Analysis of Breast Cancer Differences between China and Western Countries Based on Radiogenomics
Abstract
:1. Introduction
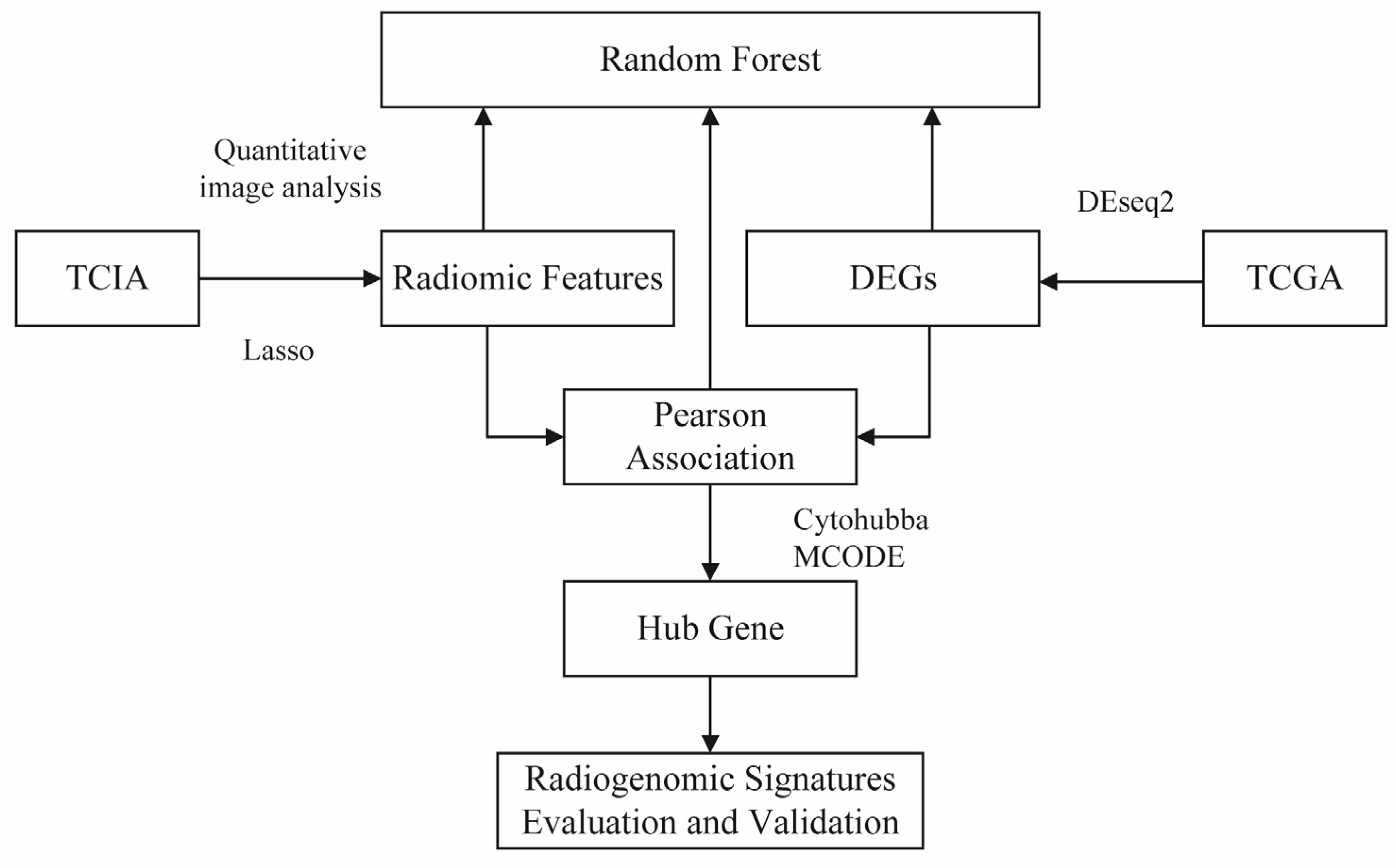
2. Materials and Methods
2.1. Genomic and Picture Datasets
- Downloaded the TCGA-BRCA dataset from the TCIA database. To reduce the image quality difference between cases in multiple institutions, we selected MRI images obtained by the same scanner, and a total of 91 patients were obtained;
- Downloaded the gene expression RNAseq data from the GDC TCGA Breast Cancer dataset from UCSC Xena [32] (http://xena.ucsc.edu/ accessed on 1 November 2021). These transcriptome data correspond to patients with imaging data;
2.2. Picture Data Analysis
2.3. Transcriptomedata Analysis
2.4. Association between Transcriptome and Image Features
2.5. Model Development and Statistical Analysis
3. Results
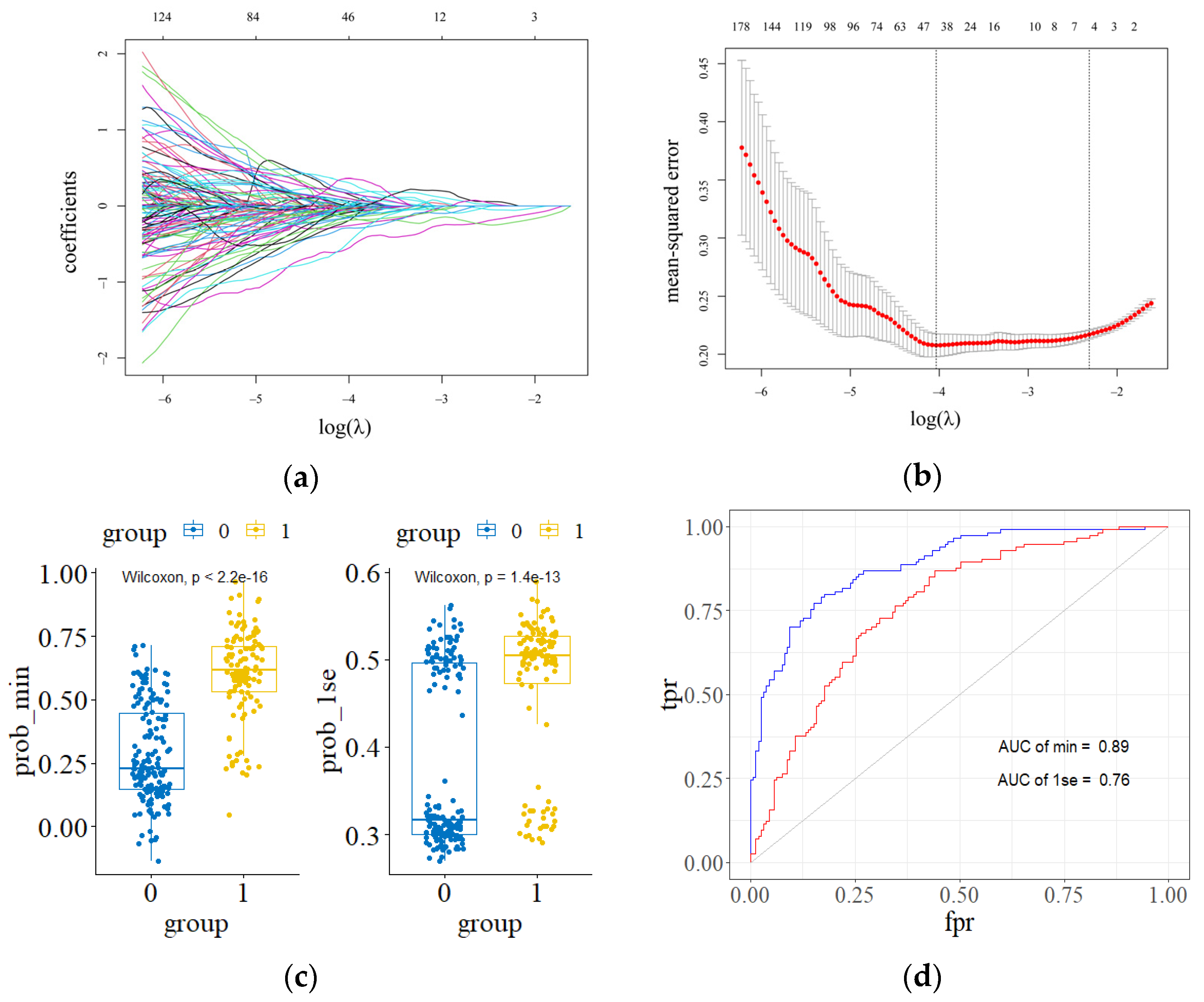
3.1. Radiomic Features
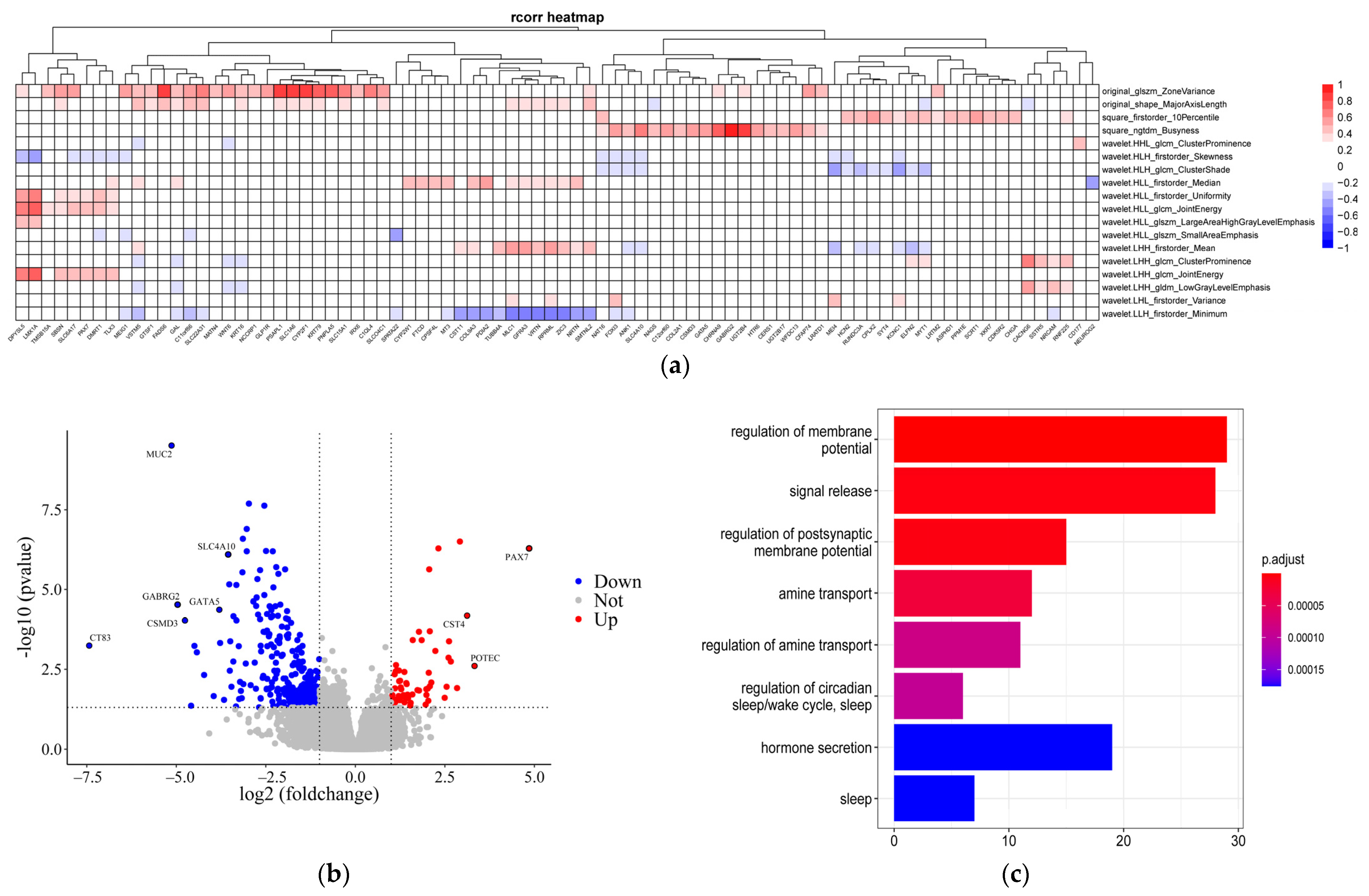
3.2. Transcriptome Data Characteristics
3.3. Association between DEGs and Radiomics Features
3.3.1. Diagnostic Role of Radiogenomics Signature
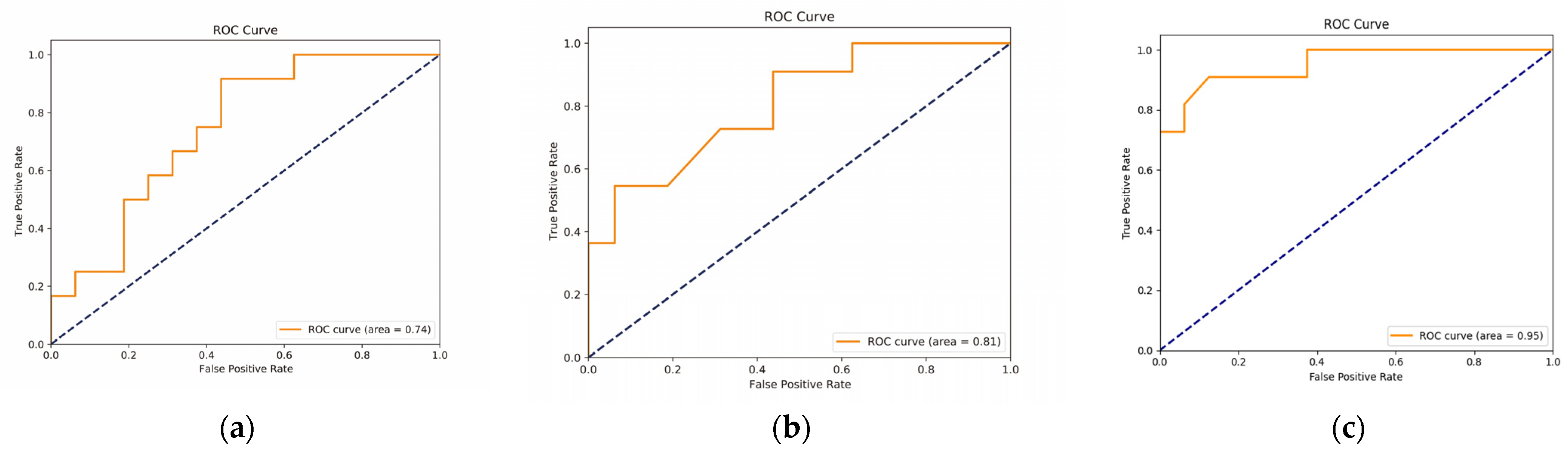
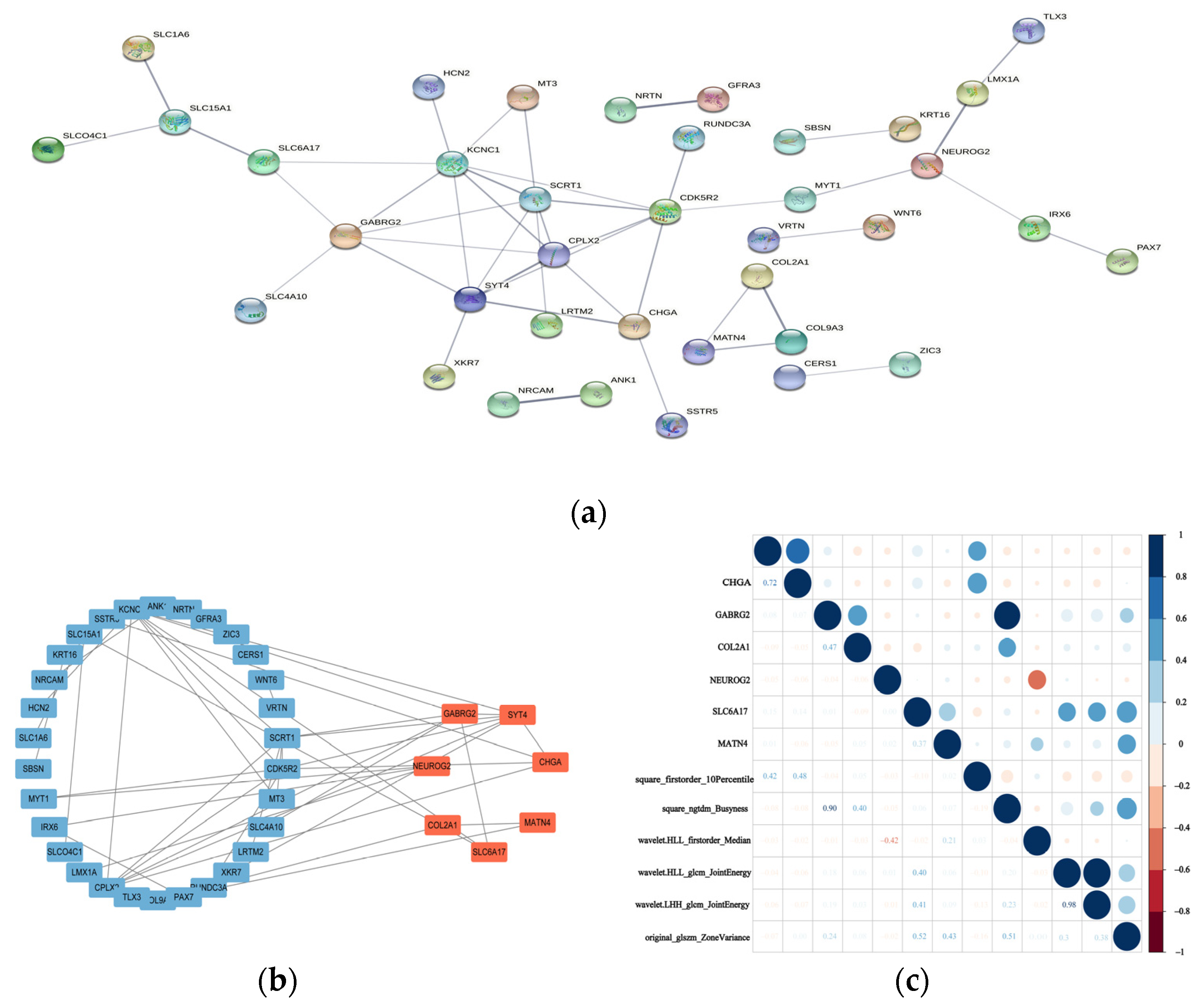
3.3.2. Validation of the Hub Genes for the Differential Diagnosis of Chinese and Western BC Samples
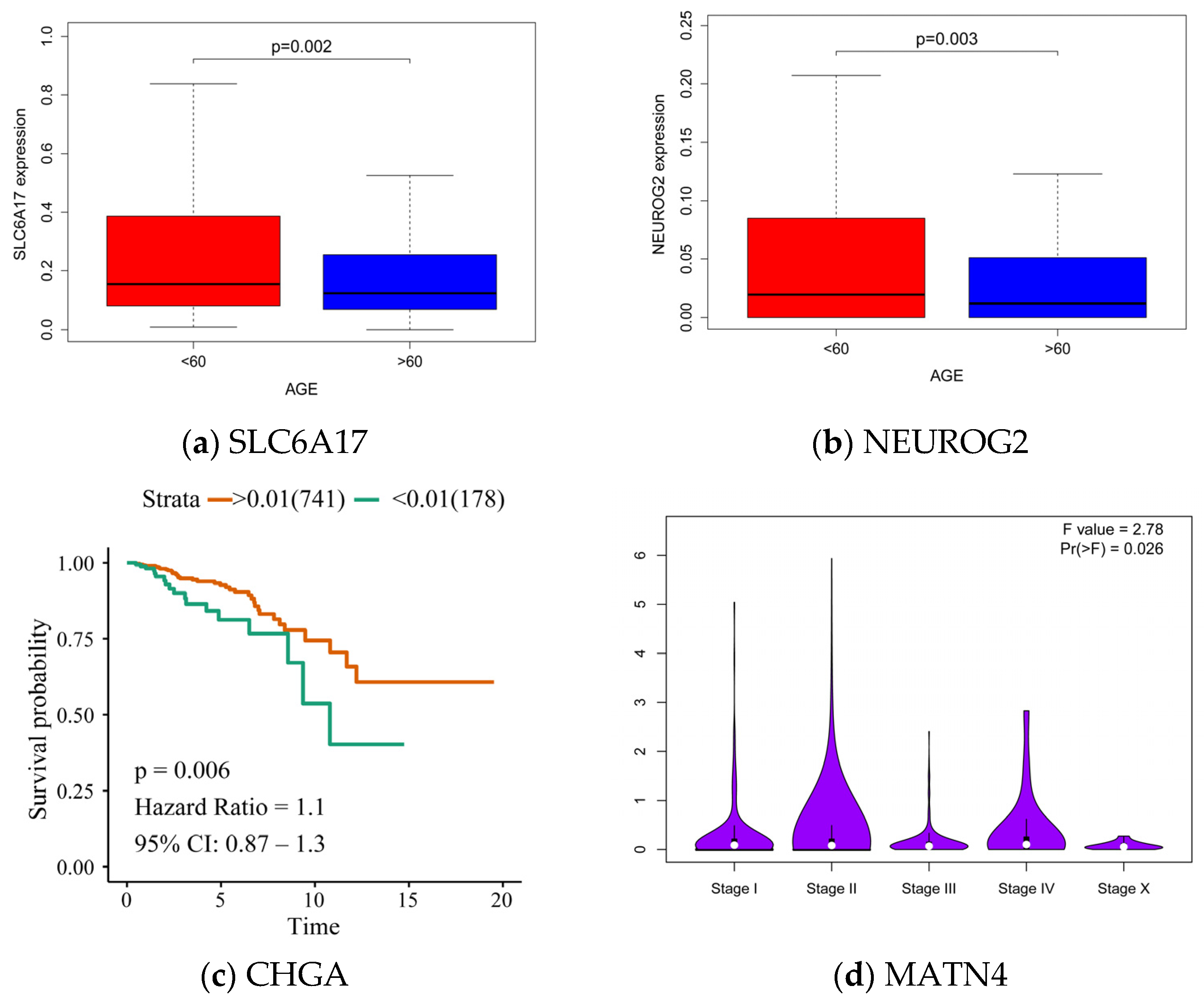
3.3.3. Association between Hub Genes and Imaging Features
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Chen, H.D.; Yu, Y.W.; Li, N.; Chen, W.Q. Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chin. Med. J. 2021, 134, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Zheng, R.; Zhang, S.; Wang, S.; Chen, R.; Sun, K.; Zeng, H.; Zhou, J.; Wei, W. Global patterns of breast cancer incidence and mortality: A population-based cancer registry data analysis from 2000 to 2020. Cancer Commun. 