The Theory of Carcino-Evo-Devo and Its Non-Trivial Predictions
Abstract
:1. Introduction
“Tumors are the source of extra cell masses, which may be used in the evolution of multicellular organisms for the expression of evolutionarily new genes, for the origin of new differentiated cell types with novel functions and for building new structures, which constitute evolutionary innovations and morphological novelties”.
Hereditary tumors may play an evolutionary role by providing conditions (space and resources) for the expression of genes newly evolving in the DNA of germ cells. As a result of expression of novel genes, tumor cells may acquire new functions and differentiate in new directions, which may lead to the origin of new cell types, tissues and organs. New cell type is inherited in progeny generations due to genetic and epigenetic mechanisms similar to those for pre-existing cell types.
2. Non-Trivial Predictions of the Carcino-Evo-Devo Theory
- (1)
- The number of cellular oncogenes should correspond to the number of cell types in the organism. Evolution of oncogenes, tumor suppressor and differentiation gene classes should proceed concurrently.
- (2)
- Evolutionarily new and evolving genes should be specifically expressed in tumors (TSEEN genes).
- (2’)
- The whole classes of genes with tumor-specific expression may be evolutionarily novel:
- CT antigen genes;
- HERVs;
- ncRNA genes;
- pan-cancer genes.
- (3)
- Human orthologs of fish TSEEN genes should acquire progressive functions connected with new cell types, tissues and organs.
- (4)
- Selection of tumors for new functions in the organism is possible. Evolutionarily novel organs should recapitulate tumor features in their development.
3. Confirmation of Non-Trivial Predictions
3.1. The Number of Cellular Oncogenes Should Correspond to the Number of Cell Types in the Organism
“The evolutionary role of cellular oncogenes, or proto-oncogenes might consist in sustaining a definite genetically determined level of autonomous proliferative processes in evolving populations of multicellular organisms and in promoting the expression of evolutionarily new genes in anaplastic cells of extra cell masses. After the origin of a new cell type, the corresponding oncogene should have turned into a cell type-specific regulator of cell division. If such scenario is true, then the number of different proto-oncogenes should be about 200—in accordance with the number of cell types in the multicellular organism”.[4]
“The evolutionary role of cellular oncogenes may consist in sustaining the definite level of autonomous proliferative processes in the evolving populations of organisms and in promoting the expression of evolutionarily new genes. After the origin of a new cell type, the corresponding oncogene should have turned into a cell type-specific regulator of cell division and gene expression. If true, the number of cellular oncogenes should correspond to the number of cell types in higher animals (10-fold higher than the 20 or so limit predicted a few years ago and now already about 70)”.[6]
Evolution of Oncogenes, Tumor Suppressor and Differentiation Gene Classes Should Proceed Concurrently
3.2. Evolutionarily New and Evolving Genes Should Be Specifically Expressed in Tumors (TSEEN Genes)
3.2.1. Single TSEEN Genes
3.2.2. TSEEN Gene Classes
Evolutionarily Novel HERVs Expressed Predominantly in Tumors
The Evolutionary Novelty of Tumor-Specifically Expressed Sequences Obtained by Global Subtraction (Pan-Cancer Genes)
3.2.3. Human TSEEN Protein-Coding Genes Database
3.3. Human Orthologs of Fish TSEEN Genes Should Acquire Progressive Functions Connected with New Cell Types, Tissues and Organs
Conclusion on Predictions 2 and 3: TSEEN Genes—A New Biological Phenomenon and the Superclass of Novel and Evolving Genes Expressed in Tumors
3.4. Selection of Tumors for New Functions in the Organism Is Possible. Evolutionarily Novel Organs Should Recapitulate Tumor Features in Their Development
3.4.1. Selection of Tumors for New Functions
3.4.2. Evolutionarily Novel Organs That Recapitulate Tumor Features in Their Development: Mammalian Tumor-like Organs
3.4.3. Tumor Features of Mammalian Adipose. Obesity as a Tumor-like Process
4. Conclusions: Towards a Comprehensive Theory of Evolutionary Oncology
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Isaeva, V.V. Heterochronies, heterotopies, and cell resources of development in ontogenetic and evolutionary transformations. Paleontol. J. 2015, 49, 1530–1537. [Google Scholar] [CrossRef]
- Kozlov, A.P. Evolution of Living Organisms as a Multilevel Process. J. Theor. Biol. 1979, 81, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.P. The principles of multilevel development of organisms. In Problems of Analysis of Biological Systems; Maximov, V.N., Ed.; Moscow University Press: Moscow, Russia, 1983; pp. 48–62. [Google Scholar]
