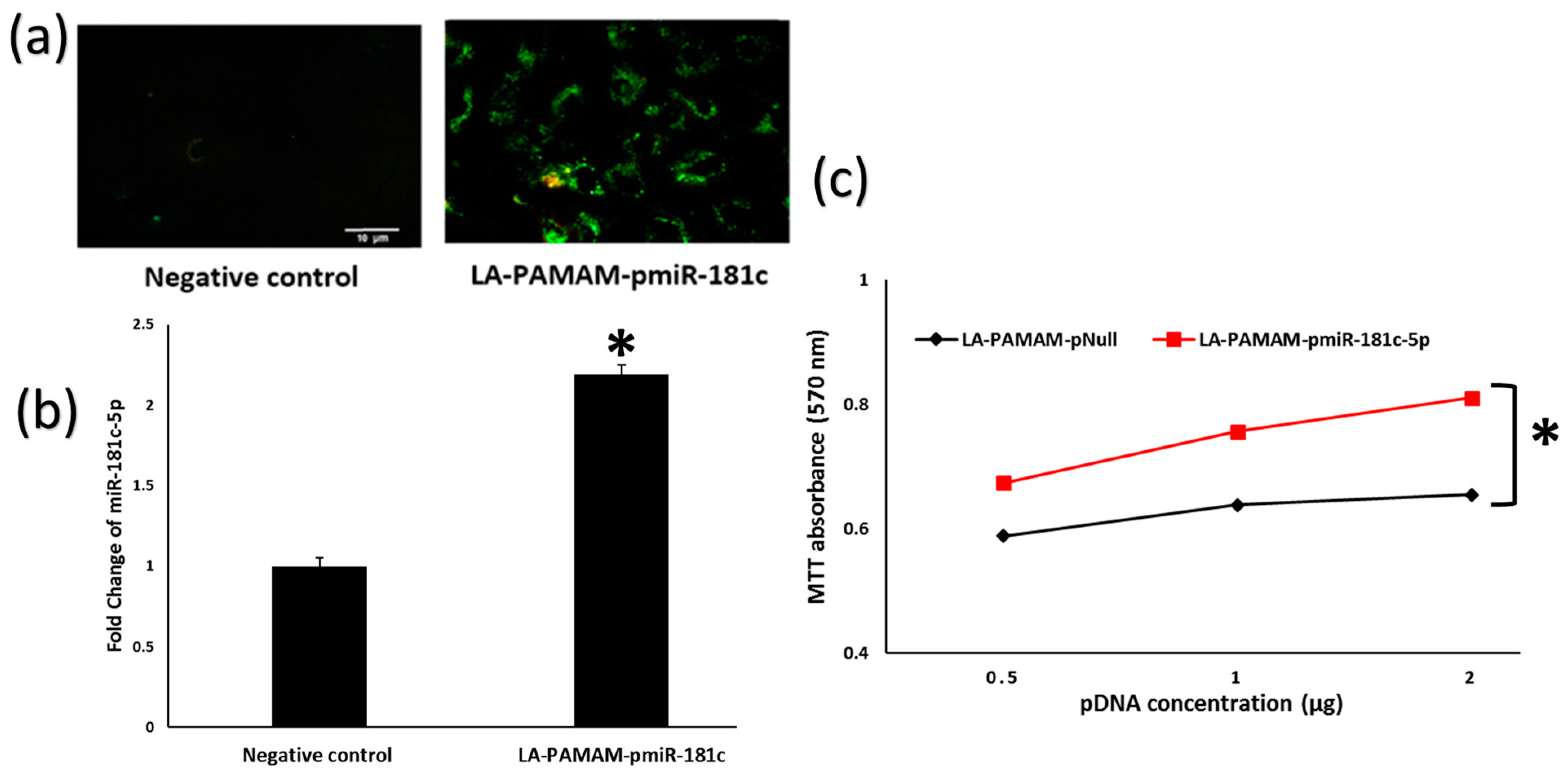
3.2. LA-PAMAM-pmiR-181c-5p Promotes HepG2 Proliferation
3.3. MiR-181c-5p Overexpression Contributes to HCC Tumor Progression
3.4. MiR-181c-5p Overexpression Deteriorates Liver Functions in the HCC Mouse Model
3.5. Bioinformatics Functional Analysis of MiR-181c-5p Downregulated Targets
3.6. Fbxl3 as a Target for MiR-181c-5p Involved in HCC
3.7. Correlation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kosmidis, C.; Koimtzis, G.; Pantos, G.; Atmatzidis, S.; Pavlidis, E.; Kosmidou, M.; Efthimiadis, C.; Anthimidis, G.; Varsamis, N.; Georgakoudi, E.; et al. Gene Therapy for Hepatocellular Carcinoma: An Update. J. Biomed. 2019, 4, 7–13. [Google Scholar] [CrossRef]
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- Akinyemiju, T.; Abera, S.; Ahmed, M.; Alam, N.; Alemayohu, M.A.; Allen, C.; Al-Raddadi, R.; Alvis-Guzman, N.; Amoako, Y.; Artaman, A.; et al. The Burden of Primary Liver Cancer and Underlying Etiologies from 1990 to 2015 at the Global, Regional, and National Level: Results from the Global Burden of Disease Study 2015. JAMA Oncol. 2017, 3, 1683–1691. [Google Scholar] [PubMed]
- Rashed, W.M.; Kandeil, M.A.M.; Mahmoud, M.O.; Ezzat, S. Hepatocellular Carcinoma (HCC) in Egypt: A comprehensive overview. J. Egypt. Natl. Cancer Inst. 2020, 32, 5. [Google Scholar] [CrossRef]
- Balogh, J.; Victor, D., 3rd; Asham, E.H.; Burroughs, S.G.; Boktour, M.; Saharia, A.; Li, X.; Ghobrial, R.M.; Monsour, H.P., Jr. Hepatocellular carcinoma: A review. J. Hepatocell. Carcinoma 2016, 3, 41–53. [Google Scholar] [CrossRef]
- Wang, X.; Liao, Z.; Bai, Z.; He, Y.; Duan, J.; Wei, L. MiR-93-5p Promotes Cell Proliferation through Down-Regulating PPARGC1A in Hepatocellular Carcinoma Cells by Bioinformatics Analysis and Experimental Verification. Genes 2018, 9, 51. [Google Scholar] [CrossRef]
- Huang, K.-W.; Hsu, F.-F.; Qiu, J.T.; Chern, G.-J.; Lee, Y.-A.; Chang, C.-C.; Huang, Y.-T.; Sung, Y.-C.; Chiang, C.-C.; Huang, R.-L.; et al. Highly efficient and tumor-selective nanoparticles for dual-targeted immunogene therapy against cancer. Sci. Adv. 2020, 6, eaax5032. [Google Scholar] [CrossRef]
- ElHefnawi, M.; Soliman, B.; Abu-Shahba, N.; Amer, M. An Integrative Meta-analysis of MicroRNAs in Hepatocellular Carcinoma. Genom. Proteom. Bioinform. 2013, 11, 354–367. [Google Scholar] [CrossRef]
- Munker, R.; Calin, G.A. MicroRNA profiling in cancer. Clin. Sci. 2011, 121, 141–158. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Soliman, B.; Salem, A.; Ghazy, M.; Abu-Shahba, N.; El Hefnawi, M. Bioinformatics functional analysis of let-7a, miR-34a, and miR-199a/b reveals novel insights into immune system pathways and cancer hallmarks for hepatocellular carcinoma. Tumor Biol. 2018, 40, 1010428318773675. [Google Scholar] [CrossRef] [PubMed]
