Gene Networks and Pathways Involved in LPS-Induced Proliferative Response of Bovine Endometrial Epithelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experiment Design
2.2. Bioinformatics Analysis (Classification of DEGs Involved in Cell Proliferation)
2.3. Transcription Factor Analysis
2.4. Functional Annotation and Pathways Analysis
3. Results
3.1. RNASeq Analysis
3.2. Results of the Comparison between Control 24 h and LPS-2 µg/mL 24 h
3.3. Classification of DEGs Based on Their Functions in Cell Cycle
3.4. Differential Expression of Multiple Transcription Factor Families
3.5. Biological Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, Y.; Van Schaik, T.; Jhamat, N.; Niazi, A.; Chanrot, M.; Charpigny, G.; Valarcher, J.F.; Bongcam-Rudloff, E.; Andersson, G.; Humblot, P. Differential gene expression in bovine endometrial epithelial cells after challenge with LPS; specific implications for genes involved in embryo maternal interactions. PLoS ONE 2019, 14, e0222081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogeveen, H.; Huijps, K.; Lam, T. Economic aspects of mastitis: New developments. N. Z. Vet. J. 2011, 59, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Sordillo, L.M.; Streicher, K.L. Mammary gland immunity and mastitis susceptibility. J. Mammary Gland. Biol. Neoplasia 2002, 7, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Arnold, L.; Stowe, C.; Harmon, R.; Bewley, J. Estimating US dairy clinical disease costs with a stochastic simulation model. J. Dairy Sci. 2017, 100, 1472–1486. [Google Scholar] [CrossRef] [Green Version]
- Cronin, J.G.; Turner, M.L.; Goetze, L.; Bryant, C.E.; Sheldon, I.M. Toll-like receptor 4 and MYD88-dependent signaling mechanisms of the innate immune system are essential for the response to lipopolysaccharide by epithelial and stromal cells of the bovine endometrium. Biol. Reprod. 2012, 86, 51–59. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, I.M.; Williams, E.J.; Miller, A.N.; Nash, D.M.; Herath, S. Uterine diseases in cattle after parturition. Vet. J. 2008, 176, 115–121. [Google Scholar] [CrossRef]
- Sheldon, I.M.; Roberts, M.H. Toll-like receptor 4 mediates the response of epithelial and stromal cells to lipopolysaccharide in the endometrium. PLoS ONE 2010, 5, e12906. [Google Scholar] [CrossRef] [Green Version]
- Herath, S.; Williams, E.J.; Lilly, S.T.; Gilbert, R.O.; Dobson, H.; Bryant, C.E.; Sheldon, I.M. Ovarian follicular cells have innate immune capabilities that modulate their endocrine function. Reproduction 2007, 134, 683–693. [Google Scholar] [CrossRef]
- Sheldon, I.; Noakes, D.; Rycroft, A.; Pfeiffer, D.; Dobson, H. Influence of uterine bacterial contamination after parturition on ovarian dominant follicle selection and follicle growth and function in cattle. Reproduction 2002, 123, 837–845. [Google Scholar] [CrossRef]