- Kozlov, A.P. Gene competition and the possible evolutionary role of tumors and cellular oncogenes. In Theoretical and Mathematical Aspects of Morphogenesis; Presnov, E.V., Maresin, V.M., Zotin, A.I., Eds.; Nauka: Moscow, Russia, 1987; pp. 136–140. [Google Scholar]
- Kozlov, A.P. Conservation principles in the system of molecular-biological laws. Trans. Leningr. Soc. Nat. Sci. 1988, 87, 4–21. [Google Scholar]
- Kozlov, A.P. Gene competition and the possible evolutionary role of tumors. Med. Hypotheses 1996, 46, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.P. Tumors and evolution. Vopr. Oncol. 2008, 54, 695–705. [Google Scholar]
- Kozlov, A.P. The Possible Evolutionary Role of Tumors in the Origin of New Cell Types. Med. Hypotheses 2010, 74, 177–185. [Google Scholar] [CrossRef]
- Kozlov, A.P. Evolution by Tumor Neofunctionalization. The Role of Tumors in the Origin of New Cell Types, Tissues and Organs; Elsevier; Academic Press: Amsterdam, The Netherlands; Boston, MA, USA; Heidelberg, Germany; London, UK; New York, NY, USA; Oxford, UK; Paris, France; San Diego, CA, USA; San Francisco, CA, USA; Singapore; Sydney, Australia; Tokyo, Japan, 2014; p. 248. [Google Scholar]
- Kozlov, A.P. Expression of evolutionarily novel genes in tumors. Infect. Agents Cancer 2016, 11, 34. [Google Scholar] [CrossRef] [Green Version]
- Kozlov, A.P. The role of heritable tumors in evolution of development: A new theory of carcino-evo-devo. Acta Nat. 2019, 11, 65–72. [Google Scholar] [CrossRef]
- Kozlov, A.P. Mammalian tumor-like organs.1. The role of tumor-like normal organs and atypical tumor organs in the evolution of development (carcino-evo-devo). Infect. Agents Cancer 2022, 17, 2. [Google Scholar] [CrossRef]
- Kozlov, A.P. Mammalian tumor-like organs. 2. Mammalian adipose has many tumor features and obesity is a tumor-like process. Infect. Agents Cancer 2022, 17, 15. [Google Scholar] [CrossRef]
- Kozlov, A.P. Biological computation and compatibility search in the possibility space as the mechanism of complexity increase during progressive evolution. Evol. Bioinform. 2022, 18, 1–5. [Google Scholar] [CrossRef]
- Makashov, A.; Malov, S.V.; Kozlov, A.P. Oncogenes, tumor suppressor and differentiation genes represent the oldest human gene classes and evolve concurrently. Sci. Rep. 2019, 9, 16410. [Google Scholar] [CrossRef]
- Valentine, J.W. Late Precambrian bilaterians: Grades and clades. Proc. Natl. Acad. Sci. USA 1994, 91, 6751–6757. [Google Scholar] [CrossRef] [Green Version]
- Bell, G.; Mooers, A.O. Size and complexity among multicellular organisms. Biol. J. Linn. Soc. 1997, 60, 345–363. [Google Scholar] [CrossRef]
- Chen, L.; Stephen, J.B.; Jaime, M.T.C.; Atahualpa, C.M.; Araxi, O.U. Correcting for Differential Transcript Coverage Reveals a Strong Relationship between Alternative Splicing and Organism Complexity. Mol. Biol. Evol. 2014, 31, 1402–1413. [Google Scholar] [CrossRef]
- Vickaryous, M.K.; Hall, B.K. Human cell type diversity, evolution, development, and classification with special reference to cells derived from neural crest. Biol. Rev. Camb. Philos. Soc. 2006, 81, 425–455. [Google Scholar] [CrossRef]
- Evtushenko, V.I.; Hanson, K.P.; Barabitskaya, O.V.; Emelyanov, A.V.; Reshetnikov, V.L.; Kozlov, A.P. Determination of the upper limit of rat genome expression. Mol. Biol. 1989, 23, 663–675. [Google Scholar]
- Baranova, A.V.; Lobashev, A.V.; Ivanov, D.V.; Krukovskaya, L.L.; Yankovsky, N.K.; Kozlov, A.P. In silico screening for tumor-specific expressed sequences in human genome. FEBS Lett. 2001, 508, 143–148. [Google Scholar] [CrossRef] [Green Version]
- Krukovskaya, L.L.; Baranova, A.; Tyezelova, T.; Polev, D.; Kozlov, A.P. Experimental study of human expressed sequences newly identified in silico as tumor specific. Tumor Biol. 2005, 26, 17–24. [Google Scholar] [CrossRef]
- Kozlov, A.P.; Galachyants, Y.P.; Dukhovlinov, I.V.; Samusik, N.A.; Baranova, A.V.; Polev, D.E.; Krukovskaya, L.L. Evolutionarily new sequences expressed in tumors. Infect. Agents Cancer 2006, 1, 8. [Google Scholar] [CrossRef] [Green Version]
- Galachyants, Y.; Kozlov, A.P. CDD as a tool for discovery of specifically-expressed transcripts. Russ. J. AIDS Cancer Public Health 2009, 13, 60–61. [Google Scholar]
- Polev, D.E.; Nosova, J.K.; Krukovskaya, L.L.; Kozlov, A.P. Expression of transcripts corresponding to cluster Hs.633957 in human healthy and tumor tissues. Mol. Biol. 2009, 43, 88–92. [Google Scholar] [CrossRef]
- Krukovskaia, L.L.; Polev, D.E.; Nosova, I.K.; Baranova, A.V.; Koliubaeva, S.N.; Kozlov, A.P. Investigation of transcription factor Brachyury (T) expression in human normal and tumor tissues. Vopr. Oncol. 2008, 54, 739–743. [Google Scholar]
- Krukovskaya, L.L.; Samusik, N.; Shilov, E.S.; Polev, D.; Kozlov, A.P. Tumor-specific expression of PBOV1, a new gene in evolution. Vopr. Onkol. 2010, 56, 327–332. [Google Scholar]
- Polev, D.E.; Krukovskaya, L.L.; Kozlov, A.P. Expression of the locus Hs.633957 in human digestive system and tumors. Vopr. Oncol. 2011, 57, 48–49. [Google Scholar]