- Elhefnawi, M.; Salah, Z.; Soliman, B. The Promise of miRNA Replacement Therapy for Hepatocellular Carcinoma. Curr. Gene Ther. 2019, 19, 290–304. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Osaki, M.; Takeshita, F.; Sugimoto, Y.; Kosaka, N.; Yamamoto, Y.; Yoshioka, Y.; Kobayashi, E.; Yamada, T.; Kawai, A.; Inoue, T.; et al. MicroRNA-143 Regulates Human Osteosarcoma Metastasis by Regulating Matrix Metalloprotease-13 Expression. Mol. Ther. 2011, 19, 1123–1130. [Google Scholar] [CrossRef]
- Qiu, T.; Zhou, X.; Wang, J.; Du, Y.; Xu, J.; Huang, Z.; Zhu, W.; Shu, Y.; Liu, P. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014, 588, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Amer, M.; Elhefnawi, M.; El-Ahwany, E.; Awad, A.F.; Gawad, N.A.; Zada, S.; Tawab, F.M.A. Hsa-miR-195 targets PCMT1 in hepatocellular carcinoma that increases tumor life span. Tumor Biol. 2014, 35, 11301–11309. [Google Scholar] [CrossRef]
- Salah, Z.; El Azeem, E.M.A.; Youssef, H.F.; Gamal-Eldeen, A.M.; Farrag, A.R.; El-Meliegy, E.; Soliman, B.; Elhefnawi, M. Effect of Tumor Suppressor MiR-34a Loaded on ZSM-5 Nanozeolite in Hepatocellular Carcinoma: In Vitro and In Vivo Approach. Curr. Gene Ther. 2019, 19, 342–354. [Google Scholar] [CrossRef]
- Ouyang, Y.-B.; Lu, Y.; Yue, S.; Giffard, R.G. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion 2012, 12, 213–219. [Google Scholar] [CrossRef]
- Belkaya, S.; Silge, R.L.; Hoover, A.R.; Medeiros, J.J.; Eitson, J.L.; Becker, A.M.; De La Morena, M.T.; Bassel-Duby, R.S.; Van Oers, N.S.C. Dynamic Modulation of Thymic MicroRNAs in Response to Stress. PLoS ONE 2011, 6, e27580. [Google Scholar] [CrossRef]
- Indrieri, A.; Carrella, S.; Carotenuto, P.; Banfi, S.; Franco, B. The Pervasive Role of the miR-181 Family in Development, Neurodegeneration, and Cancer. Int. J. Mol. Sci. 2020, 21, 2092. [Google Scholar] [CrossRef] [PubMed]
- Song, M.-K.; Park, Y.-K.; Ryu, J.-C. Polycyclic aromatic hydrocarbon (PAH)-mediated upregulation of hepatic microRNA-181 family promotes cancer cell migration by targeting MAPK phosphatase-5, regulating the activation of p38 MAPK. Toxicol. Appl. Pharmacol. 2013, 273, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hu, B.; Wang, Z.; Zhang, F.; Wei, H.; Li, L. miR-181c contributes to cisplatin resistance in non-small cell lung cancer cells by targeting Wnt inhibition factor 1. Cancer Chemother. Pharmacol. 2017, 80, 973–984. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Li, J.; Yue, W. MiR-181c modulates the proliferation, migration, and invasion of neuroblastoma cells by targeting Smad7. Acta Biochim. Biophys. Sin. 2013, 46, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Huang, J.; Han, Y.; Hao, J.; Wu, X.; Song, H.; Chen, X.; Shen, Q.; Dong, X.; Pang, H.; et al. The microRNA miR-181c enhances chemosensitivity and reduces chemoresistance in breast cancer cells via down-regulating osteopontin. Int. J. Biol. Macromol. 2018, 125, 544–556. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Liu, Z.; Peng, Y.; Yu, C. MicroRNA-181c inhibits glioblastoma cell invasion, migration and mesenchymal transition by targeting TGF-β pathway. Biochem. Biophys. Res. Commun. 