cDNA Characterization and Expression of Selenium-Dependent CqGPx3 Isoforms in the Crayfish Cherax quadricarinatus under High Temperature and Hypoxia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Bioassays for Temperature and Hypoxia
2.2. RNA Isolation, Amplification and Cloning of Partial GPx cDNA Fragments
2.3. CqGPx3a and CqGPx3b Rapid Amplification of cDNA Ends (RACE) for Nerve Cord and Pereiopods
2.4. CqGPx3a and CqGPx3b mRNA and Deduced Amino Acid Sequence Analysis
2.5. CqGPx3, CqGPx3a and CqGPx3b mRNA Isoforms Quantification
2.6. Statistics
3. Results
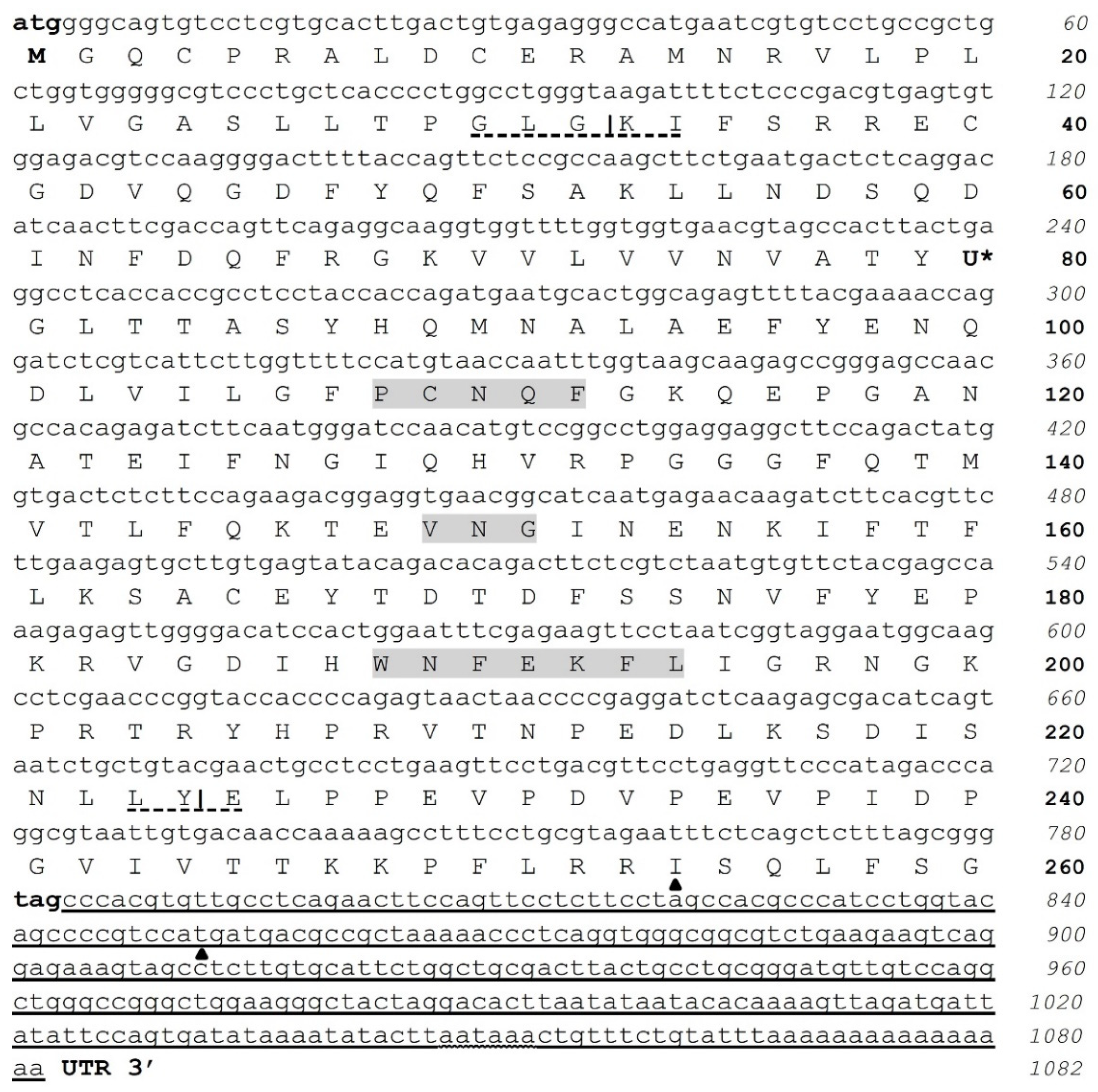
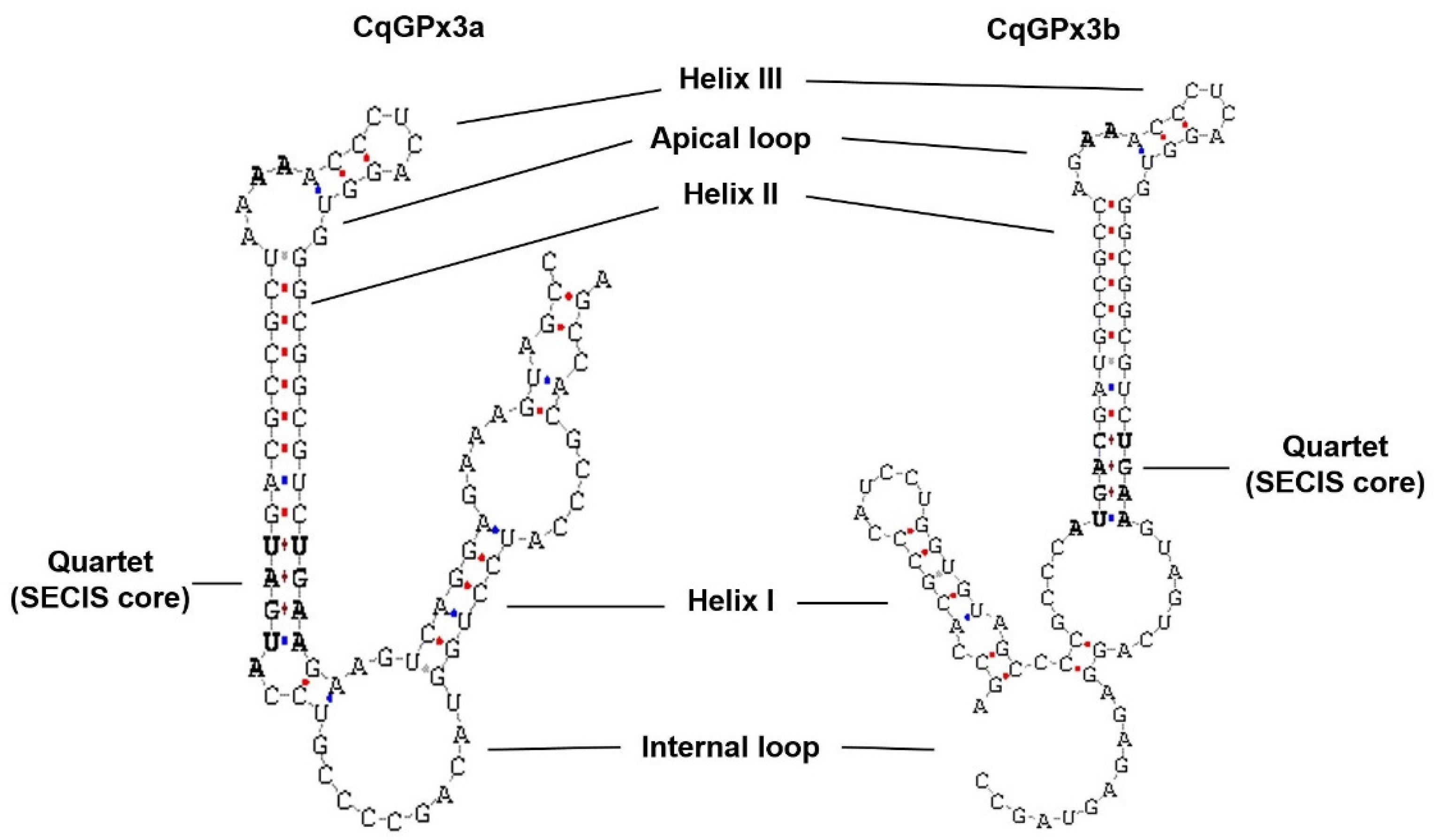
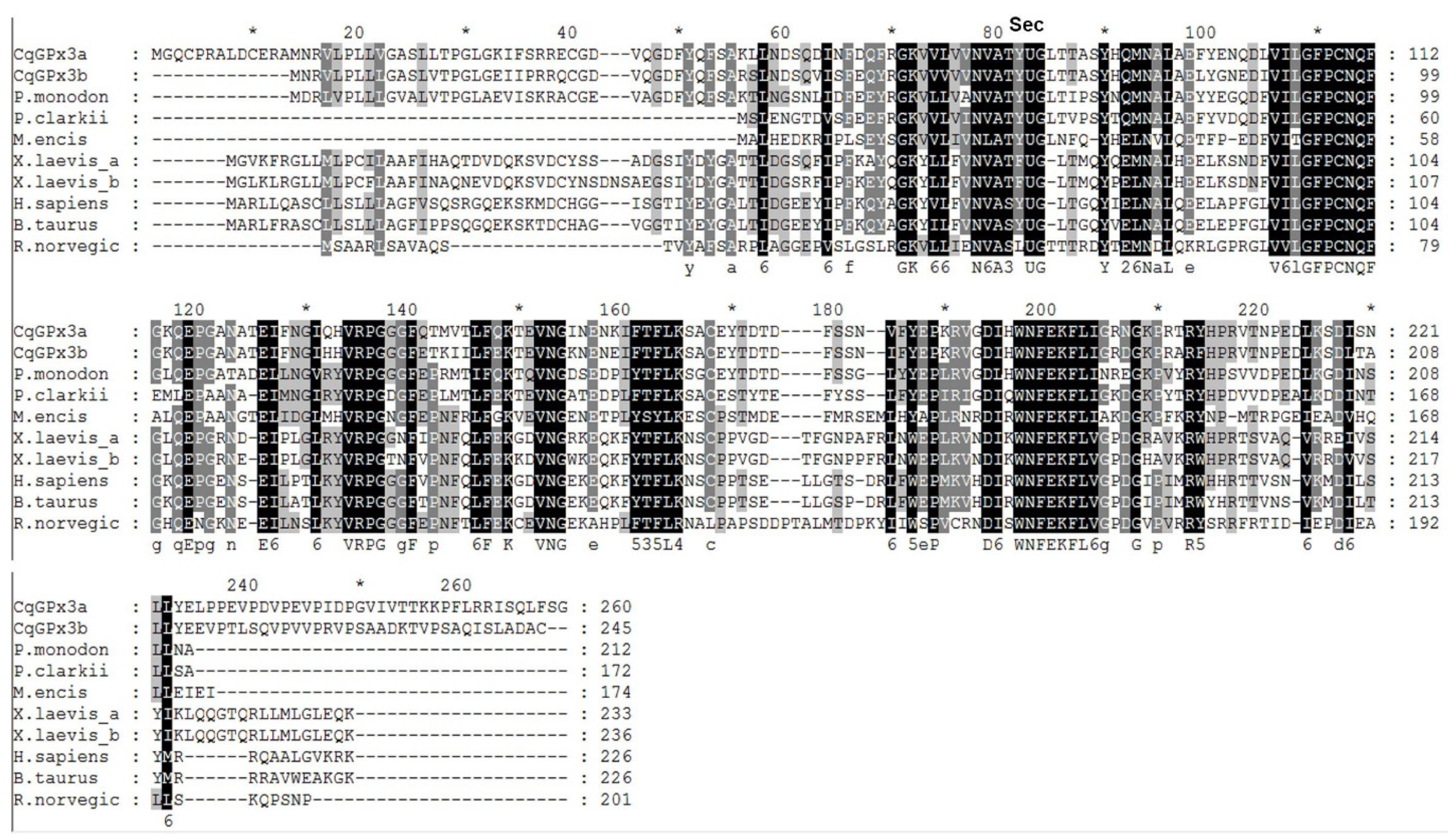
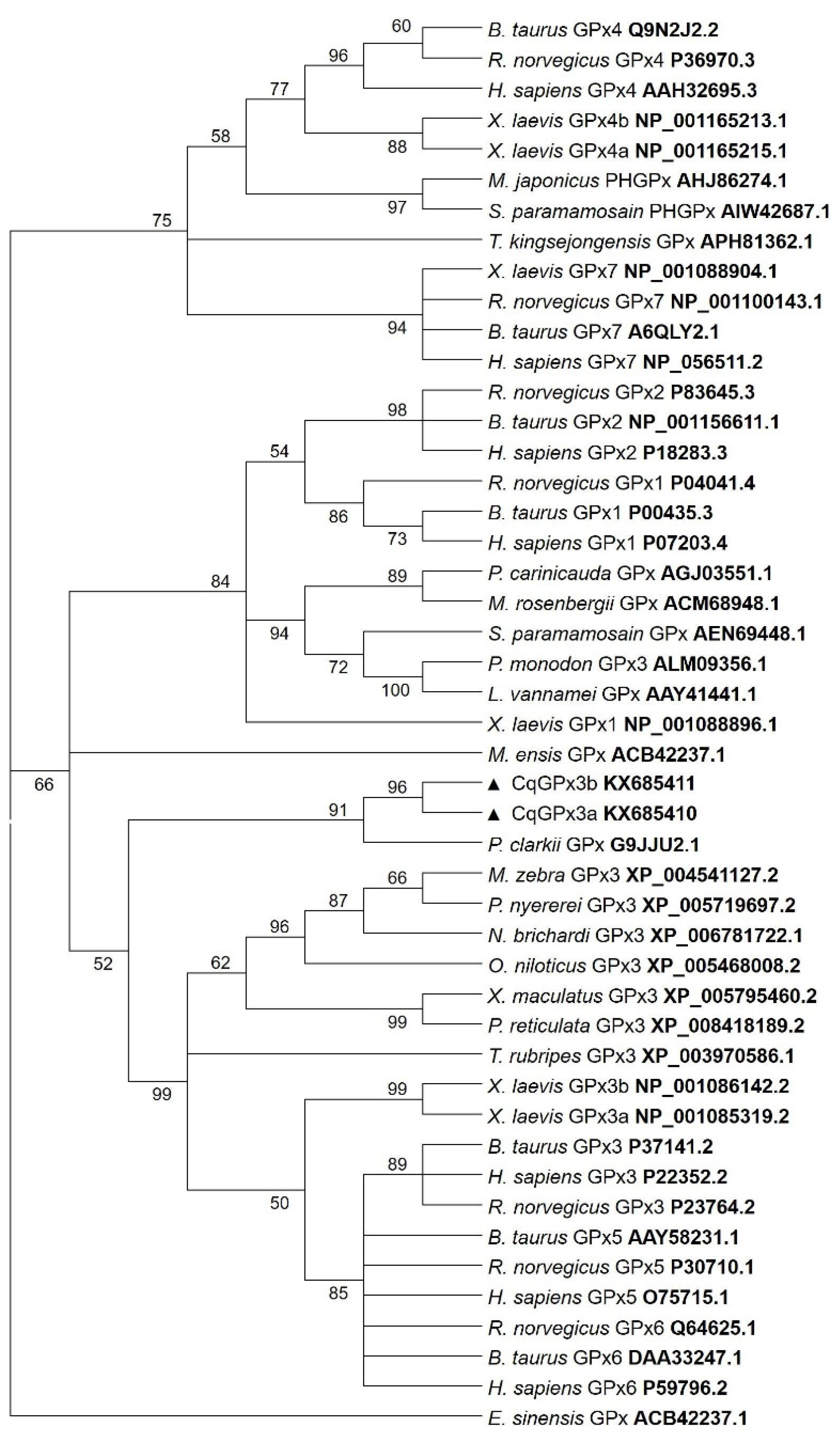
3.1. Two Isoforms of GPx3 Were Identified in Nervous System Tissues
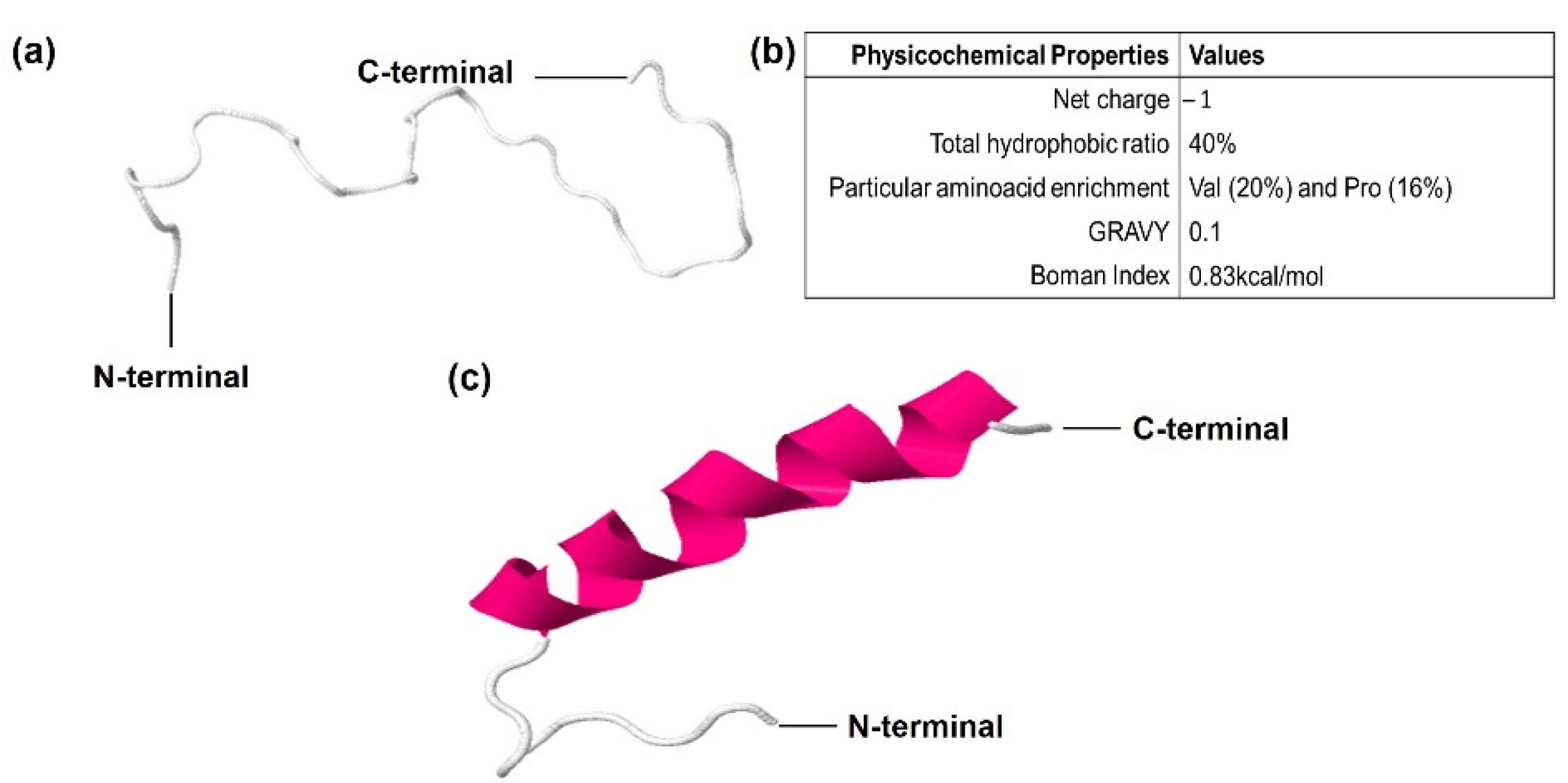
3.2. CqGPx3b C-Terminal Domain Prediction of Pro-Rich Antimicrobial Function
3.3. Expression of CqGPx3, CqGPx3a and CqGPx3b Isoforms in a Tissue-Specific Manner in the Nervous System and Muscle
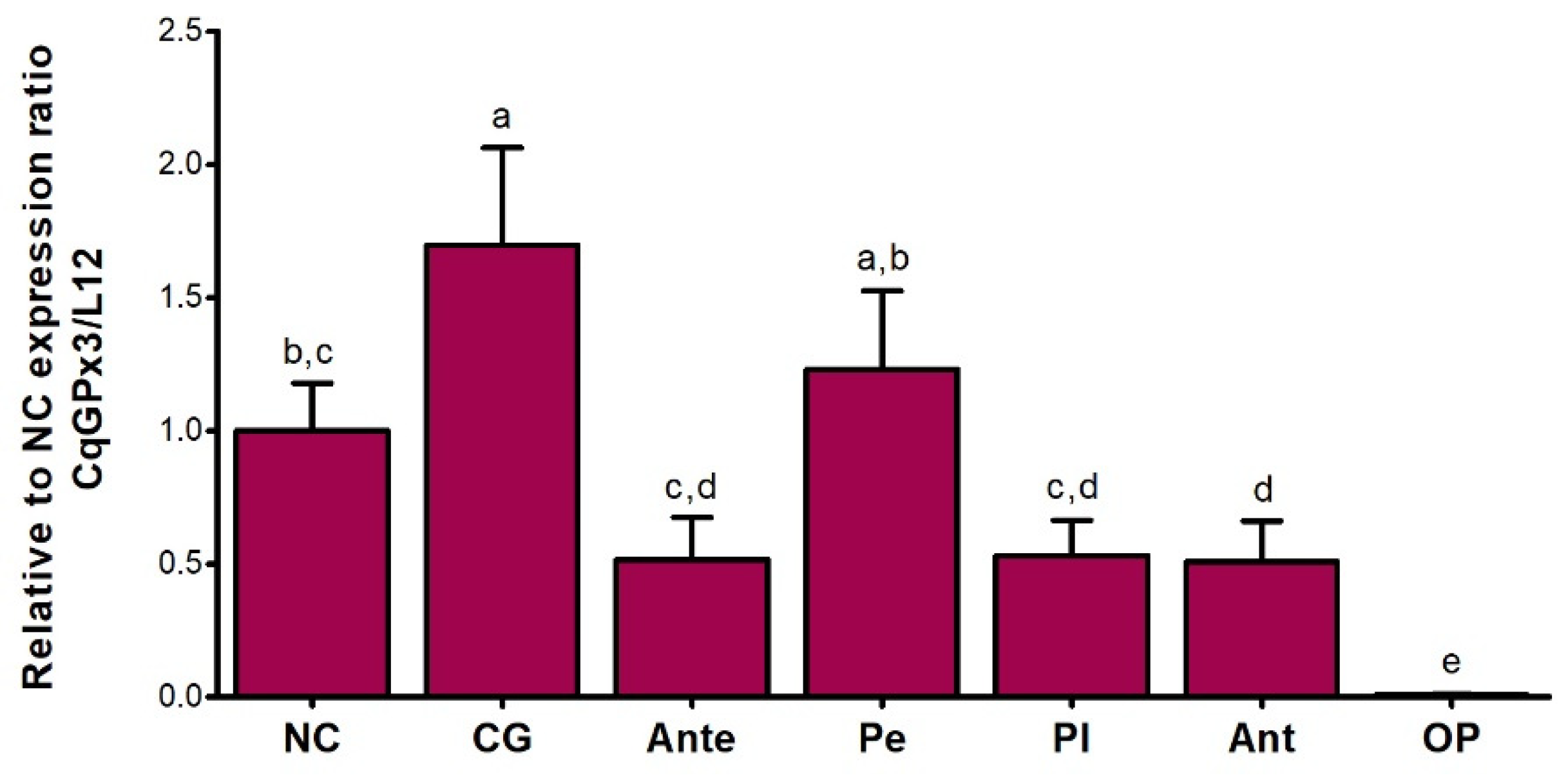
3.4. CqGPx3a and CqGPx3b Expression in Stressful Conditons
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fixsen, S.M.; Howard, M.T. Processive Selenocysteine Incorporation during Synthesis of Eukaryotic Selenoproteins. J. Mol. Biol. 2010, 399, 385–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donovan, J.; Copeland, P.R. Evolutionary History of Selenocysteine Incorporation from the Perspective of SECIS Binding Proteins. BMC Evol. Biol. 2009, 9, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ursini, F.; Maiorino, M.; Brigelius-Flohé, R.; Aumann, K.D.; Roveri, A.; Schomburg, D.; Flohé, L. Diversity of Glutathione Peroxidases. Meth. Enzymol. 1995, 252, 38–53. [Google Scholar] [CrossRef]
- Chambers, I.; Frampton, J.; Goldfarb, P.; Affara, N.; McBain, W.; Harrison, P.R. The Structure of the Mouse Glutathione Peroxidase Gene: The Selenocysteine in the Active Site Is Encoded by the “termination” Codon, TGA. EMBO J. 1986, 5, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- García-Triana, A.; Yepiz-Plascencia, G. The Crustacean Selenoproteome Similarity to Other Arthropods Homologs: A Mini Review. Electron. J. Biotechnol. 2012, 15, 17. [Google Scholar] [CrossRef]