- Williams, E.J.; Sibley, K.; Miller, A.N.; Lane, E.A.; Fishwick, J.; Nash, D.M.; Herath, S.; England, G.C.; Dobson, H.; Sheldon, I.M. The effect of Escherichia coli lipopolysaccharide and tumour necrosis factor α on ovarian function. Am. J. Reprod. Immunol. 2008, 60, 462–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavon, Y.; Leitner, G.; Klipper, E.; Moallem, U.; Meidan, R.; Wolfenson, D. Subclinical, chronic intramammary infection lowers steroid concentrations and gene expression in bovine preovulatory follicles. Domest. Anim. Endocrinol. 2011, 40, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, Y.; Lin, C.; Yan, R.; Li, Z.; Chen, Q.; Zhang, H.; Xu, H.; Chen, X.; Chen, Y. Regularity of Toll-Like Receptors in Bovine Mammary Epithelial Cells Induced by Mycoplasma bovis. Front. Vet. Sci. 2022, 9, 846700. [Google Scholar] [CrossRef] [PubMed]
- Verspohl, E.J.; Podlogar, J. LPS-induced proliferation and chemokine secretion from BEAS-2B cells. Pharmacol. Pharm. 2012, 3, 18716. [Google Scholar] [CrossRef] [Green Version]
- Ulmer, A.J.; Flad, H.-D.; Rietschel, T.; Mattern, T. Induction of proliferation and cytokine production in human T lymphocytes by lipopolysaccharide (LPS). Toxicology 2000, 152, 37–45. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2018, 9, 7204. [Google Scholar] [CrossRef] [Green Version]
- Chanrot, M.; Guo, Y.; Dalin, A.; Persson, E.; Båge, R.; Svensson, A.; Gustafsson, H.; Humblot, P. Dose related effects of LPS on endometrial epithelial cell populations from dioestrus cows. Anim. Reprod. Sci. 2017, 177, 12–24. [Google Scholar] [CrossRef]
- He, Z.; Gao, Y.; Deng, Y.; Li, W.; Chen, Y.; Xing, S.; Zhao, X.; Ding, J.; Wang, X. Lipopolysaccharide induces lung fibroblast proliferation through Toll-like receptor 4 signaling and the phosphoinositide3-kinase-Akt pathway. PLoS ONE 2012, 7, e35926. [Google Scholar] [CrossRef] [Green Version]
- Marcia Delattre, A.; Carabelli, B.; Aurélio Mori, M.; Pudell, C.; RBL da Silva, D.; Menezes, I.; RG Kempe, P.; Vinícius Staziaki, P.; Dombrowski, A.P.; da Cunha, C. Multiple intranigral unilateral LPS infusion protocol generates a persistent cognitive impairment without cumulative dopaminergic impairment. CNS Neurol. Disord. Drug Targets 2013, 12, 1002–1010. [Google Scholar] [CrossRef]
- Eslani, M.; Movahedan, A.; Afsharkhamseh, N.; Sroussi, H.; Djalilian, A.R. The role of toll-like receptor 4 in corneal epithelial wound healing. Investig. Ophthalmol. Vis. Sci. 2014, 55, 6108–6115. [Google Scholar] [CrossRef]
- Español, A.J.; Maddaleno, M.; Lombardi, M.G.; Cella, M.; Martinez Pulido, P.; Sales, M.E. Treatment with LPS plus INF-γ induces the expression and function of muscarinic acetylcholine receptors, modulating NIH 3 T 3 cell proliferation: Participation of NOS and COX. Br. J. Pharmacol. 2014, 171, 5154–5167. [Google Scholar] [PubMed] [Green Version]