2015, 469, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Lupo, G.; La Rosa, G.R.M.; Crimi, S.; Anfuso, C.D.; Salemi, R.; Rapisarda, E.; Libra, M.; Candido, S. Identification of Novel MicroRNAs and Their Diagnostic and Prognostic Significance in Oral Cancer. Cancers 2019, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Zabaglia, L.M.; Bartolomeu, N.C.; Dos Santos, M.P.; Peruquetti, R.L.; Chen, E.; de Arruda Cardoso Smith, M.; Payão, S.L.M.; Rasmussen, L.T. Decreased MicroRNA miR-181c Expression Associated with Gastric Cancer. J. Gastrointest. Cancer 2018, 49, 97–101. [Google Scholar] [CrossRef]
- Zhang, W.-L.; Zhang, J.-H. miR-181c promotes proliferation via suppressing PTEN expression in inflammatory breast cancer. Int. J. Oncol. 2015, 46, 2011–2020. [Google Scholar] [CrossRef]
- Chen, M.; Wang, M.; Xu, S.; Guo, X.; Jiang, J. Upregulation of miR-181c contributes to chemoresistance in pancreatic cancer by inactivating the Hippo signaling pathway. Oncotarget 2015, 6, 44466–44479. [Google Scholar] [CrossRef]
- Ji, J.; Yamashita, T.; Budhu, A.; Forgues, M.; Jia, H.-L.; Li, C.; Deng, C.; Wauthier, E.; Reid, L.M.; Ye, Q.-H.; et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology 2009, 50, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Gramantieri, L.; Ferracin, M.; Fornari, F.; Veronese, A.; Sabbioni, S.; Liu, C.-G.; Calin, G.A.; Giovannini, C.; Ferrazzi, E.; Grazi, G.L.; et al. Cyclin G1 Is a Target of miR-122a, a MicroRNA Frequently Down-regulated in Human Hepatocellular Carcinoma. Cancer Res. 2007, 67, 6092–6099. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hao, F.; Fei, X.; Chen, Y. SPP1 functions as an enhancer of cell growth in hepatocellular carcinoma targeted by miR-181c. Am. J. Transl. Res. 2019, 11, 6924–6937. [Google Scholar] [PubMed]
- Ai, J.; Gong, C.; Wu, J.; Gao, J.; Liu, W.; Liao, W.; Wu, L. MicroRNA-181c suppresses growth and metastasis of hepatocellular carcinoma by modulating NCAPG. Cancer Manag. Res. 2019, 11, 3455–3467. [Google Scholar] [CrossRef]
- Du, J.; Sun, Y.; Shi, Q.-S.; Liu, P.-F.; Zhu, M.-J.; Wang, C.-H.; Du, L.-F.; Duan, Y.-R. Biodegradable Nanoparticles of mPEG-PLGA-PLL Triblock Copolymers as Novel Non-Viral Vectors for Improving siRNA Delivery and Gene Silencing. Int. J. Mol. Sci. 2012, 13, 516–533. [Google Scholar] [CrossRef]
- Dizaj, S.M.; Jafari, S.; Khosroushahi, A.Y. A sight on the current nanoparticle-based gene delivery vectors. Nanoscale Res. Lett. 2014, 9, 252. [Google Scholar] [CrossRef]
- Abedi-Gaballu, F.; Dehghan, G.; Ghaffari, M.; Yekta, R.; Abbaspour-Ravasjani, S.; Baradaran, B.; Ezzati Nazhad Dolatabadi, J.; Hamblin, M.R. PAMAM dendrimers as efficient drug and gene delivery nanosystems for cancer therapy. Appl. Mater. Today 2018, 12, 177–190. [Google Scholar] [CrossRef]