2021, 41, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Shu, X.; Pal, T.; Berlin, J.; Nguyen, S.M.; Zheng, W.; Bailey, C.E.; Shu, X.O. Racial/Ethnic Disparities in Mortality Related to Access to Care for Major Cancers in the United States. Cancers 2022, 14, 3390. [Google Scholar] [CrossRef]
- Leong, S.P.; Shen, Z.Z.; Liu, T.J.; Agarwal, G.; Tajima, T.; Paik, N.S.; Sandelin, K.; Derossis, A.; Cody, H.; Foulkes, W.D. Is breast cancer the same disease in Asian and Western countries? World J. Surg. 2010, 34, 2308–2324. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Sun, S.; Yuan, J.P.; Wang, Y.H.; Cao, T.Z.; Zheng, H.M.; Jiang, X.Q.; Gong, Y.P.; Tu, Y.; Yao, F.; et al. Characteristics of breast cancer in Central China, literature review and comparison with USA. Breast 2016, 30, 208–213. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.; Zhang, F.; Chen, D.; Ye, G.; Li, Y.; Zha, Y.; Chen, W.; Liu, D.; Liao, X.; Huang, Q.; et al. A comparison of Chinese multicenter breast cancer database and SEER database. Sci. Rep. 2022, 12, 10395. [Google Scholar] [CrossRef]
- Han, Y.-Q.; Yi, Z.-B.; Yu, P.; Wang, W.-N.; Ouyang, Q.-C.; Yan, M.; Wang, X.-J.; Hu, X.-C.; Jiang, Z.-F.; Huang, T.; et al. Comparisons of Treatment for HER2-Positive Breast Cancer between Chinese and International Practice: A Nationwide Multicenter Epidemiological Study from China. J. Oncol. 2021, 2021, 6621722. [Google Scholar] [CrossRef]
- Zeng, H.; Zheng, R.; Guo, Y.; Zhang, S.; Zou, X.; Wang, N.; Zhang, L.; Tang, J.; Chen, J.; Wei, K.; et al. Cancer survival in China, 2003-2005: A population- based study. Int. J. Cancer 2015, 136, 1921–1930. [Google Scholar] [CrossRef] [Green Version]
