Conserved Signatures in Protein Sequences Reliably Demarcate Different Clades of Rodents/Glires Species and Consolidate Their Evolutionary Relationships
Abstract
:1. Introduction
2. Materials and Methods
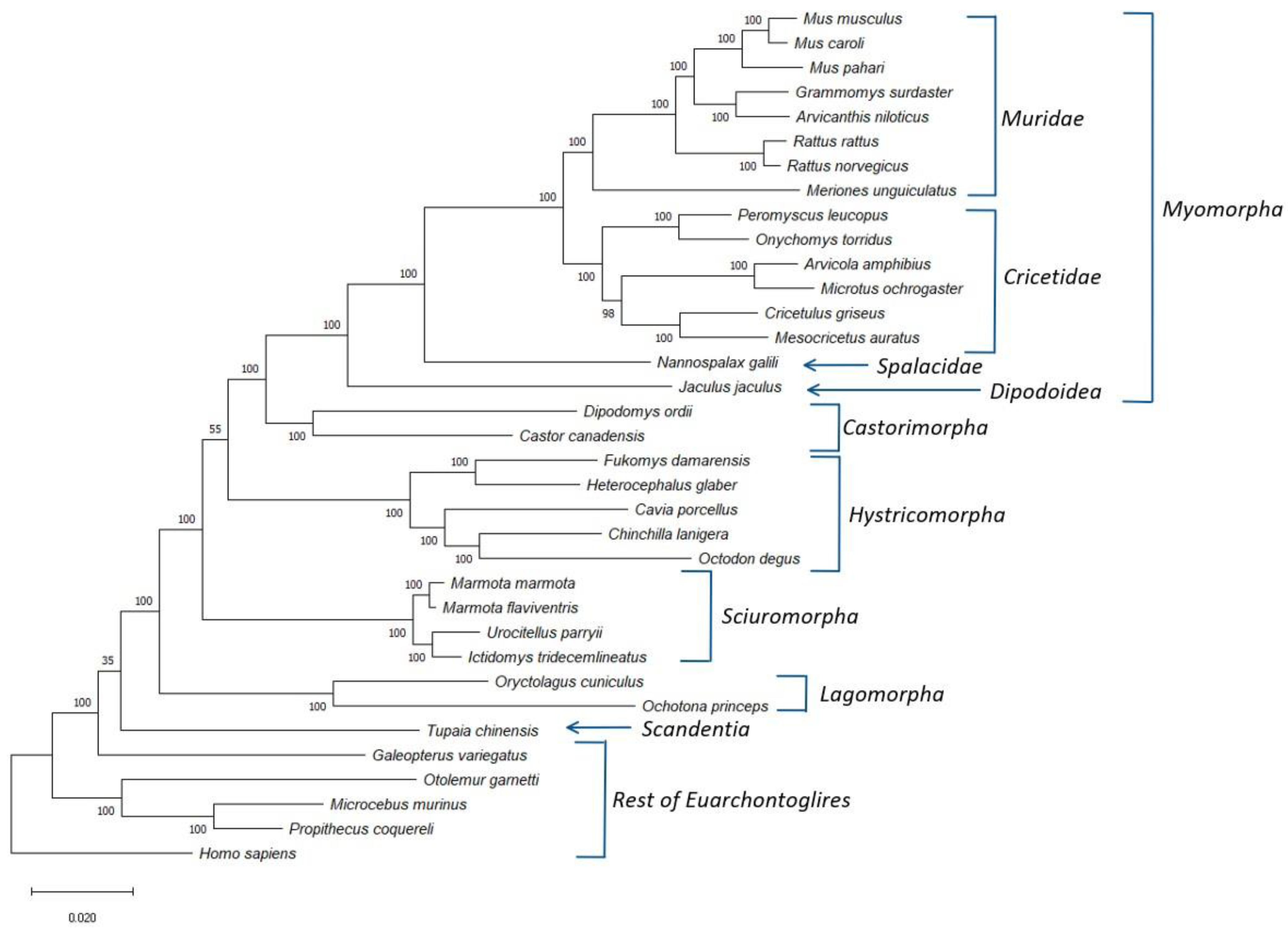
2.1. Construction of Phylogenetic Tree
2.2. Identification of Conserved Signature Indels (CSIs)
3. Results
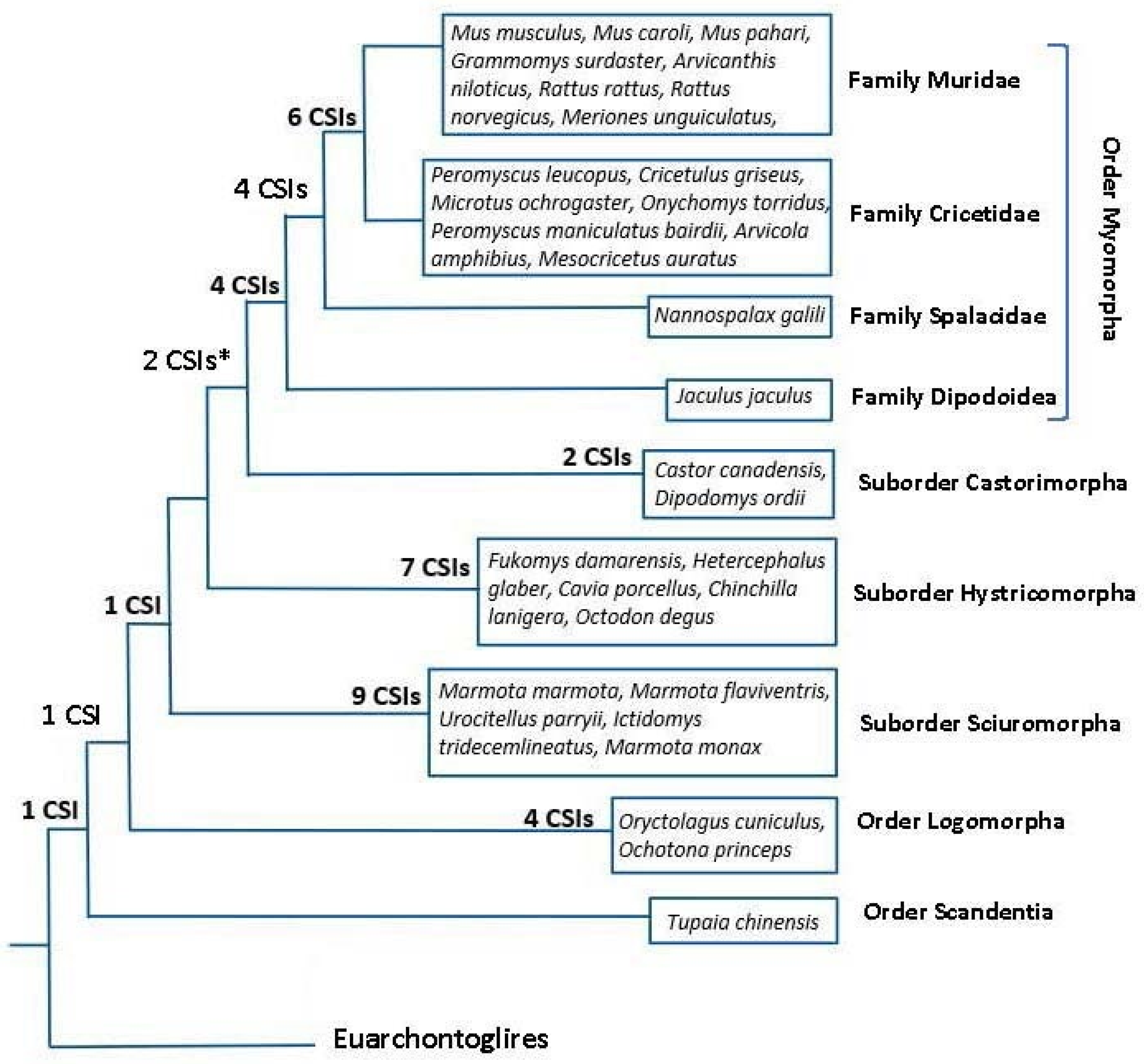
3.1. Phylogenetic Analysis of Rodentia
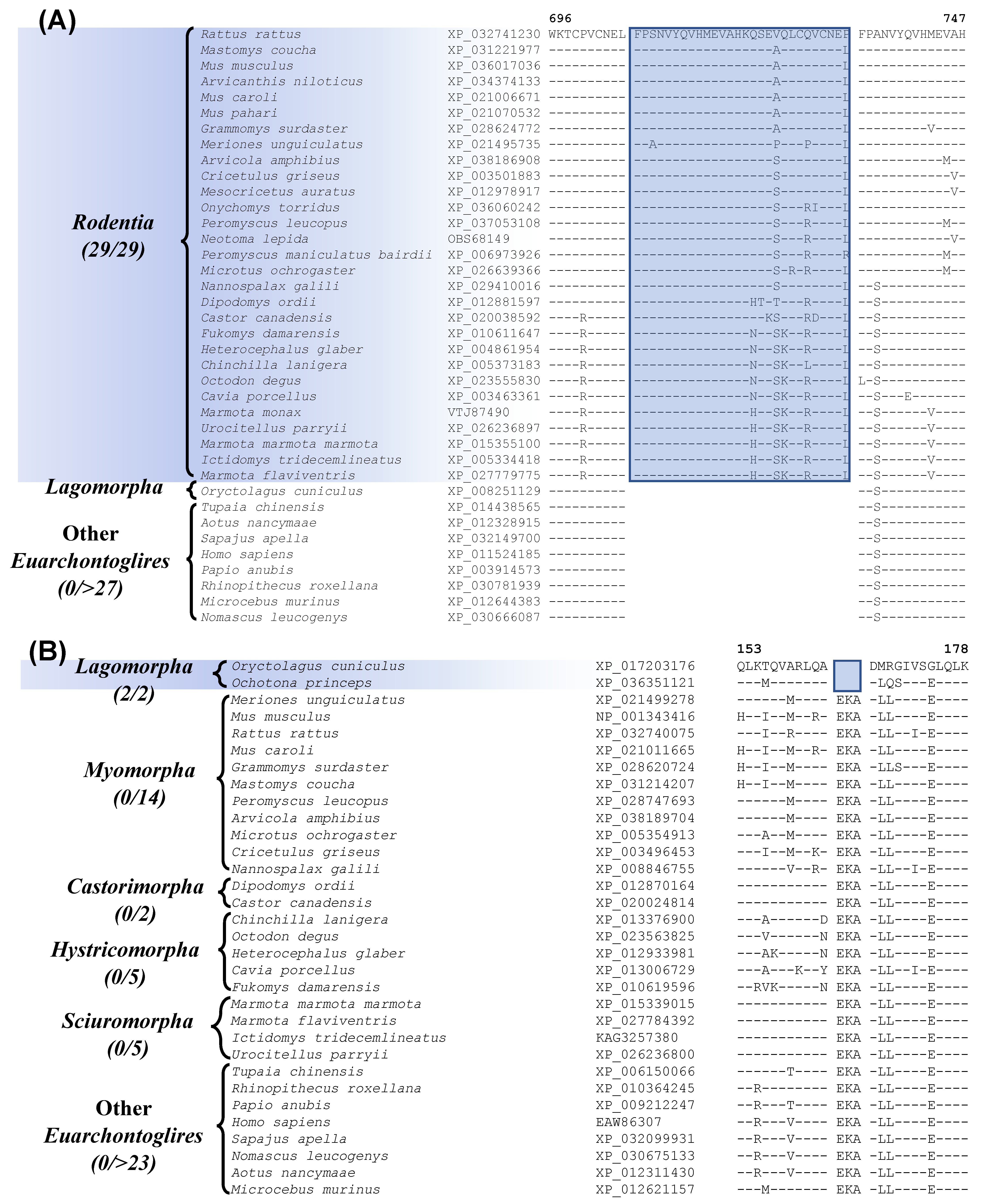
3.2. Identification of Molecular Markers Specific for Different Main Groups within the Glires
3.3. Molecular Signatures Specific for the Glires, Rodentia and Lagomorpha
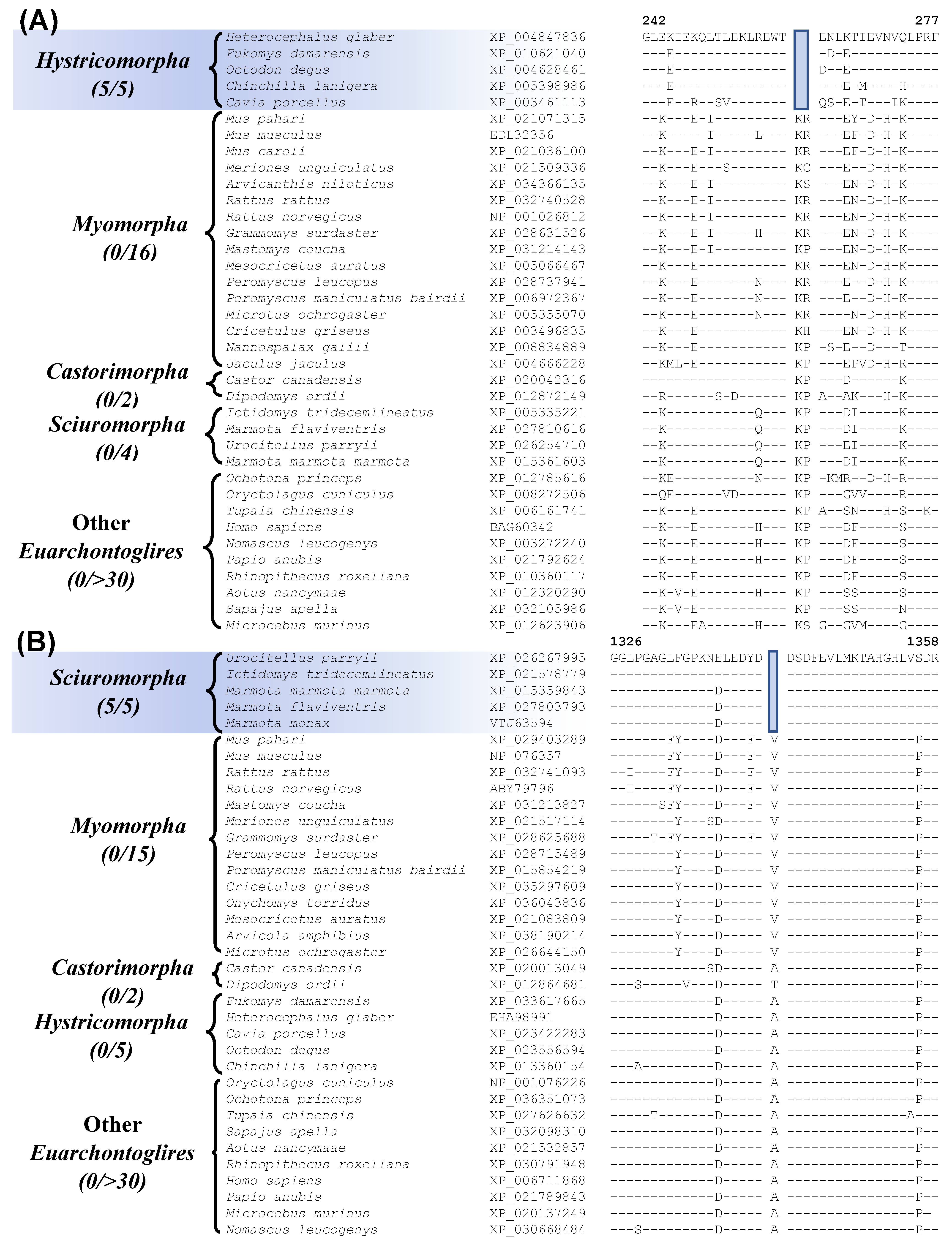
3.4. Molecular Signatures Specific for the Rodentia Suborders
| Protein Name | Accession No. | Figure Number | Indel Size | Indel Location | Specificity |
|---|---|---|---|---|---|
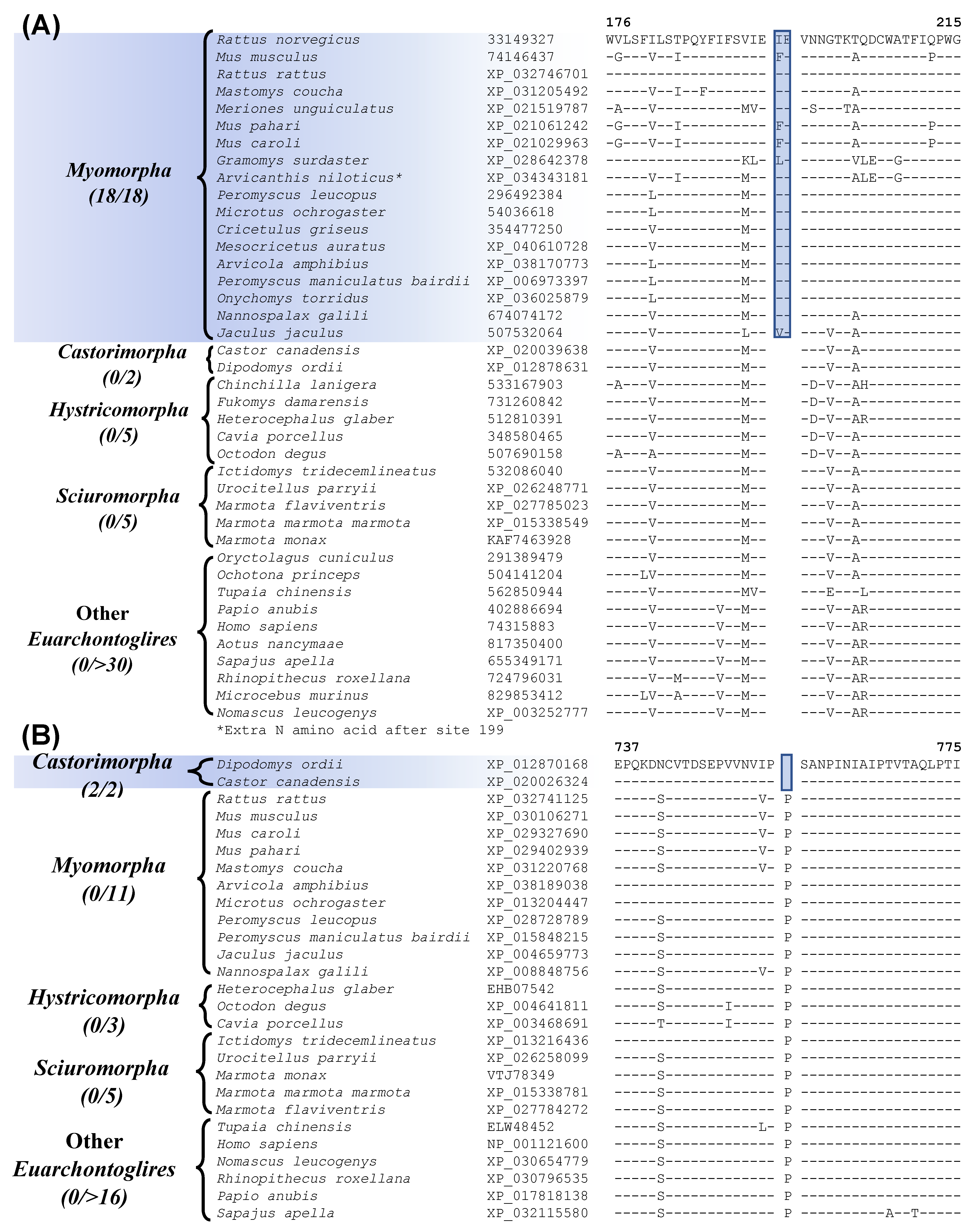