- Bono, N.; Pennetta, C.; Bellucci, M.C.; Sganappa, A.; Malloggi, C.; Tedeschi, G.; Candiani, G.; Volonterio, A. Role of Generation on Successful DNA Delivery of PAMAM–(Guanidino)Neomycin Conjugates. ACS Omega 2019, 4, 6796–6807. [Google Scholar] [CrossRef]
- Guo, R.; Yao, Y.; Cheng, G.; Wang, S.H.; Li, Y.; Shen, M.; Zhang, Y.; Baker, J.R.; Wang, J.; Shi, X. Synthesis of glycoconjugated poly(amindoamine) dendrimers for targeting human liver cancer cells. RSC Adv. 2011, 2, 99–102. [Google Scholar] [CrossRef]
- Fu, F.; Wu, Y.; Zhu, J.; Wen, S.; Shen, M.; Shi, X. Multifunctional Lactobionic Acid-Modified Dendrimers for Targeted Drug Delivery to Liver Cancer Cells: Investigating the Role Played by PEG Spacer. ACS Appl. Mater. Interfaces 2014, 6, 16416–16425. [Google Scholar] [CrossRef]
- Iacobazzi, R.M.; Porcelli, L.; Lopedota, A.A.; Laquintana, V.; Lopalco, A.; Cutrignelli, A.; Altamura, E.; Di Fonte, R.; Azzariti, A.; Franco, M.; et al. Targeting human liver cancer cells with lactobionic acid-G(4)-PAMAM-FITC sorafenib loaded dendrimers. Int. J. Pharm. 2017, 528, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, W.; Wang, B.; Gao, Y.; Song, Z.; Zheng, Q.C. Ligand-based targeted therapy: A novel strategy for hepatocellular carcinoma. Int. J. Nanomed. 2016, 11, 5645–5669. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, A.M.; Mohamed, R.H.; El-Hakam, F.E.-Z.A.; Yassin, M.A.; ElHefnawi, M. Targeted delivery of miR-218 via decorated hyperbranched polyamidoamine for liver cancer regression. Int. J. Pharm. 2021, 610, 121256. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wang, Y.; Ling, Y.; Zhou, C.; Wang, H.; Teschendorff, A.E.; Zhao, Y.; Zhao, H.; He, Y.; Zhang, G.; et al. dbDEMC 3.0: Functional Exploration of Differentially Expressed miRNAs in Cancers of Human and Model Organisms. Genom. Proteom. Bioinform. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Shi, J.; Gao, Y.; Cui, C.; Zhang, S.; Li, J.; Zhou, Y.; Cui, Q. HMDD v3.0: A database for experimentally supported human microRNA–disease associations. Nucleic Acids Res. 2018, 47, D1013–D1017. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009, 37, D98–D104. [Google Scholar] [CrossRef]
- Xie, B.; Ding, Q.; Han, H.; Wu, D. miRCancer: A microRNA-cancer association database constructed by text mining on literature. Bioinformatics 2013, 29, 638–644. [Google Scholar] [CrossRef]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013, 42, D92–D97. [Google Scholar] [CrossRef]
- Titford, M. The long history of hematoxylin. Biotech. Histochem. 2005, 80, 73–78. [Google Scholar] [CrossRef]
- Paradis, V. Histopathology of hepatocellular carcinoma. In Multidisciplinary Treatment of Hepatocellular Carcinoma; Recent Results Cancer Research; Springer Nature: Cham, Switzerland, 2013; Volume 190, pp. 21–32. [Google Scholar] [CrossRef]
- Dweep, H.; Gretz, N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef]