- Sivasubramaniam, P.G.; Zhang, B.-L.; Zhang, Q.; Smith, J.S.; Zhang, B.; Tang, Z.-H.; Chen, G.-J.; Xie, X.-M.; Xu, X.-Z.; Yang, H.-J.; et al. Breast Cancer Disparities: A Multicenter Comparison of Tumor Diagnosis, Characteristics, and Surgical Treatment in China and the US. Oncologist 2015, 20, 1044–1050. [Google Scholar] [CrossRef]
- Lin, J.; Hu, H.; Shriver, C.D.; Zhu, K. Survival among Breast Cancer Patients: Comparison of the U.S. Military Health System with the Surveillance, Epidemiology and End Results Program. Clin. Breast Cancer 2022, 22, e506–e516. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.; Villa, D.; Woods, R.; Yerushalmi, R.; Gelmon, K. Breast Cancer Subtype Variation by Race and Ethnicity in a Diverse Population in British Columbia. Clin. Breast Cancer 2016, 16, e49–e55. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. Methods for RNA Modification Mapping Using Deep Sequencing: Established and New Emerging Technologies. Genes 2019, 10, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurian, A.W.; Gong, G.D.; Chun, N.M.; Mills, M.A.; Staton, A.D.; Kingham, K.E.; Crawford, B.B.; Lee, R.; Chan, S.; Donlon, S.S.; et al. Performance of BRCA1/2 mutation prediction models in Asian Americans. J. Clin. Oncol. 2008, 26, 4752–4758. [Google Scholar] [CrossRef] [Green Version]
- Yap, Y.S.; Lu, Y.S.; Tamura, K.; Lee, J.E.; Ko, E.Y.; Park, Y.H.; Cao, A.Y.; Lin, C.H.; Toi, M.; Wu, J.; et al. Insights Into Breast Cancer in the East vs the West: A Review. JAMA Oncol. 2019, 5, 1489–1496. [Google Scholar] [CrossRef]
- Chen, L.; Fu, F.; Huang, M.; Lv, J.; Zhang, W.; Wang, C. The spectrum of BRCA1 and BRCA2 mutations and clinicopathological characteristics in Chinese women with early-onset breast cancer. Breast Cancer Res. Treat. 2020, 180, 759–766. [Google Scholar] [CrossRef]