- Wu, L.T.; Chu, K.H. Characterization of an Ovary-Specific Glutathione Peroxidase from the Shrimp Metapenaeus Ensis and Its Role in Crustacean Reproduction. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010, 155, 26–33. [Google Scholar] [CrossRef]
- Ren, Q.; Sun, R.-R.; Zhao, X.-F.; Wang, J.-X. A Selenium-Dependent Glutathione Peroxidase (Se-GPx) and Two Glutathione S-Transferases (GSTs) from Chinese Shrimp (Fenneropenaeus Chinensis). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2009, 149, 613–623. [Google Scholar] [CrossRef]
- Liu, C.-H.; Tseng, M.-C.; Cheng, W. Identification and Cloning of the Antioxidant Enzyme, Glutathione Peroxidase, of White Shrimp, Litopenaeus Vannamei, and Its Expression Following Vibrio Alginolyticus Infection. Fish. Shellfish Immunol. 2007, 23, 34–45. [Google Scholar] [CrossRef]
- Xia, X.-F.; Zheng, J.-J.; Shao, G.-M.; Wang, J.-L.; Liu, X.-S.; Wang, Y.-F. Cloning and Functional Analysis of Glutathione Peroxidase Gene in Red Swamp Crayfish Procambarus Clarkii. Fish. Shellfish Immunol. 2013, 34, 1587–1595. [Google Scholar] [CrossRef]
- Liu, W.; Zhao, C.; Wang, P.; Wang, S.; Lin, H.; Qiu, L. The Response of Glutathione Peroxidase 1 and Glutathione Peroxidase 7 under Different Oxidative Stresses in Black Tiger Shrimp, Penaeus Monodon. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2018, 217, 1–13. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Maiorino, M. Glutathione Peroxidases. Biochim. Biophys. Acta 2013, 1830, 3289–3303. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.S.; Kim, M.; Youn, B.-S.; Lee, N.S.; Park, J.W.; Lee, I.K.; Lee, Y.S.; Kim, J.B.; Cho, Y.M.; Lee, H.K.; et al. Glutathione Peroxidase 3 Mediates the Antioxidant Effect of Peroxisome Proliferator-Activated Receptor γ in Human Skeletal Muscle Cells. Mol. Cell. Biol. 2009, 29, 20–30. [Google Scholar] [CrossRef] [Green Version]
- Uittenbogaard, M.; Baxter, K.K.; Chiaramello, A. The Neurogenic Basic Helix-Loop-Helix Transcription Factor NeuroD6 Confers Tolerance to Oxidative Stress by Triggering an Antioxidant Response and Sustaining the Mitochondrial Biomass. ASN Neuro 2010, 