| vasopressin V1a receptor | 74146437 | Figure 4A Figure S8 | 2 aa Ins | 176–215 | Myomorpha |
| nck-associated protein 5-like isoform X1 | XP_006521185 | Figure S9 | 1 aa Del | 584–619 | |
| ATP-dependent DNA helicase DDX11 isoform 1 | NP_001335221 | Figure S10 | 3 aa Ins | 481–514 | |
| F-actin-uncapping protein LRRC16A | BAC31591 | Figure S11 | 1 aa Del | 1150–1174 | |
| zinc finger E-box-binding homeobox 1 | XP_030106271 | Figure 4B Figure S12 | 1 aa Del | 737–775 | Castorimorpha |
| cAMP-responsive element modulator | XP_030106165 | Figure S13 | 1 aa Del | 150–186 | |
| leukocyte elastase inhibitor A | EDL32356 | Figure 5A Figure S14 | 2 aa Del | 242–277 | Hystricomorpha |
| sterol regulatory element-binding protein cleavage-activating protein | AAH70437 | Figure S15 | 1 aa Del | 1040–1077 | |
| early endosome antigen 1 isoform X1 | XP_006513587 | Figure S16 | 1 aa Del | 58–91 | |
| tudor domain-containing protein 1 | NP_001002238 | Figure S17 | 1 aa Del | 42–74 | |
| tudor domain-containing protein 1 | AAI29955 | Figure S18 | 6 aa Del | 669–703 | |
| autophagy-related protein 9A isoform a | XP_011236992 | Figure S19 | 2 aa Ins | 659–687 | |
| probable small intestine urate exporter | XP_006516763 | Figure S20 | 2 aa Ins | 429–469 | |
| ryanodine receptor 2 | NP_076357 | Figure 5B Figure S21 | 1 aa Del | 1326–28 | Sciuromorpha |
| A disintegrin and metallo-proteinase with thrombospondin motifs 13 isoform 1 preproprotein | NP_001001322 | Figure S22 | 2 aa Del | 1072–1109 | |
| telomerase-binding protein EST1A | EDL12790 | Figure S23 | 1 aa Del | 472–503 | |
| oxysterol-binding protein-related protein 8 isoform b | XP_006513700 | Figure S24 | 1 aa Ins | 816–844 | |
| rab-3A-interacting protein isoform 2 | NP_001003950 | Figure S25 | 1 aa Del | 36–67 | |