- Samusik, N.A.; Galachyants, Y.P.; Kozlov, A.P. Analysis of evolutionary novelty of tumor-specifically expressed sequences. Russ. J. Genet. Appl. Res. 2011, 1, 138–148. [Google Scholar] [CrossRef]
- Dobrynin, P.; Matyunina, E.; Malov, S.V.; Kozlov, A.P. The novelty of human cancer/testis antigen encoding genes in evolution. Int. J. Genom. 2013, 2013, e105108. [Google Scholar] [CrossRef]
- Samusik, N.; Krukovskaya, L.; Meln, I.; Shilov, E.; Kozlov, A.P. PBOV1 is a human de novo gene with tumor-specific expression that is associated with a positive clinical outcome of cancer. PLoS ONE 2013, 8, e56162. [Google Scholar] [CrossRef]
- Polev, D.E.; Karnaukhova, J.K.; Krukovskaya, L.L.; Kozlov, A.P. ELFN1-AS1, a novel primate gene with possible microRNA function expressed predominantly in tumors. BioMed Res. Int. 2014, 2014, e398097. [Google Scholar] [CrossRef] [Green Version]
- Matyunina, E.; Emelyanov, A.; Kozlov, A. Evolutionarily novel genes expressed in fish tumors determine progressive evolutionary characters. In Proceedings of the 106th Annual Meeting of the American Association for Cancer Research, Philadelphia, PA, USA, 18–22 April 2015; AACR: Philadelphia, PA, USA, 2015. Abstract Number 1927. [Google Scholar]
- Krukovskaya, L.L.; Polev, D.E.; Kurbatova, T.V.; Karnaukhova, Y.K.; Kozlov, A.P. The study of the tumor specificity of expression of some evolutionarily novel genes. Vopr. Oncol. 2016, 62, 495–500. [Google Scholar]
- Matyunina, E.A.; Emelyanov, A.V.; Kurbatova, T.V.; Makashov, A.A.; Mizgirev, I.V.; Kozlov, A.P. Evolutionary novel genes are expressed in transgenic fish tumors and their orthologs are involved in development of progressive traits in humans. Infect. Agents Cancer 2019, 14, 46. [Google Scholar] [CrossRef] [Green Version]
- Karnaukhova, I.K.; Polev, D.E.; Krukovskaya, L.L.; Makashov, A.A.; Masharsky, A.E.; Nazarenko, O.V.; Poverennaya, I.V.; Makeev, V.J.; Kozlov, A.P. A new cancer/testis long noncoding RNA, the OTP-AS1 RNA. Sci. Rep. 2022. [Google Scholar] [CrossRef]
- Kozlov, A.P. Mammalian adipose has many tumor features and obesity is the tumor-like process. In Proceedings of the Annual Meeting of the American Association for Cancer Research 2022, New Orleans, LA, USA, 8–13 April 2022; AACR: New Orleans, LA, USA, 2022. Abstract Number 3371/6077. [Google Scholar]
- Koonin, E.V. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005, 39, 309–338. [Google Scholar] [CrossRef] [Green Version]
- McLysaght, A.; Hurst, L.D. Open questions in the study of de novo genes: What, how and why? Nat. Rev. Genet. 2016, 17, 567. [Google Scholar] [CrossRef]
- Clamp, M.; Fry, B.; Kamal, M.; Xie, X.; Cuff, J.; Linn, M.F.; Kellis, M.; Lindblad-Toh, K.; Lander, E.S. Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl. Acad. Sci. USA 2007, 104, 19428–19433. [Google Scholar] [CrossRef] [Green Version]
- An, G.; Ng, A.Y.; Meka, C.S.R.; Luo, G.; Bright, S.P.; Cazares, L.; Wright, G.L., Jr.; Veltri, R.W. Cloning and characterization UROC28, a novel gene overexpressed in prostate, breast and bladder cancer. Cancer Res. 2000, 60, 7014–7020. [Google Scholar]
- Wang, L.; Niu, C.H.; Wu, S.; Wu, H.M.; Ouyang, F.; He, M.; He, S.Y. PBOV1 correlates with progression of ovarian cancer and inhibits proliferation of ovarian cancer calls. Oncol. Rep. 2016, 35, 488–496. [Google Scholar] [CrossRef] [Green Version]
- Pan, T.; Wu, R.; Liu, B.; Wen, H.; Tu, Z.; Guo, J.; Yang, J.; Shen, G. PBOV1 promotes prostate cancer proliferation by promoting G1/S transition. Onco. Targets Ther. 2016, 9, 787–795. [Google Scholar] [CrossRef] [Green Version]
- Carleton, N.M.; Zhu, G.; Gorbunov, M.; Miller, M.C.; Pienta, K.J.; Resar, L.M.S.; Veltri, R.W. PBOV1 as a potential biomarker for more advanced prostate cancer based on protein and digital histomorphometric analysis. Prostate 2018, 78, 547–559. [Google Scholar] [CrossRef]
- Guo, Y.; Wu, Z.; Shen, S.; Guo, R.; Wang, J.; Wang, W.; Zhao, K.; Kuang, M.; Shuai, X. Nanomedicines reveal how PBOV1 promotes hepatocellular carcinoma for effective gene therapy. Nat. Commun. 2018, 9, 3430. [Google Scholar] [CrossRef]
- Xue, C.; Zhong, Z.; Ye, S.; Wang, Y.; Ye, O. Association between the overexpression of PBOV1 and the prognosis of patients with hepatocellular carcinoma. Oncol. Lett. 2018, 16, 3401–3407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, Q.; Han, J.; Deng, H.; Wu, F.; Guo, S.; Ye, Z. miR-431-5p alters the epithelial-to-mesenchimal transition markers targeting UROC28 in hepatoma cells. Onco. Targets Ther. 2018, 11, 6489–6503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loizidou, M.A.; Cariolou, M.A.; Neuhausen, S.L.; Newbold, R.F.; Bashiardes, E.; Marcou, Y.; Michael, T.; Daniel, M.; Kakouri, E.; Papadopoulos, P.; et al. Genetic variation in genes interacting with BRCA1/2 and risk of breast cancer in the Cypriot population. Breast Cancer Res. Treat. 2010, 121, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Y.; Gao, F.; Peng, C.G.; Zheng, C.J.; Wu, M.F. Has-miR-203 inhibits fracture healing via targeting PBOV1. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5797–5803. [Google Scholar] [PubMed]
