Cloning and Functional Characterization of SpZIP2
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Analyses of Expression Pattern for SpZIP in S. plumbizincicola and S. alfredii
2.3. Sequence Characterization and Transmembrane Region Prediction
2.4. Phylogenetic Analysis and Sequence Alignment
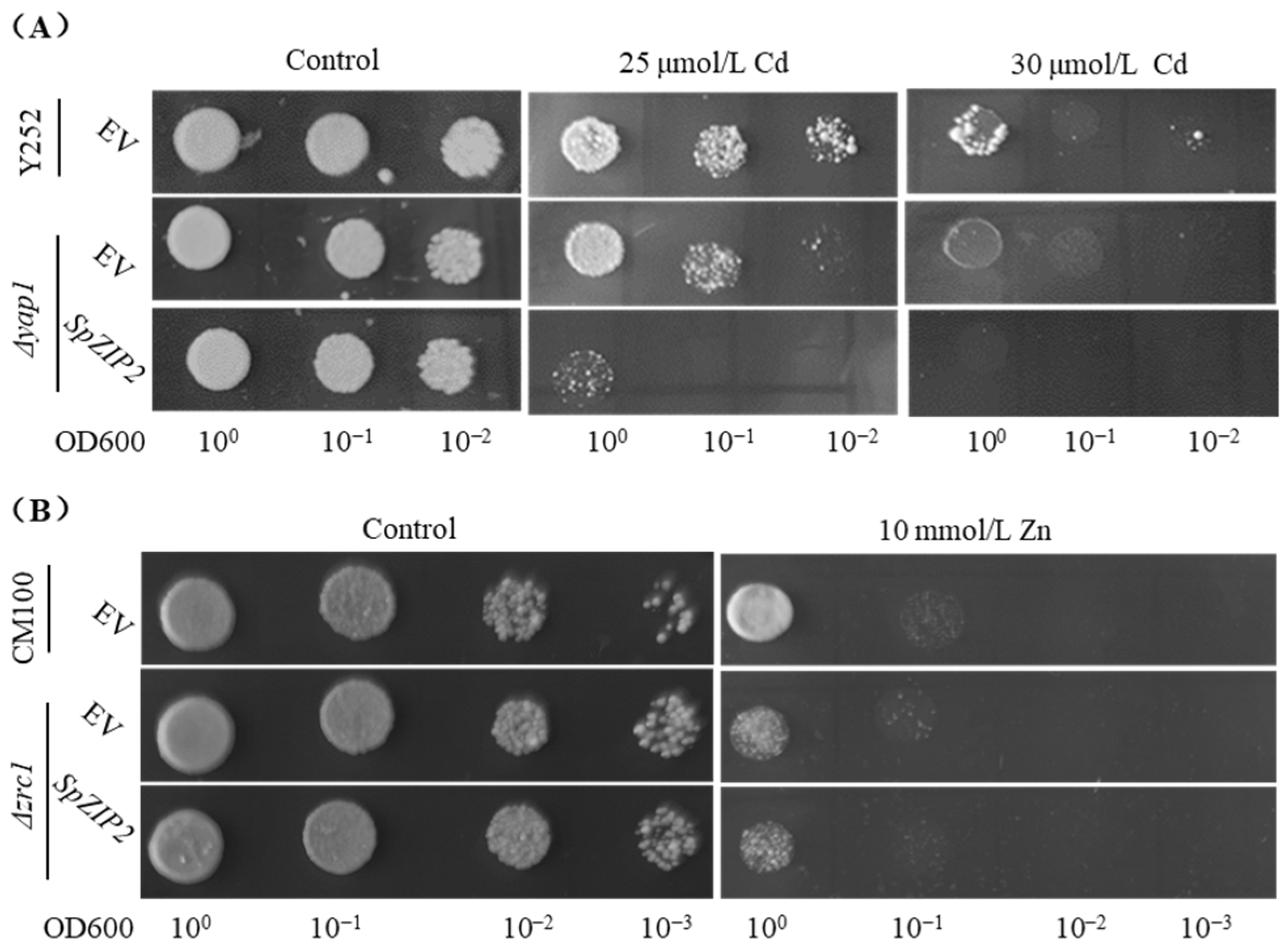
2.5. Cd Tolerance and Accumulation Analyses Using Yeast Strain
2.6. Subcellular Localization Analyses
2.7. Statistical Analyses
3. Results
3.1. Expression Pattern of SpZIP2
3.2. Structure and Characteristics of SpZIP2
3.3. Phylogenetic Analyses and Sequence Alignment of SpZIP2 with Other ZIP Members
3.4. SpZIP2-Mediated Cd Tolerance and Accumulation in Yeast
3.5. SpZIP2 Localized to Membrane Systems Include Plasma Membrane
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhao, F.J.; Tang, Z.; Song, J.J.; Huang, X.Y.; Wang, P. Toxic metals and metalloids: Uptake, transport, detoxification, phytoremediation, and crop improvement for safer food. Mol. Plant 2022, 15, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.H.; Liu, Y.J.; Zhou, S.B.; Guo, F.G.; Bi, D.; Guo, X.H.; Baker, A.J.M.; Smith, J.A.C.; Luo, Y.M. Sedum plumbizincicola X.H. Guo et S.B. Zhou ex L.H. Wu (Crassulaceae): A new species from Zhejiang Province, China. Plant Syst. Evol. 2013, 299, 487–498. [Google Scholar] [CrossRef]
- Peng, J.S.; Wang, Y.J.; Ding, G.; Ma, H.L.; Zhang, Y.J.; Gong, J.M. A Pivotal role of cell wall in cadmium accumulation in the crassulaceae hyperaccumulator Sedum plumbizincicola. Mol. Plant 2017, 10, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Gurajala, H.K.; Wu, L.H.; van der Ent, A.; Qiu, R.L.; Baker, A.J.M.; Tang, Y.T.; Yang, X.E.; Shu, W.S. Hyperaccumulator Plants from China: A Synthesis of the Current State of Knowledge. Environ. Sci. Technol. 2018, 52, 11980–11994. [Google Scholar] [CrossRef] [PubMed]
- Verbruggen, N.; Hermans, C.; Schat, H. Molecular mechanisms of metal hyperaccumulation in plants. N. Phytol. 2009, 181, 759–776. [Google Scholar] [CrossRef]
- Ma, T.T.; Zhou, L.Q.; Chen, L.K.; Li, Z.; Wu, L.H.; Christie, P.; Luo, Y.M. Oxytetracycline toxicity and its effect on phytoremediation by Sedum plumbizincicola and Medicago sativa in metal-contaminated soil. J. Agric. Food Chem. 2016, 64, 8045–8053. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.J.; Zhang, Y.; Dong, B.; Gao, W.Y.; Cheng, C.; Luo, Y.M.; Christie, P.; Wu, L.H. Assessment of phytoextraction using Sedum plumbizincicola and rice production in Cd-polluted acid paddy soils of south China: A field study. Agr. Ecosyst. Environ. 2019, 286, 106651. [Google Scholar] [CrossRef]
- Deng, Y.Q.; Cao, X.Y.; Tan, C.Y.; Sun, L.J.; Peng, X.; Bai, J.; Huang, S.P. Strengthening the effect of Bacillus megaterium on remediation of Cd-contaminated farmland soil by Sedum plumbizincicola. Ying Yong Sheng Tai Xue Bao 2020, 31, 3111–3118. [Google Scholar]
- Bian, F.; Zhong, Z.; Wu, S.; Zhang, X.; Yang, C.; Xiong, X. Comparison of heavy metal phytoremediation in monoculture and intercropping systems of Phyllostachys praecox and Sedum plumbizincicola in polluted soil. Int. J. Phytoremediat. 2018, 20, 490–498. [Google Scholar] [CrossRef]