2, e00034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Haddad, M.; Jean, E.; Turki, A.; Hugon, G.; Vernus, B.; Bonnieu, A.; Passerieux, E.; Hamade, A.; Mercier, J.; Laoudj-Chenivesse, D.; et al. Glutathione Peroxidase 3, a New Retinoid Target Gene, Is Crucial for Human Skeletal Muscle Precursor Cell Survival. J. Cell. Sci. 2012, 125, 6147–6156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fain, J.N.; Buehrer, B.; Bahouth, S.W.; Tichansky, D.S.; Madan, A.K. Comparison of Messenger RNA Distribution for 60 Proteins in Fat Cells vs. the Nonfat Cells of Human Omental Adipose Tissue. Metab. Clin. Exp. 2008, 57, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Elabd, C.; Ichim, T.E.; Miller, K.; Anneling, A.; Grinstein, V.; Vargas, V.; Silva, F.J. Comparing Atmospheric and Hypoxic Cultured Mesenchymal Stem Cell Transcriptome: Implication for Stem Cell Therapies Targeting Intervertebral Discs. J. Transl. Med. 2018, 16, 222. [Google Scholar] [CrossRef] [PubMed]
- Aboouf, M.A.; Armbruster, J.; Thiersch, M.; Gassmann, M.; Gödecke, A.; Gnaiger, E.; Kristiansen, G.; Bicker, A.; Hankeln, T.; Zhu, H.; et al. Myoglobin, Expressed in Brown Adipose Tissue of Mice, Regulates the Content and Activity of Mitochondria and Lipid Droplets. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2021, 1866, 159026. [Google Scholar] [CrossRef] [PubMed]
- Nalvarte, I.; Damdimopoulos, A.E.; Rüegg, J.; Spyrou, G. The Expression and Activity of Thioredoxin Reductase 1 Splice Variants v1 and v2 Regulate the Expression of Genes Associated with Differentiation and Adhesion. Biosci. Rep. 2015, 35, e00269. [Google Scholar] [CrossRef] [Green Version]
- Eissa, N.; Wang, H.-P.; Yao, H.; Shen, Z.-G.; Shaheen, A.A.; Abou-ElGheit, E.N. Expression of Hsp70, Igf1, and Three Oxidative Stress Biomarkers in Response to Handling and Salt Treatment at Different Water Temperatures in Yellow Perch, Perca Flavescens. Front. Physiol. 2017, 8, 683. [Google Scholar] [CrossRef]
- Sharma, A.; Paul, A.; Parida, S.; Pattanayak, S.; Mohapatra, A.; Rajesh Kumar, P.; Sahoo, M.K.; Sundaray, J.K.; Sahoo, P.K. Dynamics of Expression of Antibacterial and Antioxidant Defence Genes in Indian Major Carp, Labeo Rohita in Response to Aeromonas Hydrophila Infection. Microb. Pathog. 2018, 125, 108–115. [Google Scholar] [CrossRef]