- Niranjan, R.; Nagarajan, R.; Hanif, K.; Nath, C.; Shukla, R. LPS induces mediators of neuroinflammation, cell proliferation, and GFAP expression in human astrocytoma cells U373MG: The anti-inflammatory and anti-proliferative effect of guggulipid. Neurol. Sci. 2014, 35, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Wiktorowska-Owczarek, A.; Namiecińska, M.; Owczarek, J. The effect of ibuprofen on bFGF, VEGF secretion and cell proliferation in the presence of LPS in HMEC-1 cells. Acta Pol. Pharm. 2015, 72, 889–894. [Google Scholar] [PubMed]
- Tian, M.-Y.; Fan, J.-H.; Zhuang, Z.-W.; Dai, F.; Wang, C.-Y.; Hou, H.-T.; Ma, Y.-Z. Effects of silymarin on p65 NF-κB, p38 MAPK and CYP450 in LPS-induced hoof dermal inflammatory cells of dairy cows. BMC Vet. Res. 2019, 15, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farber, E. Cell proliferation as a major risk factor for cancer: A concept of doubtful validity. Cancer Res. 1995, 55, 3759–3762. [Google Scholar]
- Jhamat, N.; Niazi, A.; Guo, Y.; Chanrot, M.; Ivanova, E.; Kelsey, G.; Bongcam-Rudloff, E.; Andersson, G.; Humblot, P. LPS-treatment of bovine endometrial epithelial cells causes differential DNA methylation of genes associated with inflammation and endometrial function. BMC Genom. 2020, 21, 385. [Google Scholar] [CrossRef]
- Yokota, S.-i.; Okabayashi, T.; Rehli, M.; Fujii, N.; Amano, K.-i. Helicobacter pylori lipopolysaccharides upregulate toll-like receptor 4 expression and proliferation of gastric epithelial cells via the MEK1/2-ERK1/2 mitogen-activated protein kinase pathway. Infect. Immun. 2010, 78, 468–476. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Zhang, W.; Hu, X. Different concentrations of lipopolysaccharide regulate barrier function through the PI3K/Akt signalling pathway in human pulmonary microvascular endothelial cells. Sci. Rep. 2018, 8, 9963. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Sun, S.-P.; Zhu, H.-S.; Jiao, X.-Q.; Zhong, K.; Guo, Y.-J.; Zha, G.-M.; Han, L.-Q.; Yang, G.-Y.; Li, H.-P. GABA regulates the proliferation and apoptosis of MAC-T cells through the LPS-induced TLR4 signaling pathway. Res. Vet. Sci. 2018, 118, 395–402. [Google Scholar] [CrossRef]
- Cohn, Z.J.; Kim, A.; Huang, L.; Brand, J.; Wang, H. Lipopolysaccharide-induced inflammation attenuates taste progenitor cell proliferation and shortens the life span of taste bud cells. BMC Neurosci. 2010, 11, 72. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Lin, Y.; Liu, L.; Bian, Y.; Zhang, L.; Gao, X.; Li, Q. 14-3-3γ regulates lipopolysaccharide-induced inflammatory responses and lactation in dairy cow mammary epithelial cells by inhibiting NF-κB and MAPKs and up-regulating mTOR signaling. Int. J. Mol. Sci. 2015, 16, 16622–16641. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Fan, C.-W.; Maa, M.-C.; Leu, T.-H. Lipopolysaccharide-promoted proliferation of Caco-2 cells is mediated by c-Src induction and ERK activation. Biomedicine 2015, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Butchar, J.P.; Parsa, K.V.; Marsh, C.B.; Tridandapani, S. Negative regulators of toll-like receptor 4-mediated macrophage inflammatory response. Curr. Pharm. Des. 2006, 12, 4143–4153. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Zhou, A.; Xu, L.; Zhang, X. The role of TLR4-mediated PTEN/PI3K/AKT/NF-κB signaling pathway in neuroinflammation in hippocampal neurons. Neuroscience 2014, 269, 93–101. [Google Scholar] [CrossRef]