| dual specificity protein phosphatase CDC14B | XP_036013890 | Figure S26 | 2 aa Ins | 335–370 | |
| zinc finger protein 385A | NP_038894 | Figure S27 | 1 aa Del | 7–44 | |
| rho family-interacting cell polarization regulator 2 | BAE37527 | Figure S28 | 1 aa Ins | 336–371 | |
| rho family-interacting cell polarization regulator 2 | XP_006516650 | Figure S29 | 5 aa Del | 587–625 | |
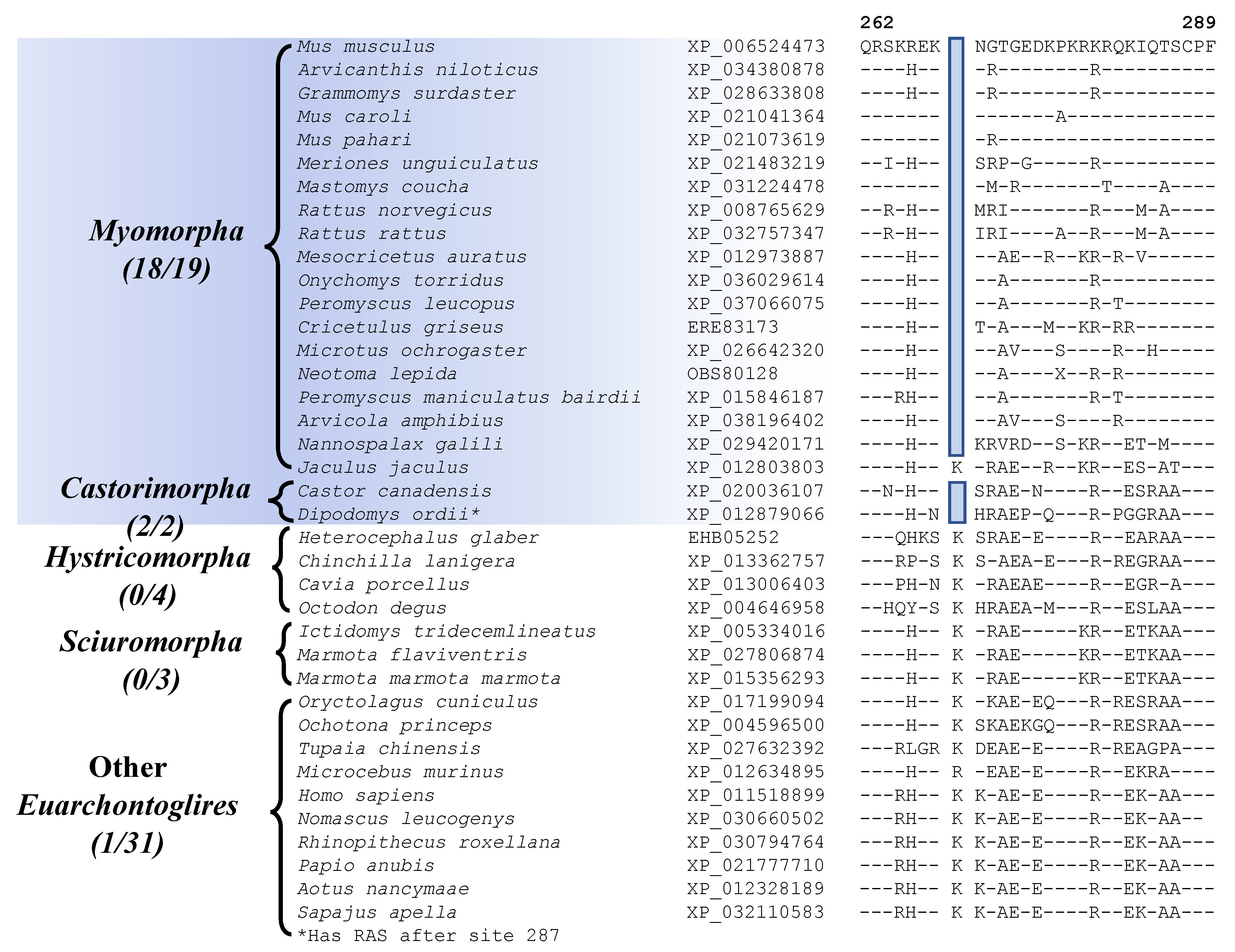
| ATP-dependent DNA helicase DDX11 isoform 1 | XP_006524473 | Figure 6 Figure S30 | 1 aa Del | 262–289 | Myomorpha and Castorimorpha |
| voltage-dependent L-type calcium channel subunit beta-2 | XP_006497377 | Figure S31 | 1 aa Ins | 375–401 |
3.5. Molecular Signatures Specific for the Family Level Clades in Myomorpha
| Protein Name | Accession No. | Figure Number | Indel Size | Indel Location | Specificity |
|---|---|---|---|---|---|