- Zeng, C.; Guo, X.; Wen, W.; Shi, J.; Long, J.; Cai, Q.; Shu, X.O.; Xiang, Y.; Zheng, W. Evaluation of pathogenetic mutations in breast cancer predisposition genes in population-based studies conducted among Chinese women. Breast Cancer Res. Treat. 2020, 181, 465–473. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Yang, L.; Yao, L.; Kuang, X.Y.; Zuo, W.J.; Li, S.; Qiao, F.; Liu, Y.R.; Cao, Z.G.; Zhou, S.L.; et al. Characterization of PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer patients. Nat. Commun. 2018, 9, 1357. [Google Scholar] [CrossRef] [Green Version]
- Habel, L.A.; Capra, A.M.; Oestreicher, N.; Greendale, G.A.; Cauley, J.A.; Bromberger, J.; Crandall, C.J.; Gold, E.B.; Modugno, F.; Salane, M.; et al. Mammographic density in a multiethnic cohort. Menopause 2007, 14, 891–899. [Google Scholar] [CrossRef]
- Zhao, H.; Zou, L.; Geng, X.; Zheng, S. Limitations of mammography in the diagnosis of breast diseases compared with ultrasonography: A single-center retrospective analysis of 274 cases. Eur. J. Med. Res. 2015, 20, 49. [Google Scholar] [CrossRef]
- Zeng, J.; Lin, L.; Deng, F. Infrared thermal imaging as a nonradiation method for detecting thermal expression characteristics in normal female breasts in China. Infrared Phys. Technol. 2020, 104, 103125. [Google Scholar] [CrossRef]
- Pinker, K.; Shitano, F.; Sala, E.; Do, R.K.; Young, R.J.; Wibmer, A.G.; Hricak, H.; Sutton, E.J.; Morris, E.A. Background, Current Role, and Potential Applications of Radiogenomics. J. Magn. Reson. Imaging 2018, 47, 604–620. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiri, I.; Amini, M.; Nazari, M.; Hajianfar, G.; Haddadi Avval, A.; Abdollahi, H.; Oveisi, M.; Arabi, H.; Rahmim, A.; Zaidi, H. Impact of feature harmonization on radiogenomics analysis: Prediction of EGFR and KRAS mutations from non-small cell lung cancer PET/CT images. Comput. Biol. Med. 2022, 142, 105230. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Albadawy, E.; Saha, A.; Zhang, J.; Harowicz, M.R.; Mazurowski, M.A. Deep learning for identifying radiogenomic associations in breast cancer. Comput. Biol. Med. 2019, 109, 85–90. [Google Scholar] [CrossRef] [Green Version]