- Hlaing, S.M.M.; Lou, J.; Cheng, J.; Xun, X.; Li, M.; Lu, W.; Hu, X.; Bao, Z. Tissue-Biased and Species-Specific Regulation of Glutathione Peroxidase (GPx) Genes in Scallops Exposed to Toxic Dinoflagellates. Toxins 2021, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- Yamindago, A.; Lee, N.; Lee, N.; Jo, Y.; Woo, S.; Yum, S. Fluoxetine in the Environment May Interfere with the Neurotransmission or Endocrine Systems of Aquatic Animals. Ecotoxicol. Environ. Saf. 2021, 227, 112931. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Gallo, R.L. AMPed up Immunity: How Antimicrobial Peptides Have Multiple Roles in Immune Defense. Trends Immunol. 2009, 30, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Scocchi, M.; Mardirossian, M.; Runti, G.; Benincasa, M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr. Top. Med. Chem. 2016, 16, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Krizsan, A.; Volke, D.; Weinert, S.; Sträter, N.; Knappe, D.; Hoffmann, R. Insect-Derived Proline-Rich Antimicrobial Peptides Kill Bacteria by Inhibiting Bacterial Protein Translation at the 70S Ribosome. Angew. Chem. Int. Ed. Engl. 2014, 53, 12236–12239. [Google Scholar] [CrossRef]
- Shi, X.-Z.; Zhao, X.-F.; Wang, J.-X. A New Type Antimicrobial Peptide Astacidin Functions in Antibacterial Immune Response in Red Swamp Crayfish Procambarus Clarkii. Dev. Comp. Immunol. 2014, 43, 121–128. [Google Scholar] [CrossRef]
- Bone, J.W.P.; Renshaw, G.M.C.; Furse, J.M.; Wild, C.H. Using Biochemical Markers to Assess the Effects of Imposed Temperature Stress on Freshwater Decapod Crustaceans: Cherax Quadricarinatus as a Test Case. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2015, 185, 291–301. [Google Scholar] [CrossRef]
- Sun, S.; Xuan, F.; Fu, H.; Zhu, J.; Ge, X.; Gu, Z. Transciptomic and Histological Analysis of Hepatopancreas, Muscle and Gill Tissues of Oriental River Prawn (Macrobrachium Nipponense) in Response to Chronic Hypoxia. BMC Genom. 2015, 16, 491. [Google Scholar] [CrossRef] [Green Version]