- Deng, L.; Li, Z.; Wang, J.; Liu, H.; Li, N.; Wu, L.; Hu, P.; Luo, Y.; Christie, P. Long-term field phytoextraction of zinc/cadmium contaminated soil by Sedum plumbizincicola under different agronomic strategies. Int. J. Phytoremediat. 2016, 18, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhou, J.; Zhou, T.; Li, Z.; Jiang, J.; Zhu, D.; Hou, J.; Wang, Z.; Luo, Y.; Christie, P. Estimating cadmium availability to the hyperaccumulator Sedum plumbizincicola in a wide range of soil types using a piecewise function. Sci. Total Environ. 2018, 637–638, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Dong, M.; Mao, P.; Zhuang, P.; Paz-Ferreiro, J.; Li, Y.; Li, Y.; Hu, X.; Netherway, P.; Li, Z. Evaluation of phytoremediation potential of five Cd (hyper)accumulators in two Cd contaminated soils. Sci. Total Environ. 2020, 721, 137581. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Li, Z.; Zhou, T.; Zhou, S.; Wu, L.; Luo, Y.; Christie, P. Phytoextraction potential of soils highly polluted with cadmium using the cadmium/zinc hyperaccumulator Sedum plumbizincicola. Int. J. Phytoremediat. 2019, 21, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Zhu, D.; Wu, L.; Xing, W.; Luo, Y.; Christie, P. Repeated phytoextraction of metal contaminated calcareous soil by hyperaccumulator Sedum plumbizincicola. Int. J. Phytoremediat. 2018, 20, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jia, M.; Wu, L.; Christie, P.; Luo, Y. Changes in metal availability, desorption kinetics and speciation in contaminated soils during repeated phytoextraction with the Zn/Cd hyperaccumulator Sedum plumbizincicola. Environ. Pollut. 2016, 209, 123–131. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, H.; Wu, L.; Xu, W. A genetic transformation method for cadmium hyperaccumulator Sedum plumbizincicola and Non-hyperaccumulating ecotype of Sedum alfredii. Front. Plant Sci. 2017, 8, 1047. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, L.; Zhao, F.J.; Wu, L.; Liu, A.; Xu, W. SpHMA1 is a chloroplast cadmium exporter protecting photochemical reactions in the Cd hyperaccumulator Sedum plumbizincicola. Plant Cell Environ. 2019, 42, 1112–1124. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, H.; Wu, L.; Liu, A.; Zhao, F.J.; Xu, W. Heavy metal ATPase 3 (HMA3) confers cadmium hypertolerance on the cadmium/zinc hyperaccumulator Sedum plumbizincicola. New Phytol. 2017, 215, 687–698. [Google Scholar] [CrossRef]
- Lu, Z.; Qiu, W.; Jin, K.; Yu, M.; Han, X.; He, X.; Wu, L.; Wu, C.; Zhuo, R. Identification and analysis of bZIP family genes in Sedum plumbizincicola and their potential roles in response to cadmium stress. Front. Plant Sci. 2022, 13, 859386. [Google Scholar] [CrossRef]
- Huang, Q.; Qiu, W.; Yu, M.; Li, S.; Lu, Z.; Zhu, Y.; Kan, X.; Zhuo, R. Genome-wide characterization of Sedum plumbizincicola HMA gene family provides functional implications in cadmium response. Plants 2022, 11, 215. [Google Scholar] [CrossRef]