- Goodier, M.R.; Londei, M. Lipopolysaccharide stimulates the proliferation of human CD56+ CD3− NK cells: A regulatory role of monocytes and IL-10. J. Immunol. 2000, 165, 139–147. [Google Scholar] [CrossRef] [Green Version]
- Tulic, M.K.; Manoukian, J.J.; Eidelman, D.H.; Hamid, Q. T-cell proliferation induced by local application of LPS in the nasal mucosa of nonatopic children. J. Allergy Clin. Immunol. 2002, 110, 771–776. [Google Scholar] [CrossRef]
- Toda, K.; Kumagai, N.; Tsuchimoto, K.; Inagaki, H.; Suzuki, T.; Oishi, T.; Atsukawa, K.; Saito, H.; Morizane, T.; Hibi, T. Induction of hepatic stellate cell proliferation by LPS-stimulated peripheral blood mononuclear cells from patients with liver cirrhosis. J. Gastroenterol. 2000, 35, 214–220. [Google Scholar] [CrossRef]
- Knapp, M.; Jones, P.; Black, S.; Vitetta, E.; Slavin, S.; Strober, S. Characterization of a Spontaneous Murine B Cell Leukemia (BCL1): I. Cell Surface Expression of IgM, IgD, Ia, and FcR. J. Immunol. 1979, 123, 992–999. [Google Scholar]
- Zhong, C.; Cui, K.; Wilhelm, C.; Hu, G.; Mao, K.; Belkaid, Y.; Zhao, K.; Zhu, J. Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment. Nat. Immunol. 2016, 17, 169–178. [Google Scholar] [CrossRef] [Green Version]
- Chakravortty, D.; KS, N.K. Modulation of barrier function of small intestinal epithelial cells by lamina propria fibroblasts in response to lipopolysaccharide: Possible role of tnfα in inducing barrier dysfunction. Microbiol. Immunol. 1999, 43, 527–533. [Google Scholar] [CrossRef]
- Park, J.; Gores, G.J.; Patel, T. Lipopolysaccharide induces cholangiocyte proliferation via an interleukin-6–mediated activation of p44/p42 mitogen-activated protein kinase. Hepatology 1999, 29, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.-Y.; Tang, J.; Zheng, P.; Liu, Y. CD24 and Siglec-10 selectively repress tissue damage–induced immune responses. Science 2009, 323, 1722–1725. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, S.P.; Splinter, P.L.; Trussoni, C.E.; Gajdos, G.B.; Lineswala, P.N.; LaRusso, N.F. Cholangiocyte N-Ras protein mediates lipopolysaccharide-induced interleukin 6 secretion and proliferation. J. Biol. Chem. 2011, 286, 30352–30360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.-X.; Li, M.; Chen, X.-O.; Lian, Q.-Q.; Wang, Q.; Gao, F.; Jin, S.-W.; Zheng, S.