- Liang, S.; Zhang, R.; Liang, D.; Song, T.; Ai, T.; Xia, C.; Xia, L.; Wang, Y. Multimodal 3D DenseNet for IDH Genotype Prediction in Gliomas. Genes 2018, 9, 382. [Google Scholar] [CrossRef] [Green Version]
- Segal, E.; Sirlin, C.B.; Ooi, C.; Adler, A.S.; Gollub, J.; Chen, X.; Chan, B.K.; Matcuk, G.R.; Barry, C.T.; Chang, H.Y.; et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat. Biotechnol. 2007, 25, 675–680. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, H.; Guo, W.; Drukker, K.; Lan, L.; Giger, M.L.; Ji, Y. Deciphering Genomic Underpinnings of Quantitative MRI-based Radiomic Phenotypes of Invasive Breast Carcinoma. Sci. Rep. 2015, 5, 17787. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Li, X.; Teng, X.; Rubin, D.L.; Napel, S.; Daniel, B.L.; Li, R. Magnetic resonance imaging and molecular features associated with tumor- infiltrating lymphocytes in breast cancer. Breast Cancer Res. 2018, 20, 101. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N.; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Goldman, M.; Craft, B.; Hastie, M.; Repečka, K.; Kamath, A.; McDade, F.; Rogers, D.; Brooks, A.N.; Zhu, J.; Haussler, D. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. biorXiv 2019, 326470. [Google Scholar] [CrossRef] [Green Version]
- Tong, M.; Deng, Z.; Yang, M.; Xu, C.; Zhang, X.; Zhang, Q.; Liao, Y.; Deng, X.; Lv, D.; Zhang, X.; et al. Transcriptomic but not genomic variability confers phenotype of breast cancer stem cells. Cancer Commun. 2018, 38, 56. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Wang, H.; Duan, J.; Luo, W.; Zhao, R.; Shen, Y.; Wang, B.; Tao, S.; Sun, Y.; Ye, Q.; et al. An Alternatively Spliced p62 Isoform Confers Resistance to Chemotherapy in Breast Cancer. Cancer Res. 2022, 82, 4001–4015. [Google Scholar] [CrossRef]