- Yang, C.A.; Li, J.P.; Yen, J.C.; Lai, I.L.; Ho, Y.C.; Chen, Y.V.; Lan, J.L.; Chang, J.G. lncRNA NTT/PBOV1 axis promotes monocyte differentiation and is elevated in rheumatoid arthritis. Int. J. Mol. Sci. 2018, 19, 2806. [Google Scholar] [CrossRef] [Green Version]
- Jie, Y.; Ye, L.; Chen, H.; Yu, X.; Cai, L.; He, W.; Fu, Y. ELFN1-AS1 accelerates cell proliferation, invasion and migration via regulating miR-497-3p/CLDN4 axis in ovarian cancer. Bioengineered 2020, 11, 872–882. [Google Scholar] [CrossRef]
- Du, Y.; Hou, Y.; Shi, Y.; Liu, J.; Li, T. Long non-coding RNA ELFN1-AS1 promoted colon cancer cell growth and migration via the miR-191-5p/special AT-rich sequence-binding protein axis. Front. Oncol. Sec. Cancer Genet. 2021, 10, 588360. [Google Scholar] [CrossRef]
- Ma, G.; Li, G.; Gou, A.; Xiao, Z.; Xu, Y.; Song, S.; Guo, K.; Liu, Z. Long non-coding RNA ELFN1-AS1 in the pathogenesis of pancreatic cancer. Ann. Transl. Med. 2021, 9, 10. [Google Scholar] [CrossRef]
- Li, Y.; Gan, Y.; Liu, J.; Li, J.; Zhou, Z.; Tian, R.; Sun, R.; Liu, J.; Xiao, Q.; Li, Y.; et al. Downregulation of MEIS1 mediated by ELFN1-AS1/EZH2/DNMT3a axis promotes tumorigenesis and oxaliplatin resistance in colorectal cancer. Signal Transduct. Target. Ther. 2022, 7, 87. [Google Scholar] [CrossRef]
- Weisman, C.M. The origins and functions of de novo genes: Against all odds? J. Mol. Evol. 2022, 90, 244–257. [Google Scholar] [CrossRef]
- Casola, C. From de novo to “de nono”: The majority of novel protein-coding genes identified with phylostratigraphy are old genes or recent duplicates. Genome Biol. Res. 2018, 10, 2906–2918. [Google Scholar] [CrossRef]
- Zhang, Y.E.; Long, M. New genes contribute to genetic and phenotypic novelties in human evolution. Curr. Opin. Genet. Dev. 2014, 29, 90–96. [Google Scholar] [CrossRef] [Green Version]
- Sverdlov, E.D. Retroviruses and primate evolution. Bioessays 2000, 22, 161–171. [Google Scholar] [CrossRef]
- Khodosevich, K.; Lebedev, Y.; Sverdlov, E. Endogenous retroviruses and human evolution. Comp. Funct. Genom. 2002, 3, 494–498. [Google Scholar] [CrossRef] [Green Version]
- Brinzevich, D.; Young, G.R.; Sebra, R.; Ayllon, J.; Maio, S.M.; Deikus, G.; Chen, B.K.; Fernandez-Sesma, A.; Simon, V.; Mulder, L.C. HIV-1 interacts with human endogenous retrovirus K (HML-2) envelopes derived from human primary lymphocytes. J. Virol. 2014, 88, 6213–6223. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.E.; Vibranovsky, M.D.; Landback, P.; Marais, G.A.B.; Long, M. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on X chromosome. PLoS Biol. 2010, 8, e1000494. [Google Scholar] [CrossRef]
- Makashov, A.A.; Malov, S.V.; Kozlov, A.P. Expression of HERV-K HML-2 in Tumor Tissues. Symposium “Evolutionary Oncology and Evolutionary Virology” within the Framework of the VII St. Petersburg International Oncology Forum “White Nights 2021”; Russian Federation: St. Petersburg, Russia, 2021. [Google Scholar]
- Kim, T.; Jeon, Y.J.; Cui, R.; Lee, J.H.; Peng, Y.; Kim, S.H.; Tili, E.; Alder, H.; Croce, C.M. Role of MYC-regulated long noncoding RNAs in cell cycle regulation and tumorigenesis. J. Natl. Cancer Inst. 2015, 107, dju505. [Google Scholar] [CrossRef]
- Wu, S.M.; Liu, H.; Huang, P.J.; Chang, I.Y.; Lee, C.C.; Yang, C.Y.; Tsai, W.S.; Tan, B.C. circlncRNAnet: An integrated web-based resource for mapping functional networks of long or circular forms of noncoding RNAs. GigaScience 2018, 7, gix 118. [Google Scholar] [CrossRef] [Green Version]
- Karnaukhova, Y.K.; Polev, D.E.; Krukovskaya, L.L.; Kozlov, A.P. Study of the expression of gene Orthopedia homeobox in various tumor and normal human tissues. Vopr. Oncol. 2017, 63, 128–134. [Google Scholar]
- Carvunis, A.R.; Rolland, T.; Wapinski, I.; Calderwood, M.A.; Yildirim, M.A.; Simonis, M.; Charloteaux, B.; Hidalgo, C.A.; Barbette, J.; Santhanam, B.; et al. Proto-genes and de novo gene birth. Nature 2012, 487, 370–374. [Google Scholar] [CrossRef]
- Palena, C.; Polev, D.E.; Tsang, K.Y.; Fernando, R.I.; Litzinger, M.; Krukovskaya, L.L.; Baranova, A.V.; Kozlov, A.P.; Schlom, J. The human T-box mesodermal transcription factor Brachyury is a candidate target for T-cell mediated cancer immunotherapy. Clin. Cancer Res. 2007, 13, 2471–2478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlom, J.; Palena, C.M.; Kozlov, A.P.; Tsang, K.Y. Brachyury Polypeptides and Methods for Use. U.S. Patent No. 8,188,214 B2; Application Granted, 29 May 2012. Available online: https://patents.google.com/patent/US8188214B2/en (accessed on 7 October 2022).
- Kozlov, A.P.; Matyunina, E.A.; Makashov, A.A. The Biomedical Center TSEEN Genes Database. Certificate about Federal Registration of the Database №2021621840. 2021. Available online: https://tseendb.org/#/ (accessed on 7 October 2022).