-X. Lipoxin A4 ameliorates lipopolysaccharide-induced lung injury through stimulating epithelial proliferation, reducing epithelial cell apoptosis and inhibits epithelial–mesenchymal transition. Respir. Res. 2019, 20, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basso, F.G.; Soares, D.; Pansani, T.; Turrioni, A.; Scheffel, D.; de Souza Costa, C.; Hebling, J. Effect of LPS treatment on the viability and chemokine synthesis by epithelial cells and gingival fibroblasts. Arch. Oral Biol. 2015, 60, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shoji, W.; Takano, H.; Nishimura, N.; Aoki, Y.; Takahashi, R.; Goto, S.; Kaifu, T.; Takai, T.; Obinata, M. Increased susceptibility of MER5 (peroxiredoxin III) knockout mice to LPS-induced oxidative stress. Biochem. Biophys. Res. Commun. 2007, 355, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Klunker, L.; Kahlert, S.; Panther, P.; Diesing, A.-K.; Reinhardt, N.; Brosig, B.; Kersten, S.; Dänicke, S.; Rothkötter, H.-J.; Kluess, J. Deoxynivalenol and E. coli lipopolysaccharide alter epithelial proliferation and spatial distribution of apical junction proteins along the small intestinal axis. J. Anim. Sci. 2013, 91, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Freitag, A.; Reimann, A.; Wessler, I.; Racké, K. Effects of bacterial lipopolysaccharides (LPS) and tumour necrosis factor-α (TNFα) on rat tracheal epithelial cells in culture: Morphology, proliferation and induction of nitric oxide (NO) synthase. Pulm. Pharmacol. 1996, 9, 149–156. [Google Scholar] [CrossRef]
- Hei, Z.; Zhang, A.; Wei, J.; Gan, X.; Wang, Y.; Luo, G.; Li, X. Lipopolysaccharide effects on the proliferation of NRK52E cells via alternations in gap-junction function. J. Trauma Acute Care Surg. 2012, 73, 67–72. [Google Scholar] [CrossRef]
- Zhang, L.; Rees, M.; Bicknell, R. The isolation and long-term culture of normal human endometrial epithelium and stroma. Expression of mRNAs for angiogenic polypeptides basally and on oestrogen and progesterone challenges. J. Cell Sci. 1995, 108, 323–331. [Google Scholar] [CrossRef]
- Müller-Decker, K.; Manegold, G.; Butz, H.; Hinz, D.E.; Hüttner, D.; Richter, K.H.; Tremmel, M.; Weißflog, R.; Marks, F. Inhibition of cell proliferation by bacterial lipopolysaccharides in TLR4-positive epithelial cells: Independence of nitric oxide and cytokine release. J. Investig. Dermatol. 2005, 124, 553–561. [Google Scholar] [CrossRef] [Green Version]
- Daly, K.A.; Mailer, S.L.; Digby, M.R.; Lefévre, C.; Thomson, P.; Deane, E.; Nicholas, K.R.; Williamson, P. Molecular analysis of tammar (Macropus eugenii) mammary epithelial cells stimulated with lipopolysaccharide and lipoteichoic acid. Vet. Immunol. Immunopathol. 2009, 129, 36–48. [Google Scholar] [CrossRef]
- Calvinho, L.F.; Almeida, R.A.; Oliver, S. Influence of bacterial factors on proliferation of bovine mammary epithelial cells. Rev. Argent. Microbiol. 2001, 33, 28–35. [Google Scholar]
- Piotrowska-Tomala, K.; Siemieniuch, M.; Szóstek, A.; Korzekwa, A.; Woclawek-Potocka, I.; Galváo, A.; Okuda, K.; Skarzynski, D. Lipopolysaccharides, cytokines, and nitric oxide affect secretion of prostaglandins and leukotrienes by bovine mammary gland epithelial cells. Domest. Anim. Endocrinol. 2012, 43, 278–288. [Google Scholar] [CrossRef]