- Datta, J.; Willingham, N.; Manouchehri, J.M.; Schnell, P.; Sheth, M.; David, J.J.; Kassem, M.; Wilson, T.A.; Radomska, H.S.; Coss, C.C.; et al. Activity of Estrogen Receptor beta Agonists in Therapy-Resistant Estrogen Receptor-Positive Breast Cancer. Front. Oncol. 2022, 12, 857590. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Kahle, D.; Wickham, H. ggmap: Spatial Visualization with ggplot2. R J. 2013, 5, 144–161. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Mueller, M. pROC: An open-source package for R and S plus to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Tan, R.; Ong, W.S.; Lee, K.H.; Lim, A.H.; Park, S.; Park, Y.H.; Lin, C.H.; Lu, Y.S.; Ono, M.; Ueno, T.; et al. HER2 expression, copy number variation and survival outcomes in HER2-low non-metastatic breast cancer: An international multicentre cohort study and TCGA-METABRIC analysis. BMC Med. 2022, 20, 105. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Chen, X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019, 2, 141–160. [Google Scholar] [CrossRef] [Green Version]
- Salimimoghadam, S.; Taefehshokr, S.; Loveless, R.; Teng, Y.; Bertoli, G.; Taefehshokr, N.; Musaviaroo, F.; Hajiasgharzadeh, K.; Baradaran, B. The role of tumor suppressor short non-coding RNAs on breast cancer. Crit. Rev. Oncol. Hematol. 2021, 158, 103210. [Google Scholar] [CrossRef]
- Yang, M.F.; Long, X.X.; Hu, H.S.; Bin, Y.L.; Chen, X.M.; Wu, B.H.; Peng, Q.Z.; Wang, L.S.; Yao, J.; Li, D.F. Comprehensive analysis on the expression profile and prognostic values of Synaptotagmins (SYTs) family members and their methylation levels in gastric cancer. Bioengineered 2021, 12, 3550–3565. [Google Scholar] [CrossRef]