- Ouyang, G.; Yi, B.; Pan, G.; Chen, X. A robust twelve-gene signature for prognosis prediction of hepatocellular carcinoma. Cancer Cell Int. 2020, 20, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Yamashita, T.; Wang, X.W. Wnt/beta-catenin signaling activates microRNA-181 expression in hepatocellular carcinoma. Cell Biosci. 2011, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Hatano, E.; Kitamura, K.; Myomoto, A.; Fujiwara, T.; Takizawa, S.; Tsuchiya, S.; Tsujimoto, G.; Uemoto, S.; Shimizu, K. MicroRNA Profile Predicts Recurrence after Resection in Patients with Hepatocellular Carcinoma within the Milan Criteria. PLoS ONE 2011, 6, e16435. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yu, K.; Zheng, J.; Lin, H.; Zhao, Q.; Zhang, X.; Feng, W.; Wang, L.; Xu, J.; Xie, D.; et al. Dysregulation, functional implications, and prognostic ability of the circadian clock across cancers. Cancer Med. 2019, 8, 1710–1720. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Pogribny, I.P.; Rusyn, I. The DEN and CCl4-Induced Mouse Model of Fibrosis and Inflammation-Associated Hepatocellular Carcinoma. Curr. Protoc. Pharmacol. 2014, 66, 14.30.1–14.30.10. [Google Scholar] [CrossRef]
- Zhang, W.; Ge, Y.; Cheng, Q.; Zhang, Q.; Fang, L.; Zheng, J. Decorin is a pivotal effector in the extracellular matrix and tumour microenvironment. Oncotarget 2018, 9, 5480–5491. [Google Scholar] [CrossRef]
- Horváth, Z.; Kovalszky, I.; Fullár, A.; Kiss, K.; Schaff, Z.; Iozzo, R.V.; Baghy, K. Decorin deficiency promotes hepatic carcinogenesis. Matrix Biol. 2014, 35, 194–205. [Google Scholar] [CrossRef]
- Karin, M.; Dhar, D. Liver carcinogenesis: From naughty chemicals to soothing fat and the surprising role of NRF2. Carcinogenesis 2016, 37, 541–546. [Google Scholar] [CrossRef]
- Pang, Y.; Li, J.; Gao, L.; Yang, X.; Dang, Y.; Lai, Z.; Liu, L.; Yang, J.; Wu, H.; He, R.; et al. The clinical value and potential molecular mechanism of the downregulation of MAOA in hepatocellular carcinoma tissues. Cancer Med. 2020, 9, 8004–8019. [Google Scholar] [CrossRef]
- Jiang, X.; Hao, Y. Analysis of expression profile data identifies key genes and pathways in hepatocellular carcinoma. Oncol. Lett. 2017, 15, 2625–2630. [Google Scholar] [CrossRef]
- Yang, Z.; Tao, Y.; Xu, X.; Cai, F.; Yu, Y.; Ma, L. Bufalin inhibits cell proliferation and migration of hepatocellular carcinoma cells via APOBEC3F induced intestinal immune network for IgA production signaling pathway. Biochem. Biophys. Res. Commun. 2018, 503, 2124–2131. [Google Scholar] [CrossRef] [PubMed]
- Thakur, C.; Chen, F. Connections between metabolism and epigenetics in cancers. Semin. Cancer Biol. 2019, 57, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Vettore, L.; Westbrook, R.; Tennant, D.A. New aspects of amino acid metabolism in cancer. Br. J. Cancer 2019, 122, 150–156. [Google Scholar] [CrossRef]