- Gold, P.; Freedman, S.O. Demonstration of tumor-specific antigens in human colonic carcinomata by immunological tolerance and absorption techniques. J. Exp. Med. 1965, 121, 439–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domazet-Loso, T.; Brajkovic, J.; Tautz, D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007, 23, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Van Oss, S.B.; Carvunis, A.R. De novo gene birth. PLoS Genet. 2019, 15, e1008160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozlov, A.P.; Zabezhinski, M.A.; Popovich, I.G.; Polev, D.E.; Shilov, E.S.; Murashev, B.V. Hyperplastic skin growth on the head of goldfish-comparative oncology aspects. Vopr. Oncol. 2012, 58, 387–393. [Google Scholar]
- Vorontsov, N.N. Macromutations and evolution: Fixation of Goldschmidt’s macromutations as species and genus characters. Papillomatosis and appearance of macrovilli in the rodent stomach. Russ. J. Genet. 2003, 39, 422–426. [Google Scholar] [CrossRef]
- Fernandez, A.A.; Morris, M.R. Mate choice for more melanin as a mechanism to maintain functional oncogene. Proc. Natl. Acad. Sci. USA 2008, 105, 13503–13507. [Google Scholar] [CrossRef] [Green Version]
- Harris, J.R. The evolution of placental mammals. FEBS Lett. 1991, 295, 3–4. [Google Scholar] [CrossRef] [Green Version]
- Blond, J.L.; Beseme, F.; Duret, L. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J. Virol. 1999, 73, 1175–1185. [Google Scholar] [CrossRef] [Green Version]
- Mi, S.; Lee, X.; Li, X.; Veldman, G.M.; Finnerty, H.; Racie, L.; La Vallie, E.; Tang, X.Y.; Edouard, P.; Howes, S.; et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef]
- Kurlak, L.O.; Knofler, M.; Mistry, H.D. Lumps & Bumps: Common features between placental development and cancer growth. Placenta 2017, 56, 2–4. [Google Scholar]
- Lala, P.K.; Lee, B.P.; Xu, G.; Chakraborty, C. Human placental trophoblast as an in vitro model for tumor progression. Can. J. Physiol. Pharmacol. 2002, 80, 142–149. [Google Scholar] [CrossRef]
- Lala, P.K.; Nandi, P.; Hadi, A.; Halari, C. A crossroad between placental and tumor biology: What have we learnt? Placenta 2021, 116, 12–30. [Google Scholar] [CrossRef]
- Davies, J.A. Inverse correlation between an organ’s cancer rate and its evolutionary antiquity. Organogenesis 2004, 1, 60–63. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Ovejero, D.; Arevalo-Martin, A.; Paniaga-Torija, B.; Florensa-Vila, J.; Ferrer, I.; Grassner, L.; Molina-Holgado, E. The ependymal region of the adult human spinal cord differs from other species and shows ependymoma-like features. Brain 2015, 138, 1583–1597. [Google Scholar] [CrossRef] [Green Version]
- Egeblad, M.; Nakasone, E.S.; Werb, Z. Tumors as organs: Complex tissues that interface with the entire organism. Dev. Cell 2010, 18, 884–901. [Google Scholar] [CrossRef] [Green Version]
- Cinti, S. The Adipose Organ; Editrice Kurtis: Milan, Italy, 2001; plates 94. [Google Scholar]
- Frayn, K.N.; Karpe, F.; Fielding, B.A.; Macdonald, I.A.; Coppack, S.W. Integrative physiology of human adipose tissue. Int. J. Obes. 2003, 27, 875–888. [Google Scholar] [CrossRef] [Green Version]
- Kadereit, B.; Kumar, P.; Wang, W.J.; Miranda, D.; Snapp, E.; Severina, N.; Torregroza, I.; Evans, T.; Silver, D. Evolutionarily conserved gene family important for fat storage. Proc. Natl. Acad. Sci. USA 2008, 105, 94–99. [Google Scholar] [CrossRef] [Green Version]