- Jiang, S.; Zhu, L.; Jiang, C.; Yu, S.; Wang, B.; Ren, Y. Prognosis and immune function of Synaptotagmin-4 in gastric cancer and brain low-grade glioma. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Annaratone, L.; Medico, E.; Rangel, N.; Castellano, I.; Marchio, C.; Sapino, A.; Bussolati, G. Search for neuro-endocrine markers (chromogranin A, synaptophysin and VGF) in breast cancers. An integrated approach using immunohistochemistry and gene expression profiling. Endocr. Pathol. 2014, 25, 219–228. [Google Scholar] [CrossRef]
- Yan, L.; Gong, Y.Z.; Shao, M.N.; Ruan, G.T.; Xie, H.L.; Liao, X.W.; Wang, X.K.; Han, Q.F.; Zhou, X.; Zhu, L.C.; et al. Distinct diagnostic and prognostic values of gamma-aminobutyric acid type A receptor family genes in patients with colon adenocarcinoma. Oncol. Lett. 2020, 20, 275–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullah, S.; Ali, N.; Ali, S.; Khan, A.; Ahmad, S.; Uddin, Z. Distribution of Different Genotypes MTHFR and GABRG2 Genes in Epileptic Population of Khyber Pakhtunkhwa Pakistan. Clin. Schizophr. Relat. Psychoses 2020, 14, 34–38. [Google Scholar] [CrossRef]
- Ganapathi, M.K.; Jones, W.D.; Sehouli, J.; Michener, C.M.; Braicu, I.E.; Norris, E.J.; Biscotti, C.V.; Vaziri, S.A.; Ganapathi, R.N. Expression profile of COL2A1 and the pseudogene SLC6A10P predicts tumor recurrence in high-grade serous ovarian cancer. Int. J. Cancer 2016, 138, 679–688. [Google Scholar] [CrossRef]
- Shi, W.; Gerster, K.; Alajez, N.M.; Tsang, J.; Waldron, L.; Pintilie, M.; Hui, A.B.; Sykes, J.; P’ng, C.; Miller, N.; et al. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011, 71, 2926–2937. [Google Scholar] [CrossRef] [PubMed]
| Type of Feature | Feature |
|---|---|
| Gray level co-occurrence matrix features (glcm) | Inverse Variance |
| Correlation | |
| Informational Measure of Correlation | |