| cyclin-dependent kinase-like 2 | 74177560 | Figure 7A Figure S32 | 2 aa Ins | 232–263 | Muridae and Cricetidae |
| nck-associated protein 5-like | NP_001001884 | Figure S33 | 1 aa Del | 782–817 | |
| lysosomal acid glucosylceramidase | 568921788 | Figure S34 | 1 aa Del | 276–308 | |
| cAMP-responsive element modulator | NP_001104322 | Figure S35 | 1 aa Del | 61–91 | |
| cyclin-dependent kinase 13 | XP_006516830 | Figure S36 | 1 aa Del | 549–582 | |
| voltage-dependent L-type calcium channel subunit beta-2 | XP_036013681 | Figure S37 | 1 aa Del | 444–474 | |
| CREB-regulated transcription coactivator 1 | XP_006509763 | Figure 7B Figure S38 | 4 aa Del | 270–297 | Muroidea |
| striatin-interacting proteins 2 | 148681817 | Figure S39 | 1 aa Ins | 86–119 | |
| disco-interacting protein 2 homolog C | BAC29340 | Figure S40 | 4 aa Ins | 953–988 | |
| zinc finger protein 40 | XP_006516902 | Figure S41 | 1 aa Del | 2511–2535 |
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huchon, D.E.; Madsen, O.; Sibbald, M.J.J.B.; Ament, K.; Stanhope, M.J.; Catzeflis, F.O.; De Jong, W.W.; Douzery, E.J.P. Rodent phylogeny and a timescale for the evolution of glires: Evidence from an extensive taxon sampling using three nuclear genes. Mol. Biol. Evol. 2002, 19, 1053–1065. [Google Scholar] [CrossRef] [PubMed]
- D’Elía, G.; Fabre, P.-H.; Lessa, E.P. Rodent systematics in an age of discovery: Recent advances and prospects. J. Mammal. 2019, 100, 852–871. [Google Scholar] [CrossRef]
- Single, G.D.; MacDonald, D.W. Rodents. In The Encyclopedia of Mammals, 2nd ed.; MacDonald, D.W., Ed.; Oxford University Press: Oxford, UK, 2001; pp. 578–587. [Google Scholar]