- Zwick, R.K.; Guerrero-Juarez, C.F.; Horsley, V.; Plikus, M.V. Anatomical, physiological and functional diversity of adipose tissue. Cell Metab. 2018, 27, 63–83. [Google Scholar] [CrossRef] [Green Version]
- Cinti, S. The adipose organ. In Adipose Tissue and Adipokines in Health and Disease, Nutrition and Health; Fantuzzi, G., Mazzone, T., Eds.; Humana Press Inc.: Totowa, NJ, USA, 2007; pp. 3–19. [Google Scholar]
- Pond, C.M. The Evolution of Mammalian Adipose Tissue. In Adipose Tissue Biology; Symonds, M., Ed.; Springer: New York, NY, USA, 2011; pp. 227–269. [Google Scholar]
- Cannon, B.; Nedergaard, J. Brown adipose tissue: Function and physiological significance. Physiol. Rev. 2004, 84, 277–359. [Google Scholar] [CrossRef]
- Gesta, S.; Tseng, Y.H.; Kahn, C.R. Developmental origin of fat: Tracking obesity to its source. Cell 2007, 131, 242–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mezentseva, N.V.; Kumaratilake, J.S.; Newman, S.A. The brown adipocyte differentiation pathway in birds: An evolutionary road not taken. BMC Biol. 2008, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Foulds, L. The experimental study of tumor progression: A review. Cancer Res. 1954, 14, 327–339. [Google Scholar] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.J.; Ding, H.H.; Deng, J.J.; Pan, H.; Wang, L.; Li, N.; Wang, X.; Shi, Y.; Gong, F. Inhibition of preadipocyte differentiation and adipogenesis by zinc-a2-glycoprotein treatment in 3T3-L1 cells. J. Diabet. Investig. 2013, 4, 252–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Liu, X.; Tan, C.; Wang, H.; Peng, X.; Deng, F.; Chen, L. Expression and function of zinc-a2-glycoprotein. Neurosci. Bull. 2019, 35, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Banaszak, M.; Gorna, I.; Przyslawski, J. Zink and the innovative zink-a2-plycoprotein adipokine play an important role in lipid metabolism: A critical review. Nutrients 2021, 13, 2023. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, W.; Sheng, T.; He, X.; Xiong, X. Overview of molecular mechanisms contributing to the formation of cancer-associated adipocytes. Mol. Med. Rep. 2021, 24, 768. [Google Scholar] [CrossRef]
- Hassan, M.I.; Waheed, A.; Yadav, S.; Singh, T.P.; Ahmad, F. Zinc-a2-glycoprotein: A multidisciplinary protein. Mol. Cancer Res. 2008, 6, 892–906. [Google Scholar] [CrossRef] [Green Version]
- Kong, B.; Michalsky, C.W.; Hong, X.; Valkovskaya, N.; Rieder, S.; Abiatari, I.; Streit, S.; Erkan, M.; Esposito, I.; Friess, H.; et al. AZP1 is a tumor suppressor in pancreatic cancer inducing mesenchymal-to-epithelial transdifferentiation by inhibiting TGF-beta-mediated ERK signaling. Oncogene 2010, 29, 5146–5158. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Zhao, J.; Lv, L.; Chen, Y.; Li, Y.; Jiang, S.; Wang, W.; Pan, K.; Zheng, Y.; Zhao, B.; et al. Decreased expression of AZGP1 is associated with poor prognosis in primary gastric cancer. PLoS ONE 2013, 8, e69155. [Google Scholar] [CrossRef] [Green Version]
- Tian, H.; Ge, C.; Zhao, F.; Zhu, M.; Zhang, L.; Huo, Q.; Li, H.; Chen, T.; Xie, H.; Cui, Y.; et al. Down regulation of AZGP1 by Ikaros and histone deacetylase promotes tumor progression through the PTEN/Akt and CD44s pathways in hepatocellular carcinoma. Carcinogenesis 2017, 38, 207–217. [Google Scholar]