- Lant, B.; Storey, K.B. Glucose-6-Phosphate Dehydrogenase Regulation in Anoxia Tolerance of the Freshwater Crayfish Orconectes Virilis. Enzym. Res. 2011, 2011, 524906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, M.Y.; Pavasovic, A.; Mather, P.B.; Prentis, P.J. Transcriptome Analysis and Characterisation of Gill-Expressed Carbonic Anhydrase and Other Key Osmoregulatory Genes in Freshwater Crayfish Cherax Quadricarinatus. Data Brief. 2015, 5, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Sacristán, H.J.; Ansaldo, M.; Franco-Tadic, L.M.; Fernández Gimenez, A.V.; López Greco, L.S. Long-Term Starvation and Posterior Feeding Effects on Biochemical and Physiological Responses of Midgut Gland of Cherax Quadricarinatus Juveniles (Parastacidae). PLoS ONE 2016, 11, e0150854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholas, K.B.; Deerfield, D.W.; Nicholas, H.B.J.; Deerfield, D.W.I.; Nicholas, K.B.; Nicholas, H.B.; Deerfield, D.W.; Nicholas, H.B., Jr. GeneDoc: Analysis and Visualization of Genetic Variation. Embnew. News 1997, 4, 14. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allmang, C.; Krol, A. Selenoprotein Synthesis: UGA Does Not End the Story. Biochimie 2006, 88, 1561–1571. [Google Scholar] [CrossRef]
- García-Triana, A.; Gómez-Jiménez, S.; Peregrino-Uriarte, A.B.; López-Zavala, A.; González-Aguilar, G.; Sotelo-Mundo, R.R.; Valenzuela-Soto, E.M.; Yepiz-Plascencia, G. Expression and Silencing of Selenoprotein M (SelM) from the White Shrimp Litopenaeus Vannamei: Effect on Peroxidase Activity and Hydrogen Peroxide Concentration in Gills and Hepatopancreas. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010, 155, 200–204. [Google Scholar] [CrossRef]
- Zhang, H.; Li, X.-J.; Martin, D.B.; Aebersold, R. Identification and Quantification of N-Linked Glycoproteins Using Hydrazide Chemistry, Stable Isotope Labeling and Mass Spectrometry. Nat. Biotechnol. 2003, 21, 660–666. [Google Scholar] [CrossRef]
- Cao, C.; Leng, Y.; Huang, W.; Liu, X.; Kufe, D. Glutathione Peroxidase 1 Is Regulated by the C-Abl and Arg Tyrosine Kinases. J. Biol. Chem. 2003, 278, 39609–39614. [Google Scholar] [CrossRef] [Green Version]
- El-Benna, J.; Dang, P.M.-C.; Gougerot-Pocidalo, M.-A.; Marie, J.-C.; Braut-Boucher, F. P47phox, the Phagocyte NADPH Oxidase/NOX2 Organizer: Structure, Phosphorylation and Implication in Diseases. Exp. Mol. Med. 2009, 41, 217–225. [Google Scholar] [CrossRef] [Green Version]