- Wang, X.; Dai, Y.; Zhang, X.; Pan, K.; Deng, Y.; Wang, J.; Xu, T. CXCL6 regulates cell permeability, proliferation, and apoptosis after ischemia–reperfusion injury by modulating Sirt3 expression via AKT/FOXO3a activation. Cancer Biol. Ther. 2021, 22, 30–39. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, K.; Liang, X.; Li, Y.; Zhang, Y.; Zhang, C.; Wei, H.; Luo, R.; Ge, S.; Xu, G. Complement C3 produced by macrophages promotes renal fibrosis via IL-17A secretion. Front. Immunol. 2018, 9, 2385. [Google Scholar] [CrossRef] [Green Version]
- Murphy-Ullrich, J.E. The de-adhesive activity of matricellular proteins: Is intermediate cell adhesion an adaptive state? J. Clin. Investig. 2001, 107, 785–790. [Google Scholar] [CrossRef] [Green Version]
- Bisello, A.; Horwitz, M.J.; Stewart, A.F. Parathyroid hormone-related protein: An essential physiological regulator of adult bone mass. Endocrinology 2004, 145, 3551–3553. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Liu, Q.; Zhang, Y.; Li, S.; Yi, S. Leukemia inhibitory factor regulates Schwann cell proliferation and migration and affects peripheral nerve regeneration. Cell Death Dis. 2021, 12, 417. [Google Scholar] [CrossRef]
- Laitaoja, M.; Valjakka, J.; Jäis, J. Zinc coordination spheres in protein structures. Inorg. Chem. 2013, 52, 10983–10991. [Google Scholar] [CrossRef]
- Susemihl, A.; Nagel, F.; Grabarczyk, P.; Schmidt, C.A.; Delcea, M. Easy Expression and Purification of Fluorescent N-Terminal BCL11B CCHC Zinc Finger Domain. Molecules 2021, 26, 7576. [Google Scholar] [CrossRef]
- Li, X.; Han, M.; Zhang, H.; Liu, F.; Pan, Y.; Zhu, J.; Liao, Z.; Chen, X.; Zhang, B. Structures and biological functions of zinc finger proteins and their roles in hepatocellular carcinoma. Biomark. Res. 2022, 10, 2. [Google Scholar] [CrossRef]
- Li, K.; Chen, C.; Gao, R.; Yu, X.; Huang, Y.; Chen, Z.; Liu, Z.; Chen, S.; Luo, G.; Huang, X. Inhibition of BCL11B induces downregulation of PTK7 and results in growth retardation and apoptosis in T-cell acute lymphoblastic leukemia. Biomark. Res. 2021, 9, 17. [Google Scholar] [CrossRef]
- Koo, J.H.; Plouffe, S.W.; Meng, Z.; Lee, D.-H.; Yang, D.; Lim, D.-S.; Wang, C.-Y.; Guan, K.-L. Induction of AP-1 by YAP/TAZ contributes to cell proliferation and organ growth. Genes Dev. 2020, 34, 72–86. [Google Scholar] [CrossRef]
- Miao, R.; Dai, C.C.; Mei, L.; Xu, J.; Sun, S.W.; Xing, Y.L.; Wu, L.S.; Wang, M.H.; Wei, J.F. KIAA1429 regulates cell proliferation by targeting c-jun messenger RNA directly in gastric cancer. J. Cell. Physiol. 2020, 235, 7420–7432. [Google Scholar] [CrossRef]


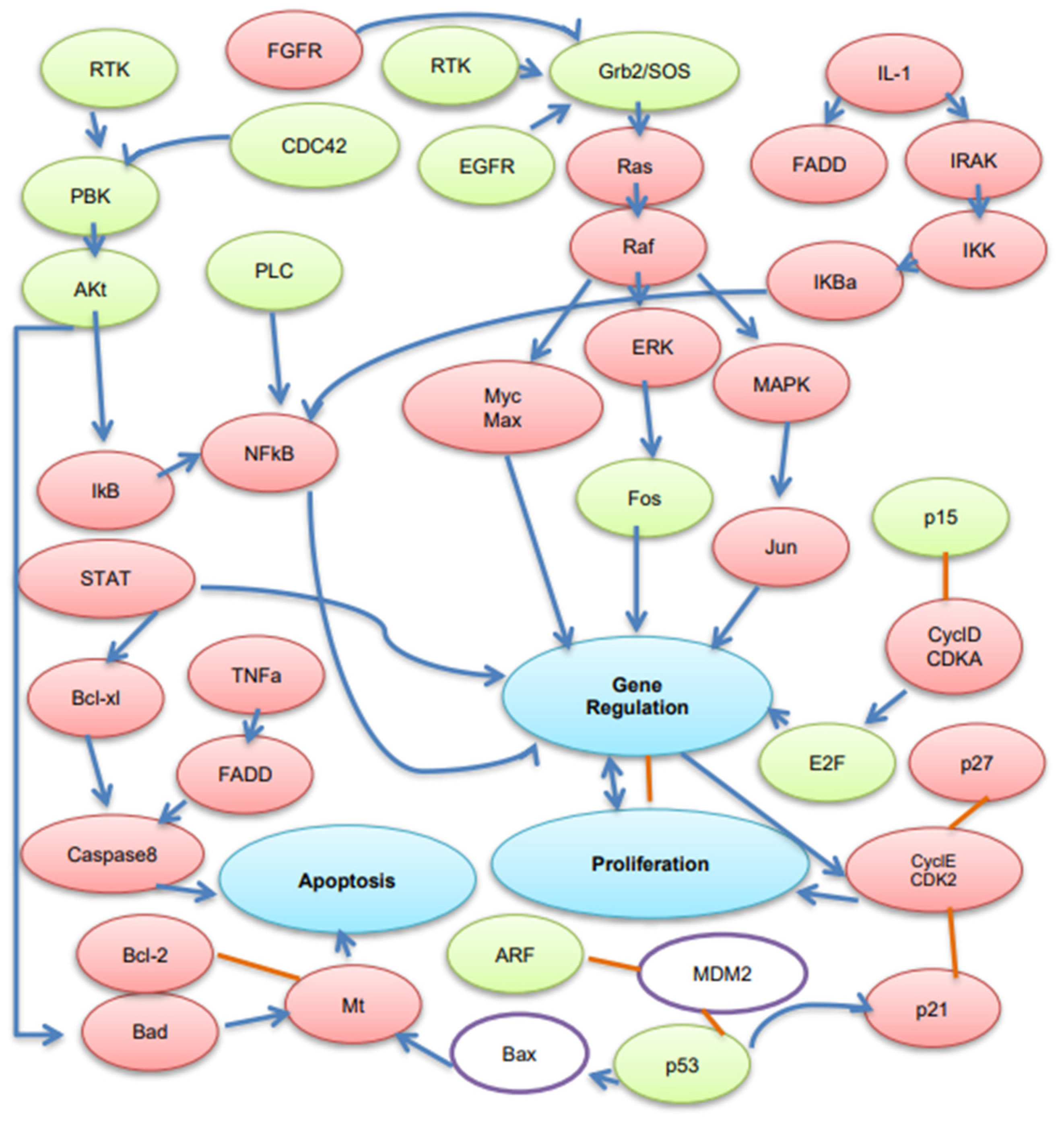
 ) show DEGs in our RNASeq reads. The red font in the figure shows tumor suppressor genes.
) show DEGs in our RNASeq reads. The red font in the figure shows tumor suppressor genes.
 ) show DEGs in our RNASeq reads. The red font in the figure shows tumor suppressor genes.
) show DEGs in our RNASeq reads. The red font in the figure shows tumor suppressor genes.