- Ai, Y.; Wang, B.; Xiao, S.; Luo, S.; Wang, Y. Tryptophan Side-Chain Oxidase Enzyme Suppresses Hepatocellular Carcinoma Growth through Degradation of Tryptophan. Int. J. Mol. Sci. 2021, 22, 12428. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Zuo, Q.; Ruan, H.; Wang, H.; Jin, H.; Cheng, Z.; Lv, Y.; Qin, W.; Wang, C. Argininosuccinate synthase 1 suppresses cancer cell invasion by inhibiting STAT3 pathway in hepatocellular carcinoma. Acta Biochim. Biophys. Sin. 2019, 51, 263–276. [Google Scholar] [CrossRef]
- Pettinelli, P.; Arendt, B.M.; Teterina, A.; McGilvray, I.; Comelli, E.M.; Fung, S.K.; Fischer, S.E.; Allard, J.P. Altered hepatic genes related to retinol metabolism and plasma retinol in patients with non-alcoholic fatty liver disease. PLoS ONE 2018, 13, e0205747. [Google Scholar] [CrossRef]
- Shirakami, Y.; Sakai, H.; Shimizu, M. Retinoid roles in blocking hepatocellular carcinoma. Hepatobiliary Surg. Nutr. 2015, 4, 222–228. [Google Scholar] [CrossRef]
- Tsuchiya, H.; Akechi, Y.; Ikeda, R.; Nishio, R.; Sakabe, T.; Terabayashi, K.; Matsumi, Y.; Ashla, A.A.; Hoshikawa, Y.; Kurimasa, A.; et al. Suppressive Effects of Retinoids on Iron-Induced Oxidative Stress in the Liver. Gastroenterology 2009, 136, 341–350.e8. [Google Scholar] [CrossRef]
- Suzui, M.; Masuda, M.; Lim, J.T.E.; Albanese, C.; Pestell, R.G.; Weinstein, I.B. Growth inhibition of human hepatoma cells by acyclic retinoid is associated with induction of p21(CIP1) and inhibition of expression of cyclin D1. Cancer Res. 2002, 62, 3997–4006. [Google Scholar]
- Adachi, S.; Okuno, M.; Matsushima-Nishiwaki, R.; Takano, Y.; Kojima, S.; Friedman, S.L.; Moriwaki, H.; Okano, Y. Phosphorylation of retinoid X receptor suppresses its ubiquitination in human hepatocellular carcinoma. Hepatology 2002, 35, 332–340. [Google Scholar] [CrossRef]
- Nakamura, N.; Shidoji, Y.; Moriwaki, H.; Muto, Y. Apoptosis in Human Hepatoma Cell Line Induced by 4,5-Didehydro geranylgeranoic Acid (Acyclic Retinoid) via Down-Regulation of Transforming Growth Factor-α. Biochem. Biophys. Res. Commun. 1996, 219, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Shao, R.-X.; Otsuka, M.; Kato, N.; Taniguchi, H.; Hoshida, Y.; Moriyama, M.; Kawabe, T.; Omata, M. Acyclic retinoid inhibits human hepatoma cell growth by suppressing fibroblast growth factor-mediated signaling pathways. Gastroenterology 2005, 128, 86–95. [Google Scholar] [CrossRef]
- Okada, H.; Honda, M.; Campbell, J.S.; Sakai, Y.; Yamashita, T.; Takebuchi, Y.; Hada, K.; Shirasaki, T.; Takabatake, R.; Nakamura, M.; et al. Acyclic Retinoid Targets Platelet-Derived Growth Factor Signaling in the Prevention of Hepatic Fibrosis and Hepatocellular Carcinoma Development. Cancer Res. 2012, 72, 4459–4471. [Google Scholar] [CrossRef]