- Baez-Duarte, B.G.; Mendoza-Carrera, F.; García-Zapién, A.; Flores-Martínez, S.E.; Sánchez-Corona, J.; Zamora-Ginez, I.; Torres-Rasgado, E.; León-Chávez, B.A.; Pérez-Fuentes, R. Multidisciplinary Research Group on Diabetes of the Instituto Mexicano del Seguro Social Glutathione Peroxidase 3 Serum Levels and GPX3 Gene Polymorphisms in Subjects with Metabolic Syndrome. Arch. Med. Res. 2014, 45, 375–382. [Google Scholar] [CrossRef]
- Milletti, F. Cell-Penetrating Peptides: Classes, Origin, and Current Landscape. Drug Discov. Today 2012, 17, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.-Q.; Shan, A.-S.; Dong, N.; Cao, Y.-P.; Lv, Y.-F.; Wang, L. The Effects of Leu or Val Residues on Cell Selectivity of α-Helical Peptides. Protein Pept. Lett. 2011, 18, 1112–1118. [Google Scholar] [CrossRef]
- Paulmann, M.; Arnold, T.; Linke, D.; Özdirekcan, S.; Kopp, A.; Gutsmann, T.; Kalbacher, H.; Wanke, I.; Schuenemann, V.J.; Habeck, M.; et al. Structure-Activity Analysis of the Dermcidin-Derived Peptide DCD-1L, an Anionic Antimicrobial Peptide Present in Human Sweat. J. Biol. Chem. 2012, 287, 8434–8443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saidemberg, D.M.; Baptista-Saidemberg, N.B.; Palma, M.S. Chemometric Analysis of Hymenoptera Toxins and Defensins: A Model for Predicting the Biological Activity of Novel Peptides from Venoms and Hemolymph. Peptides 2011, 32, 1924–1933. [Google Scholar] [CrossRef] [Green Version]
- Bahar, A.A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.P.; Deber, C.M. Guidelines for Membrane Protein Engineering Derived from de Novo Designed Model Peptides. Biopolymers 1998, 47, 41–62. [Google Scholar] [CrossRef]
- Dziuba, B.; Dziuba, M. New Milk Protein-Derived Peptides with Potential Antimicrobial Activity: An Approach Based on Bioinformatic Studies. Int. J. Mol. Sci. 2014, 15, 14531–14545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Vidal, M.; Jayasinghe, S.; Ladokhin, A.S.; White, S.H. Folding Amphipathic Helices into Membranes: Amphiphilicity Trumps Hydrophobicity. J. Mol. Biol. 2007, 370, 459–470. [Google Scholar] [CrossRef] [Green Version]
- Bhaskaran, R.; Ponnuswamy, P.K. Positional Flexibilities of Amino Acid Residues in Globular Proteins. Int. J. Pept. Protein Res. 1988, 32, 241–255. [Google Scholar] [CrossRef]