| Function | State | Fold-Change | Number | Ratio (Up/Down) |
|---|---|---|---|---|
| Proliferation (752) | Over-expressed | >2 log2_fold-change | 28 | 1.14 |
| >1 log2_fold-change | 110 | |||
| >0 log2_fold-change | 400 | |||
| Under-expressed | >1 log2_fold-change | 13 | ||
| >0 log2_fold-change | 352 |
| Gene Symbol | Gene Name | Known Function | Fold Change |
|---|---|---|---|
| CXCL6 | Granulocyte chemotactic protein 2 | Cytokine and chemokine activity, strong antibacterial activity | 6.35 |
| C3 | Complement component 3 | Activation of complement system to form mature proteins, modulates inflammation and possesses antimicrobial activity, activation of the PLC, MAPK, and AKT signaling pathways | 4.55 |
| BCL2A1 | BCL2-related protein A1 | Anti- and pro-apoptotic regulators, lymphocyte activation as well as cell survival | 4.21 |
| SLC5A5 | Solute carrier family 5 | Thyroid hormone synthesis and metabolism pathway. Increased viability | 3.79 |
| LGALS9 | Lectin, galactoside-binding, soluble, 9 | Enhancing cell migration, inhibits angiogenesis, activates ERK1/2 phosphorylation inducing cytokines (IL-6, IL-8, and IL-12) and chemokines (CCL2) | 3.49 |
| CXCL8 | Interleukin 8 | Chemotaxis; neutrophil activation; G-protein coupled receptor protein signaling pathway; angiogenesis | 3.35 |
| CTSC | Cathepsin C | Activation of many serine proteinases in cells of the immune system, protein binding. | 3.32 |
| CX3CL1 | Chemokine (C-X3-C motif) ligand 1 | Receptor binding and chemokine activity; regulating leukocyte adhesion and migration processes | 3.32 |
| CXCL3 | Chemokine (C-X-C motif) ligand 3 | Inflammation, chemokine activity, and CXCR chemokine receptor binding | 3.23 |
| CCL5 | Chemokine (C-C motif) ligand 5 | Immune-regulatory and inflammatory processes, activation of the PI3, Akt, and MAP kinases | 3.16 |
| TNF | Tumor necrosis factor | Acute phase response, pro-inflammatory immune response, regulation of cytokine secretion, insulin signaling, and glucose metabolism | 3.06 |
| IL1A | Interleukin 1 α | Immune responses, inflammatory processes, and cell proliferation. Stimulates the release of prostaglandin and collagenase. | 2.86 |
| Functions | Database | Common Genes |
|---|---|---|
| Tumor suppressor genes | 3472 | 488 |
| Segregation problems | 829 | 111 |
| Metaphase delay | 217 | 33 |
| Cell death | 5722 | 731 |
| Metaphase alignment problem | 25 | 5 |
| Condensation followed by decondensation | 43 | 9 |
| Binuclear | 584 | 66 |
| Dynamic change | 3679 | 524 |
| Mitotic delay | 1456 | 212 |
| Migration (speed) | 2877 | 317 |
| Migration (distance) | 300 | 46 |
| Inhibition of secretion | 4027 | 583 |
| Enhanced secretion | 3135 | 479 |
| Failure in decondensation | 21 | 1 |
| Chemokine | 1258 | 201 |
| Cytokines | 3324 | 513 |
| Stay close together | 95 | 20 |
| Strange nuclear shape | 98 | 8 |
| Transcription Factor | 10943 | 1141 |
| DNA replication | 4260 | 535 |
| Spindle mitotic | 1329 | 183 |
| Cell division | 1527 | 228 |
| Cell growth | 10029 | 1147 |
| DNA damage | 5377 | 693 |
| Mitochondrial respiration | 429 | 63 |
| Electron acceptors | 203 | 36 |
| Endometrial cancer | 1503 | 288 |
| Proteasome phosphorylation | 2456 | 374 |
| Telomerase erosion | 151 | 36 |
| Chromosome duplication | 3058 | 274 |
| Centrosome duplication | 408 | 56 |
| Cell cycle | 6332 | 751 |
| Endothelial cell | 4650 | 718 |
| Cell differentiation | 10738 | 1223 |
| VEGF signaling | 2892 | 492 |
| Steroid hormone receptors | 1558 | 238 |
| Interphase | 725 | 74 |
| Prophase | 378 | 39 |
| Metaphase | 686 | 82 |
| Anaphase | 754 | 85 |
| Phenotypes | Database | DEGs |
|---|---|---|
| Enhanced secretion | 223 | 19 |