- Liu, J.; Han, H.; Fan, Z.; El Beaino, M.; Fang, Z.; Li, S.; Ji, J. AZGP1 inhibits soft tissue sarcoma cells invasion and migration. BMC Cancer 2018, 18, 89. [Google Scholar] [CrossRef]
- Ji, M.; Li, W.; He, G.; Zhu, D.; Lv, S.; Tang, W.; Jian, M.; Zheng, P.; Yang, L.; Qi, Z.; et al. Zinc-a2-glycoprotein 1 promotes EMT in colorectal cancer by filamin A mediated focal adhesion pathway. J. Cancer 2019, 10, 5557–5566. [Google Scholar] [CrossRef]
- Delort, L.; Perrier, S.; Dubois, V.; Billard, H.; Mracek, T.; Bing, C.; Vasson, M.P.; Caldefie-Chézet, F. Zinc-a2-glycoprotein: A proliferative factor for breast cancer? In vitro study and molecular mechanisms. Oncol. Rep. 2013, 29, 2025–2029. [Google Scholar] [CrossRef] [Green Version]
- Massague, J. TGFb in cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef] [Green Version]
- Tian, M.; Neil, J.R.; Schiemann, W.P. Transforming growth factor-β and the hallmarks of cancer. Cell Signal. 2011, 23, 951–962. [Google Scholar] [CrossRef] [Green Version]
- Evers, B.; Jonkers, J. Mouse models of BRCA1 and BRCA2 deficiency: Past lessons, current understanding and future prospects. Oncogene 2006, 25, 5885–5897. [Google Scholar] [CrossRef] [Green Version]
- Montfort, A.; Colacios, C.; Levade, T.; Andrieu-Abadie, N.; Meyer, N.; Ségui, B. The TNF paradox in cancer progression and immunotherapy. Front. Immunol. 2019, 10, 1818. [Google Scholar] [CrossRef] [Green Version]
- Aylon, Y.; Oren, M. The paradox of p53: What, how and why? Cold Spring Harb. Perspect. Med. 2016, 6, a026328. [Google Scholar] [CrossRef] [Green Version]
- Makino, Y.; Hikita, H.; Fukumoto, K.; Sung, J.; Sakano, Y.; Murai, K.; Sakane, S.; Kodama, T.; Sakamori, R.; Kondo, J.; et al. Constitutive activation of the tumor suppressor p53 in hepatocytes paradoxically promotes non-cell autonomous liver carcinogenesis. Cancer Res. 2022, 82, 2860–2873. [Google Scholar] [CrossRef] [PubMed]
- Graham, J. Cancer Selection. The New Theory of Evolution; Aculeus Press Inc.: Lexington, VA, USA, 1992; p. 226. [Google Scholar]
- Greaves, M. Cancer. In The Evolutionary Legacy; Oxford University Press: Oxford, USA, 2000; p. 290. [Google Scholar]
- Aktipis, A. The Cheating Cell. How Evolution Helps Us Understand and Treat Cancer; Princeton University Press: Princeton, NJ, USA, 2020; p. 238. [Google Scholar]

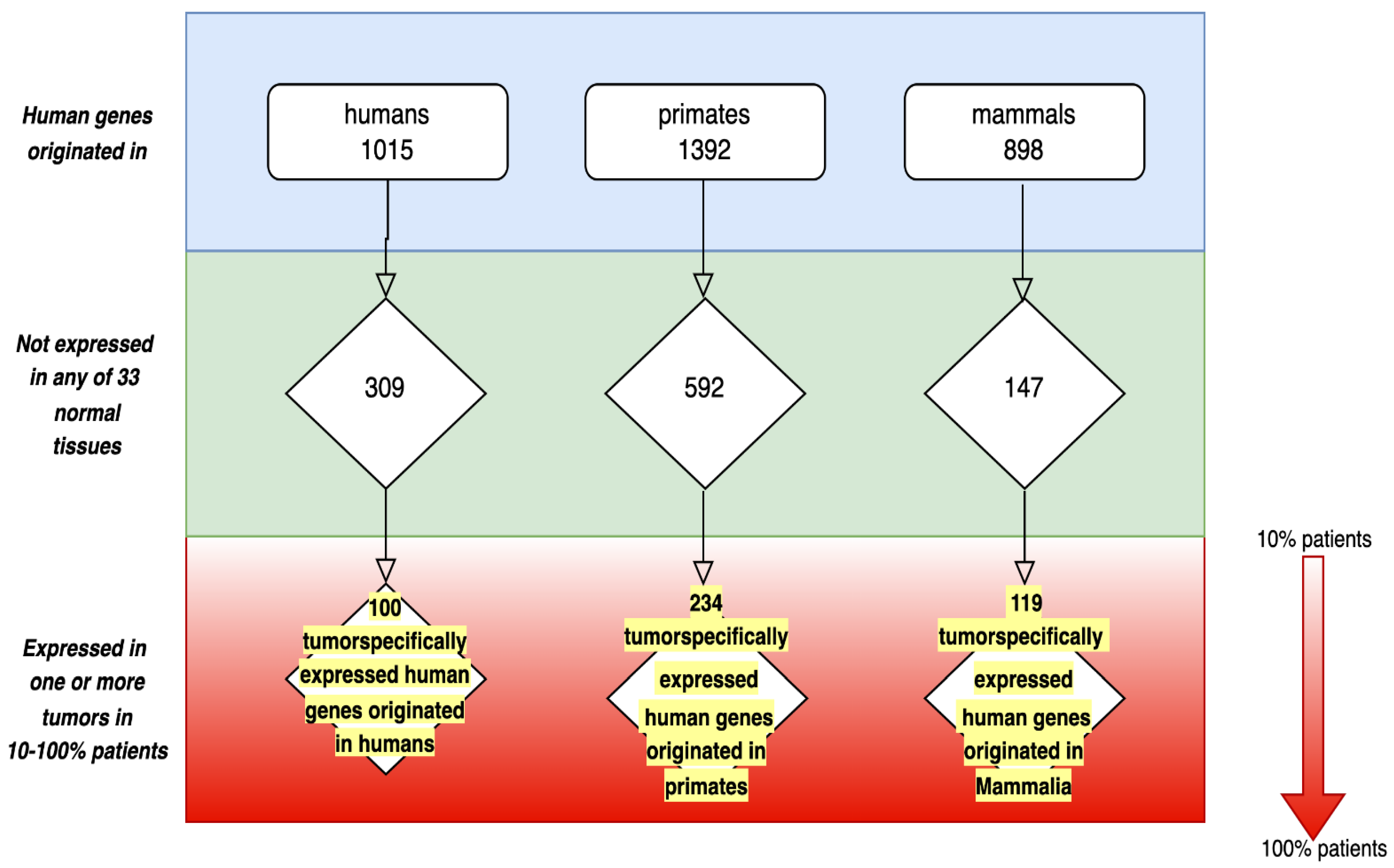
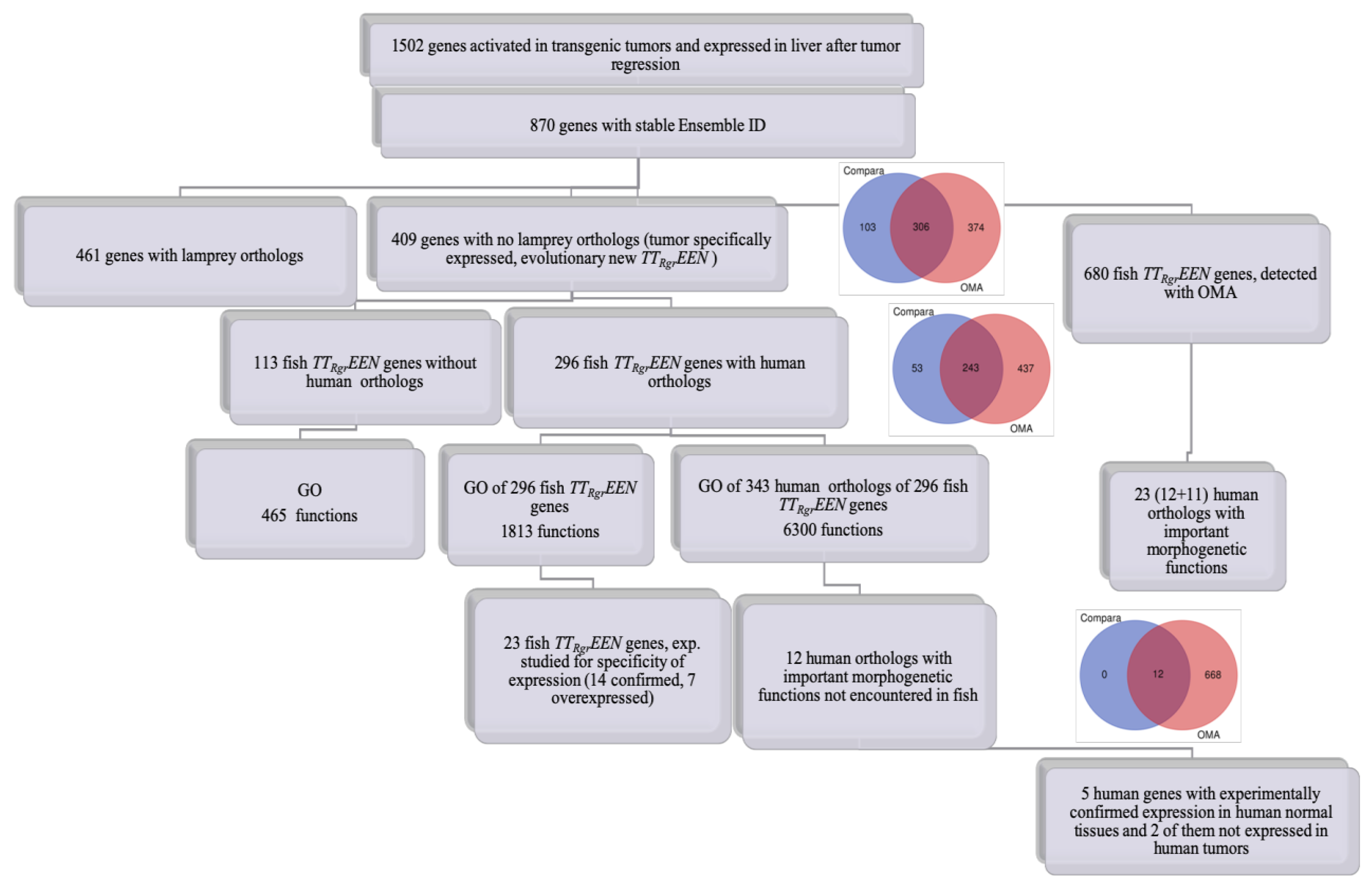
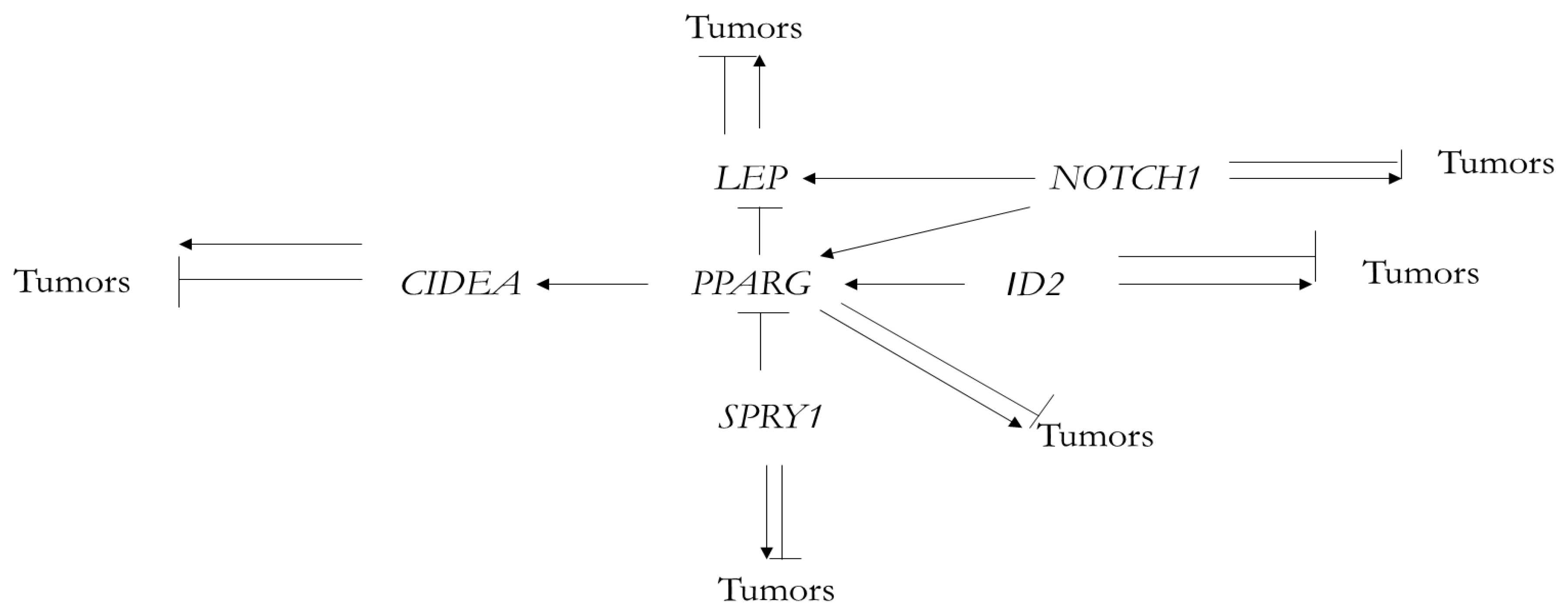

| Name of Gene (Fish Gene/Human Gene) | GO Domain | Selected GO Progressive Functions Not Encountered in Fish ([Fish Gene]/[Human Gene]) | ||
|---|---|---|---|---|
| Molecular Function (Fish Gene/Human Gene) | Cellular Component (Fish Gene/Human Gene) | Biological Process (Fish Gene/Human Gene) | ||
| Fish tgfbr2b/ Human TGFBR2 | 10/18 | 4/12 | 9/84 | [NO]/[bronchus development, bronchus morphogenesis, embryo implantation, in utero embryonic development, lung development, lung lobe morphogenesis, lung morphogenesis, mammary gland morphogenesis, ventricular septum development] |
| Fish lepa/ Human LEP | 2/4 | 3/3 | 15/106 | [NO]/[placenta development |
| Fish sema7a/ Human SEMA7A | 1/3 | 0/4 | 1/16 | [NO]/[olfactory lobe development |
| Fish klf1/Human KLF1 | 3/7 | 1/2 | 3/6 | [NO]/[maternal process involved in female pregnancy |
| Fish ephb3a/ Human EPHB3 | 7/9 | 3/8 | 5/25 | [NO]/corpus callosum development |