- Johns, M.E.; Tai, P.C.; Derby, C.D. Serine Proteases in the Spiny Lobster Olfactory Organ: Their Functional Expression along a Developmental Axis, and the Contribution of a CUB-Serine Protease. J. Neurobiol. 2004, 61, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Gardaneh, M.; Gholami, M.; Maghsoudi, N. Synergy between Glutathione Peroxidase-1 and Astrocytic Growth Factors Suppresses Free Radical Generation and Protects Dopaminergic Neurons against 6-Hydroxydopamine. Rejuvenation Res. 2011, 14, 195–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herault, O.; Hope, K.J.; Deneault, E.; Mayotte, N.; Chagraoui, J.; Wilhelm, B.T.; Cellot, S.; Sauvageau, M.; Andrade-Navarro, M.A.; Hébert, J.; et al. A Role for GPx3 in Activity of Normal and Leukemia Stem Cells. J. Exp. Med. 2012, 209, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Martindale, J.L.; Holbrook, N.J. Cellular Response to Oxidative Stress: Signaling for Suicide and Survival. J. Cell. Physiol. 2002, 192, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Cárdenas, P.; Cruz-Moreno, D.G.; González-Ruiz, R.; Peregrino-Uriarte, A.B.; Leyva-Carrillo, L.; Camacho-Jiménez, L.; Quintero-Reyes, I.; Yepiz-Plascencia, G. Combined Hypoxia and High Temperature Affect Differentially the Response of Antioxidant Enzymes, Glutathione and Hydrogen Peroxide in the White Shrimp Litopenaeus Vannamei. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2021, 254, 110909. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wei, L.; Cao, J.; Qiu, L.; Jiang, X.; Li, P.; Song, Q.; Zhou, H.; Han, Q.; Diao, X. Oxidative Stress, DNA Damage and Antioxidant Enzyme Activities in the Pacific White Shrimp (Litopenaeus Vannamei) When Exposed to Hypoxia and Reoxygenation. Chemosphere 2016, 144, 234–240. [Google Scholar] [CrossRef]
- Parrilla-Taylor, D.P.; Zenteno-Savín, T. Antioxidant Enzyme Activities in Pacific White Shrimp (Litopenaeus Vannamei) in Response to Environmental Hypoxia and Reoxygenation. Aquaculture 2011, 318, 379–383. [Google Scholar] [CrossRef]
- Bierl, C.; Voetsch, B.; Jin, R.C.; Handy, D.E.; Loscalzo, J. Determinants of Human Plasma Glutathione Peroxidase (GPx-3) Expression. J. Biol. Chem. 2004, 279, 26839–26845. [Google Scholar] [CrossRef] [Green Version]