| Inhibition of secretion | 783 | 75 |
| Mild inhibition of secretion | 2306 | 225 |
| Strong inhibition of secretion | 1524 | 146 |
| Increased proliferation | 96 | 10 |
| Migration (distance) | 144 | 15 |
| Migration (speed) | 277 | 20 |
| Grape | 153 | 14 |
| Mitotic delay | 443 | 49 |
| Dynamic changes | 741 | 71 |
| Large | 316 | 25 |
| Polylobed | 472 | 44 |
| Binuclear | 456 | 53 |
| Condensation followed by decondensation | 10 | 1 |
| Failure in decondensation | 8 | 0 |
| Metaphase alignment problems | 422 | 53 |
| Cell death | 782 | 80 |
| Metaphase delay | 275 | 34 |
| Segregation problems | 494 | 65 |
| Strange nuclear shape | 583 | 70 |
| Nuclei stay close together | 364 | 47 |
| Altered gm130 morphology | 99 | 16 |
| Altered COPI morphology | 108 | 12 |
| Altered COPII morphology | 59 | 6 |
| Retention of sh4(haspb)-gfp | 302 | 33 |
| Retention of sh4(yes)-mcherry | 126 | 11 |
| Reduction in ir-induced 53bp1 | 3 | 0 |
| Accumulation of gfp-rnf168 on | 34 | 3 |
| Transcription Factors | Interaction | Transcription Factors | Interaction |
|---|---|---|---|
| TRIM24 (transcription cofactor) | 13 | XBP1 (V$CREB) | 60 |
| LTF (V$LTFM) | 40 | PML (transcription cofactor) | 82 |
| BATF2 (V$AP1F) | 7 | HOXC4 (V$HOXF, V$HOXC) | 7 |
| PIAS1 (transcription cofactor) | 29 | OSR1 (V$OSRF) | 3 |
| EYA3 (transcription cofactor) | 2 | FOX1 (V$FKHD) | 36 |
| GZF1 (V$GZF1) | 0 | JARID2 (V$ARID) | 5 |
| CCNT1 (transcription cofactor) | 14 | CNOT8 (transcription cofactor) | 5 |
| ZNF217 (V$ZF03) | 21 | PPARGC1B (transcription cofactor) | 22 |
| CALR (transcription cofactor) | 66 | CREM (V$CREB) | 26 |
| BHLHE41 (V$HESF) | 12 | NFKB1 (V$NFKB) | 279 |
| ETV3 (V$ETSF) | 3 | RELB (V$NFKB) | 71 |
| MTA2 (transcription cofactor) | 18 | NFE2L1 (V$TCFF, AP1R) | 6 |
| CIITA (transcription cofactor) | 50 | CDCA7 | 0 |
| HIF1A (V$HIFF) | 155 | FOSL1 (V$AP1F) | 66 |
| MED17 (transcription cofactor, mediator) | 3 | FOXO1 (V$FKHD) | 84 |
| IRF1 (V$IRFF) | 111 | NRIP1 (transcription cofactor) | 28 |
| KDM2A (transcription cofactor, demethylase) | 4 | PRDM1 (V$PRDF) | 39 |
| BCL3 (transcription cofactor) | 55 | PCBD1 (transcription cofactor) | 4 |
| MEF2D (V$MEF2) | 13 | TB53 (V$P53F) | 254 |
| EHF (V$ETSF) | 17 | NFKB2 (V$NFKB) | 59 |
| STAT1 (V$IRFF, V$STAT) | 157 | SMAD3 (V$SMAD) | 98 |
| SRXN1 (V$SNAI) | 18 | SIX5 (V$MEF3) | 2 |
| ELL3 (transcription elongation cofactor) | 1 | MYCL (V$EBOX) | 13 |
| ARRB1 (transcription cofactor) | 37 | JUNB (V$AP1F) | 87 |
| PBX4 (V$PBXC, V$HOXC) | 0 | NFE2L3 (V$AP1R) | 9 |
| AHR (V$AHRR) | 84 | LITAF | 19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Najafi, M.; Guo, Y.; Andersson, G.; Humblot, P.; Bongcam-Rudloff, E. Gene Networks and Pathways Involved in LPS-Induced Proliferative Response of Bovine Endometrial Epithelial Cells. Genes 2022, 13, 2342. https://doi.org/10.3390/genes13122342
Najafi M, Guo Y, Andersson G, Humblot P, Bongcam-Rudloff E. Gene Networks and Pathways Involved in LPS-Induced Proliferative Response of Bovine Endometrial Epithelial Cells. Genes. 2022; 13(12):2342. https://doi.org/10.3390/genes13122342
Chicago/Turabian StyleNajafi, Mojtaba, Yongzhi Guo, Göran Andersson, Patrice Humblot, and Erik Bongcam-Rudloff. 2022. "Gene Networks and Pathways Involved in LPS-Induced Proliferative Response of Bovine Endometrial Epithelial Cells" Genes 13, no. 12: 2342. https://doi.org/10.3390/genes13122342
APA StyleNajafi, M., Guo, Y., Andersson, G., Humblot, P., & Bongcam-Rudloff, E. (2022). Gene Networks and Pathways Involved in LPS-Induced Proliferative Response of Bovine Endometrial Epithelial Cells. Genes, 13(12), 2342. https://doi.org/10.3390/genes13122342