- Peng, J.S.; Ding, G.; Meng, S.; Yi, H.Y.; Gong, J.M. Enhanced metal tolerance correlates with heterotypic variation in SpMTL, a metallothionein-like protein from the hyperaccumulator Sedum plumbizincicola. Plant Cell Environ. 2017, 40, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.S.; Yi, H.Y.; Gong, J.M. Isolation and characterization of cadmium tolerant gene SpMT2 in the hyperaccumulator Sedum plumbizincicola. Sheng Wu Gong Cheng Xue Bao 2020, 36, 541–548. [Google Scholar]
- Tiong, J.; McDonald, G.K.; Genc, Y.; Pedas, P.; Hayes, J.E.; Toubia, J.; Langridge, P.; Huang, C.Y. HvZIP7 mediates zinc accumulation in barley (Hordeum vulgare) at moderately high zinc supply. New Phytol. 2014, 201, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Sun, L.; Tan, L. Progress in ZIP transporter gene family in rice. Yi Chuan 2018, 40, 33–43. [Google Scholar] [PubMed]
- Guerinot, M.L. The ZIP family of metal transporters. Biochim. Biophys. Acta 2000, 1465, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Mäser, P.; Thomine, S.; Schroeder, J.I.; Ward, J.M.; Hirschi, K.; Sze, H.; Talke, I.N.; Amtmann, A.; Maathuis, F.J.; Sanders, D.; et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001, 126, 1646–1667. [Google Scholar] [CrossRef] [PubMed]
- Eide, D.; Broderius, M.; Fett, J.; Guerinot, M.L. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 5624–5628. [Google Scholar] [CrossRef]
- Varotto, C.; Maiwald, D.; Pesaresi, P.; Jahns, P.; Salamini, F.; Leister, D. The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J. 2002, 31, 589–599. [Google Scholar] [CrossRef]
- Grotz, N.; Guerinot, M.L. Molecular aspects of Cu, Fe and Zn homeostasis in plants. Biochim. Biophys. Acta 2006, 1763, 595–608. [Google Scholar] [CrossRef]
- Milner, M.J.; Seamon, J.; Craft, E.; Kochian, L.V. Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J. Exp. Bot. 2013, 64, 369–381. [Google Scholar] [CrossRef]
- Lee, S.; An, G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 2009, 32, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Connolly, E.L.; Fett, J.P.; Guerinot, M.L. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 2002, 14, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Suzuki, M.; Tsukamoto, T.; Suzuki, K.; Nakazono, M.; Kobayashi, T.; Wada, Y.; Watanabe, S.; Matsuhashi, S.; Takahashi, M.; et al. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 2006, 45, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Feng, S.J.; Zhang, B.Q.; Wang, M.Q.; Cao, H.W.; Rono, J.K.; Chen, X.; Yang, Z.M. OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol. 2019, 19, 283. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Mitani-Ueno, N.; Kashino, M.; Ma, J.F. A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J. 2015, 84, 374–384. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Suzuki, M.; Kobayashi, T.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsZIP4, a novel zinc-regulated zinc transporter in rice. J. Exp. Bot. 2005, 56, 3207–3214. [Google Scholar] [CrossRef]
- Lee, S.; Jeong, H.J.; Kim, S.A.; Lee, J.; Guerinot, M.L.; An, G. OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol. Biol. 2010, 73, 507–517. [Google Scholar] [CrossRef]