| Contrast | |
| Inverse Difference Moment Normalized | |
| Cluster Prominence | |
| Cluster Shade | |
| Joint Energy | |
| Maximal Correlation Coefficient | |
| Difference Variance | |
| Inverse Variance | |
| Gray level size zone matrix features (glszm) | Small Area Emphasis |
| Small Area, High Gray Level, Emphasis | |
| Zone Variance | |
| Zone Entropy | |
| High Gray Level Zone Emphasis | |
| Large Area, High Gray Level, Emphasis | |
| Gray Level, Non-Uniformity, Normalized | |
| Shape features (2D) | Major Axis Length |
| Maximum 2D diameter | |
| First order features | Interquartile Range |
| Robust Mean Absolute Deviation | |
| Tenth percentile | |
| Skewness | |
| Median | |
| Uniformity | |
| Mean | |
| Variance | |
| Minimum | |
| Neighboring gray tone difference matrix features (ngtdm) | Busyness |
| Complexity | |
| Coarseness | |
| Gray level run length matrix features (glrlm) | Gray Level, Non-Uniformity, Normalized |
| Low Gray Level Run Emphasis | |
| Gray Level Variance |
| GENE | AUC |
|---|---|
| SYT4 | 0.628 |
| GABRG2 | 0.865 |
| CHGA | 0.660 |
| SLC6A17 | 0.605 |
| NEUROG2 | 0.876 |
| COL2A1 | 0.789 |
| MATN4 | 0.693 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Yang, L.; Jiao, X. Analysis of Breast Cancer Differences between China and Western Countries Based on Radiogenomics. Genes 2022, 13, 2416. https://doi.org/10.3390/genes13122416
Zhang Y, Yang L, Jiao X. Analysis of Breast Cancer Differences between China and Western Countries Based on Radiogenomics. Genes. 2022; 13(12):2416. https://doi.org/10.3390/genes13122416
Chicago/Turabian StyleZhang, Yuanyuan, Lifeng Yang, and Xiong Jiao. 2022. "Analysis of Breast Cancer Differences between China and Western Countries Based on Radiogenomics" Genes 13, no. 12: 2416. https://doi.org/10.3390/genes13122416