| Primer | Sequence | Amplified DNA Fragments | |
|---|---|---|---|
| GPxF2 | 5′-CTCGTCATTCTTGGTTTTCC-3′ | qPCR CqGPx3 | |
| GPxR2 | 5′-CCGGGTTCGGGGCTTG-3′ | ||
| GPxF4 | 5′-GYAAGGTRSTDYTBRT-3′ | Internal cDNA partial amplification of CqGPx3 | |
| GPxF5 | 5′-GARRRYRBVVVCAGAAWHHWC-3′ | ||
| GPxF6 | 5′-RGRRGGYDKWYGGBVRBRYTT-3′ | ||
| GPxR3 | 5′-HBYYRRGHWSKDNBYBD-3′ | ||
| GPxR4 | 5′-SAGCCCWRMCCMDCBBAT-3′ | ||
| GPxF7 | 5′- CTCGTCATTCTTGGTTTTCCATGTAACC-3′ | RACE 3′ CqGPx3a | |
| GPxR5 | 5′- GGTTCGGGGCTTGCCATTCCTAC-3′ | RACE 5′ CqGPx3a | |
| GPxF19 | 5′-ATATTTTATGAACCCAAACGAGTTGGA-3′ | RACE 3′ CqGPx3b | qPCR CqGPc3b |
| GPxR12 | 5′-CTGAGACAGAGTAGGAACCTCTTCGT-3′ | RACE 5′ CqGPx3b | |
| GPxF21 | 5′-GTGTTCTACGAGCCAAAGAGAGTTG-3′ | qPCR CqGPx3a | |
| GPxR15 | 5′-CTACCCGCTAAAGAGCTGAGAAAT-3′ | ||
| L12F1 | 5′-CCTCTAAGTGTGTTTGCGGTGT-3′ | qPCR L12 | |
| L12R1 | 5′-AGCATCTGGTCAAGGGTCAG-3′ | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Aguirre, L.E.; Fuentes-Sidas, Y.I.; Rivera-Rangel, L.R.; Gutiérrez-Méndez, N.; Yepiz-Plascencia, G.; Chávez-Flores, D.; Zavala-Díaz de la Serna, F.J.; Peralta-Pérez, M.d.R.; García-Triana, A. cDNA Characterization and Expression of Selenium-Dependent CqGPx3 Isoforms in the Crayfish Cherax quadricarinatus under High Temperature and Hypoxia. Genes 2022, 13, 179. https://doi.org/10.3390/genes13020179
Hernández-Aguirre LE, Fuentes-Sidas YI, Rivera-Rangel LR, Gutiérrez-Méndez N, Yepiz-Plascencia G, Chávez-Flores D, Zavala-Díaz de la Serna FJ, Peralta-Pérez MdR, García-Triana A. cDNA Characterization and Expression of Selenium-Dependent CqGPx3 Isoforms in the Crayfish Cherax quadricarinatus under High Temperature and Hypoxia. Genes. 2022; 13(2):179. https://doi.org/10.3390/genes13020179
Chicago/Turabian StyleHernández-Aguirre, Laura E., Yazmin I. Fuentes-Sidas, Lizandro R. Rivera-Rangel, Néstor Gutiérrez-Méndez, Gloria Yepiz-Plascencia, David Chávez-Flores, Francisco J. Zavala-Díaz de la Serna, María del R. Peralta-Pérez, and Antonio García-Triana. 2022. "cDNA Characterization and Expression of Selenium-Dependent CqGPx3 Isoforms in the Crayfish Cherax quadricarinatus under High Temperature and Hypoxia" Genes 13, no. 2: 179. https://doi.org/10.3390/genes13020179