- Yang, X.; Huang, J.; Jiang, Y.; Zhang, H.S. Cloning and functional identification of two members of the ZIP (Zrt, Irt-like protein) gene family in rice (Oryza sativa L.). Mol. Biol. Rep. 2009, 36, 281–287. [Google Scholar] [CrossRef]
- Tan, L.; Zhu, Y.; Fan, T.; Peng, C.; Wang, J.; Sun, L.; Chen, C. OsZIP7 functions in xylem loading in roots and inter-vascular transfer in nodes to deliver Zn/Cd to grain in rice. Biochem. Biophys. Res. Commun. 2019, 512, 112–118. [Google Scholar] [CrossRef]
- Ricachenevsky, F.K.; Punshon, T.; Lee, S.; Oliveira, B.H.N.; Trenz, T.S.; Maraschin, F.D.S.; Hindt, M.N.; Danku, J.; Salt, D.E.; Fett, J.P.; et al. Elemental profiling of rice FOX lines leads to characterization of a new Zn plasma membrane transporter, OsZIP7. Front. Plant Sci. 2018, 9, 865. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.A.; Lee, J.; Guerinot, M.L.; An, G. Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol. Cells 2010, 29, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Qu, M.; Zhu, Y.; Peng, C.; Wang, J.; Gao, D.; Chen, C. ZINC TRANSPORTER5 and ZINC TRANSPORTER9 function synergistically in zinc/cadmium uptake. Plant Physiol. 2020, 183, 1235–1249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhu, Y.; Yu, L.; Yang, M.; Zou, X.; Yin, C.; Lin, Y. Research advances in cadmium uptake, transport and resistance in rice (Oryza sativa L.). Cells 2022, 11, 569. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.; Li, Y.; Wang, Y.; Lin, J.; Du, S.; Liao, X. ZAT10 plays dual roles in cadmium uptake and detoxification in Arabidopsis. Front. Plant Sci. 2022, 13, 994100. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, L.; Li, X. Arabidopsis and rice showed a distinct pattern in ZIPs genes expression profile in response to Cd stress. Bot. Stud. 2018, 59, 22. [Google Scholar] [CrossRef]
- Kavitha, P.G.; Kuruvilla, S.; Mathew, M.K. Functional characterization of a transition metal ion transporter, OsZIP6 from rice (Oryza sativa L.). Plant Physiol. Biochem. 2015, 97, 165–174. [Google Scholar]
- Tian, S.; Liang, S.; Qiao, K.; Wang, F.; Zhang, Y.; Chai, T. Co-expression of multiple heavy metal transporters changes the translocation, accumulation, and potential oxidative stress of Cd and Zn in rice (Oryza sativa). J. Hazard Mater. 2019, 380, 120853. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook, 1st ed.; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction—The Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef]
- Blom, N.; Sicheritz-Pontén, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Elble, R. A simple and efficient procedure for transformation of yeasts. Biotechniques 1992, 13, 18–20. [Google Scholar] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.K.; Ganie, S.A.; Rana, M.K.; Sharma, T.R. Genome-wide analysis of zinc transporter genes of maize (Zea mays). Plant Mol. Biol. Rep. 2013, 32, 605–616. [Google Scholar] [CrossRef]