- Li, N.; Li, L.; Chen, Y. The Identification of Core Gene Expression Signature in Hepatocellular Carcinoma. Oxidative Med. Cell. Longev. 2018, 2018, 3478305-15. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Han, X.; Li, C.; Bai, W. FBXL3 is regulated by miRNA-4735-3p and suppresses cell proliferation and migration in non-small cell lung cancer. Pathol.-Res. Pract. 2018, 215, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Tekcham, D.S.; Chen, D.; Liu, Y.; Ling, T.; Zhang, Y.; Chen, H.; Wang, W.; Otkur, W.; Qi, H.; Xia, T.; et al. F-box proteins and cancer: An update from functional and regulatory mechanism to therapeutic clinical prospects. Theranostics 2020, 10, 4150–4167. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhu, Y.; Hong, X.; Zhang, M.; Qiu, X.; Wang, Z.; Qi, Z.; Hong, X. miR-181d and c-myc-mediated inhibition of CRY2 and FBXL3 reprograms metabolism in colorectal cancer. Cell Death Dis. 2017, 8, e2958. [Google Scholar] [CrossRef]
- Sun, X.; Sit, A.; Feinberg, M.W. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc. Med. 2013, 24, 105–112. [Google Scholar] [CrossRef]






| Gene | Forward Sequence | Reverse Sequence |
|---|---|---|
| mGAPDH mFbxl3 | 5′-TCATCATCTCCGCCCCTTCT-3′ 5′-TAGACGACACCCCAGTCGAT-3′ | 5′-TGATGGCATGGACTGTGGTC-3′ 5′-CACAGAATGCCTGCTGGAGA-3′ |
| Expression Status | Design | Reference | Database |
|---|---|---|---|
| Down | Microarray on HCCs and LCs | L. Gramantieri et al. (2007) [32] | MiR2Disease Base [46] |
| Down | Hep3B vs L02 cells | J. Wang et al. (2019) [33] | |
| Down | Microarray on tumor tissues and adjacent non-tumor tissues | J. Ai et al. (2019) [34] | |
| Up | HpSC-HCC vs MH-HCC | J. Ji et al. (2009) [31] | MiR2Disease Base [46] MiR Cancer [47] |
| Up | Human HCC cell lines (Hep3B, HuH7, HuH1, MHCC97 and Smmc7721) | J. Ji et al. (2011) [53] | HMDD [45] |
| Up | Microarray on tumor and non-tumor hepatic tissue specimens of HCC patients | Sato F et al. (2011) [54] | dbDEMC2 [44] |
| Gene | Fold Change miR-181c-5p | Fold Change Fbxl3 |
|---|---|---|
| Fbxl3 | ||
| Pearson Correlation | −0.952 ** | 1 |
| p-value | 0.000 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abd ElAziz, O.N.; Elfiky, A.M.; Yassin, M.A.; Abd El-Hakam, F.E.-Z.; Saleh, E.M.; El-Hefnawi, M.; Mohamed, R.H. In Silico and In Vivo Evaluation of microRNA-181c-5p’s Role in Hepatocellular Carcinoma. Genes 2022, 13, 2343. https://doi.org/10.3390/genes13122343
Abd ElAziz ON, Elfiky AM, Yassin MA, Abd El-Hakam FE-Z, Saleh EM, El-Hefnawi M, Mohamed RH. In Silico and In Vivo Evaluation of microRNA-181c-5p’s Role in Hepatocellular Carcinoma. Genes. 2022; 13(12):2343. https://doi.org/10.3390/genes13122343
Chicago/Turabian StyleAbd ElAziz, Omnia Nasser, Asmaa M. Elfiky, Mohamed A. Yassin, Fatma El-Zahraa Abd El-Hakam, Eman M. Saleh, Mahmoud El-Hefnawi, and Rania Hassan Mohamed. 2022. "In Silico and In Vivo Evaluation of microRNA-181c-5p’s Role in Hepatocellular Carcinoma" Genes 13, no. 12: 2343. https://doi.org/10.3390/genes13122343
APA StyleAbd ElAziz, O. N., Elfiky, A. M., Yassin, M. A., Abd El-Hakam, F. E.-Z., Saleh, E. M., El-Hefnawi, M., & Mohamed, R. H. (2022). In Silico and In Vivo Evaluation of microRNA-181c-5p’s Role in Hepatocellular Carcinoma. Genes, 13(12), 2343. https://doi.org/10.3390/genes13122343