| Fish dazap1/Human DAZAP1 | 2/6 | 0/6 | 0/6 | [NO]/maternal placenta development |
| Fish spry1/ Human SPRY1 | 0/1 | 1/6 | 6/16 | [NO]/bud elongation involved in lung branching |
| Fish lmx1bb/ Human LMX1B | 3/7 | 1/1 | 13/9 | [NO]/in utero embryonic development |
| Fish nr2e1/ Human NR2E1 | 5/9 | 1/2 | 2/41 | [NO]/cerebral cortex development, cerebral cortex neuron differentiation, dentate gyrus development, layer formation in cerebral cortex |
| Fish sobpa/ Human SOBP | 0/2 | 0/1 | 0/5 | [NO]/cochlea development |
| Fish ccdc40/Human CCDC40 | 0/0 | 4/5 | 11/14 | [NO]/lung development |
| Fish fosl1a/ Human FOSL1 | 0/7 | 0/6 | 0/29 | [NO]/placenta blood vessel development |
| Fish atxn1l/Human ATXN1L | 1/3 | 1/5 | 0/10 | [NO]/lung alveolus development |
| Fish id2a/Human ID2 | 1/3 | 2/4 | 8/56 | [NO]/epithelial cell differentiation involved in mammary gland alveolus development, mammary gland epithelial cell proliferation, mammary gland alveolus development, ventricular septum development |
| Fish ccr11.1/Human CX3CR1 | 3/4 | 2/7 | 4/17 | [NO]/cerebral cortex cell migration |
| Fish cntnap2a/Human CNTNAP2 | 0/2 | 2/15 | 1/8 | [NO]/cerebral cortex development |
| Fish mycn/Human MYCN | 2/7 | 1/3 | 1/20 | [NO]/lung development |
| Fish neflb/Human NEFL | 1/10 | 2/10 | 1/29 | [NO]/cerebral cortex development |
| Fish notch1b/Human NOTCH1 | 3/15 | 1/20 | 15/162 | [NO]/lung development |
| Fish reck/Human RECK | 0/5 | 0/4 | 7/8 | [NO]/embryo implantation |
| Fish srd5a1/Human SRD5A1 | 2/7 | 2/11 | 4/40 | [NO]/cerebral cortex development |
| Fish wnt7bb/Human WNT7B | 2/3 | 3/9 | 4/42 | [NO]/trachea cartilage morphogenesis, lobar bronchus development, lung epithelium development, lung development, lung morphogenesis, chorio-allantoic fusion, embryonic placenta morphogenesis, mammary gland epithelium development |
| Fish pparg/Human PPARG | 7/30 | 2/8 | 6/81 | [NO]/placenta development |
| Name of Gene | Progressive Functions Connected with Beiging, BAT and Thermoregulation, Not Encountered in Fish |
|---|---|
| LEP | Regulation of energy metabolism in mammals Regulation of beige/brown fat cell differentiation Lipostatic function and thermoregulation |
| NOTCH1 | Regulation of adipose browning, energy metabolism and thermogenesis |
| SPRY1 | Initiation and regulation of adipogenesis Maintaining proliferation and differentiation of adipose stem cells (ASCs) |
| PPARG | Differentiation of adipocytes Activation of thermogenic gene expression in brown adipocytes The role in lipodystrophy, obesity and diabetes |
| ID2 | Stimulation of adipocyte differentiation and adipogenesis The role in obesity |
| CIDEA | Association with lipid droplets Regulation of lipid metabolism Regulation of adipocyte beiging |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozlov, A.P. The Theory of Carcino-Evo-Devo and Its Non-Trivial Predictions. Genes 2022, 13, 2347. https://doi.org/10.3390/genes13122347
Kozlov AP. The Theory of Carcino-Evo-Devo and Its Non-Trivial Predictions. Genes. 2022; 13(12):2347. https://doi.org/10.3390/genes13122347
Chicago/Turabian StyleKozlov, A. P. 2022. "The Theory of Carcino-Evo-Devo and Its Non-Trivial Predictions" Genes 13, no. 12: 2347. https://doi.org/10.3390/genes13122347
APA StyleKozlov, A. P. (2022). The Theory of Carcino-Evo-Devo and Its Non-Trivial Predictions. Genes, 13(12), 2347. https://doi.org/10.3390/genes13122347