- Li, X.B.; Suo, H.C.; Liu, J.T.; Wang, L.; Li, C.C.; Liu, W. Genome-wide identification and expression analysis of the potato ZIP gene family under Zn-deficiency. Biol. Plant 2020, 64, 845–855. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, S.; Li, D.; Xu, X.; Li, C. Genome-wide analysis of the zrt, irt-like protein (zip) family and their responses to metal stress in Populus trichocarpa. Plant Mol. Biol. Rep. 2017, 35, 534–549. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, N.; Zhang, Z.; Shi, G. Genome-wide identification and expression profile reveal potential roles of peanut ZIP family genes in Zinc/Iron-deficiency tolerance. Plants 2022, 11, 786. [Google Scholar] [CrossRef]
- Gao, F.; Li, J.; Zhang, J.; Li, N.; Tang, C.; Bakpa, E.P.; Xie, J. Genome-wide identification of the ZIP gene family in lettuce (Lactuca sativa L.) and expression analysis under different element stress. PLoS ONE 2022, 17, e0274319. [Google Scholar] [CrossRef]
- Gong, F.; Qi, T.; Hu, Y.; Jin, Y.; Liu, J.; Wang, W.; He, J.; Tu, B.; Zhang, T.; Jiang, B.; et al. Genome-wide investigation and functional verification of the ZIP family transporters in wild emmer wheat. Int. J. Mol. Sci. 2022, 23, 2866. [Google Scholar] [CrossRef]
- Evens, N.P.; Buchner, P.; Williams, L.E.; Hawkesford, M.J. The role of ZIP transporters and group F bZIP transcription factors in the Zn-deficiency response of wheat (Triticum aestivum). Plant J. 2017, 92, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Yamaji, N.; Yokosho, K.; Ma, J.F. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 2012, 24, 2155–2167. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.; Wang, W.G.; Yamaji, N.; Fukuoka, S.; Che, J.; Ueno, D.; Ando, T.; Deng, F.L.; Hori, K.; Yano, M.; et al. Duplication of a manganese/cadmium transporter gene reduces cadmium accumulation in rice grain. Nat. Food 2022, 3, 597–607. [Google Scholar] [CrossRef]
- Ueno, D.; Yamaji, N.; Kono, I.; Huang, C.F.; Ando, T.; Yano, M.; Ma, J.F. Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 16500–16505. [Google Scholar] [CrossRef]
- Chao, D.Y.; Silva, A.; Baxter, I.; Huang, Y.S.; Nordborg, M.; Danku, J.; Lahner, B.; Yakubova, E.; Salt, D.E. Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana. PLoS Genet. 2012, 8, e1002923. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, T.-L.; Tang, T.-W.; Zhang, P.-H.; Liu, M.; Zhao, J.; Peng, J.-S.; Meng, S. Cloning and Functional Characterization of SpZIP2. Genes 2022, 13, 2395. https://doi.org/10.3390/genes13122395
Han T-L, Tang T-W, Zhang P-H, Liu M, Zhao J, Peng J-S, Meng S. Cloning and Functional Characterization of SpZIP2. Genes. 2022; 13(12):2395. https://doi.org/10.3390/genes13122395
Chicago/Turabian StyleHan, Tian-Long, Ting-Wei Tang, Pei-Hong Zhang, Min Liu, Jing Zhao, Jia-Shi Peng, and Shuan Meng. 2022. "Cloning and Functional Characterization of SpZIP2" Genes 13, no. 12: 2395. https://doi.org/10.3390/genes13122395
APA StyleHan, T.-L., Tang, T.-W., Zhang, P.-H., Liu, M., Zhao, J., Peng, J.-S., & Meng, S. (2022). Cloning and Functional Characterization of SpZIP2. Genes, 13(12), 2395. https://doi.org/10.3390/genes13122395