- Carter, C.S.; Richardson, A.; Huffman, D.M.; Austad, S. Bring back the rat! J. Gerontol. Ser. A 2020, 75, 405–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morse, H.C., 3rd. The laboratory mouse—A historical perspective. In The Mouse in Biomedical Research; Foster, H.L., Small, J.D., Fox, J.G., Eds.; Academic Press: New York, NY, USA, 1981; Volume 1, p. 116. [Google Scholar]
- Kay, E.H.; Hoekstra, H.E. Rodents. Curr. Biol. 2008, 18, R406–R410. [Google Scholar] [CrossRef] [Green Version]
- Adkins, R.M.; Walton, A.H.; Honeycutt, R.L. Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol. Phylogenet. Evol. 2003, 26, 409–420. [Google Scholar] [CrossRef]
- Churakov, G.; Sadasivuni, M.K.; Rosenbloom, K.R.; Huchon, D.; Brosius, J.; Schmitz, J. Rodent evolution: Back to the root. Mol. Biol. Evol. 2010, 27, 1315–1326. [Google Scholar] [CrossRef] [Green Version]
- Blanga-Kanfi, S.; Miranda, H.; Penn, O.; Pupko, T.; Debry, R.W.; Huchon, D. Rodent phylogeny revised: Analysis of six nuclear genes from all major rodent clades. BMC Evol. Biol. 2009, 9, 71. [Google Scholar] [CrossRef] [Green Version]
- Swanson, M.T.; Oliveros, C.H.; Esselstyn, J.A. A phylogenomic rodent tree reveals the repeated evolution of masseter architectures. Proc. R. Soc. B Biol. Sci. 2019, 286, 20190672. [Google Scholar] [CrossRef] [Green Version]
- Fabre, P.-H.; Hautier, L.; Dimitrov, D.; Douzery, E.J.P. A glimpse on the pattern of rodent diversification: A phylogenetic approach. BMC Evol. Biol. 2012, 12, 88. [Google Scholar] [CrossRef] [Green Version]
- Doronina, L.; Matzke, A.; Churakov, G.; Stoll, M.; Huge, A.; Schmitz, J. The Beaver’s Phylogenetic Lineage Illuminated by Retroposon Reads. Sci. Rep. 2017, 7, 43562. [Google Scholar] [CrossRef] [Green Version]
- Asher, R.J.; Smith, M.R.; Rankin, A.; Emry, R.J. Congruence, fossils and the evolutionary tree of rodents and lagomorphs. R. Soc. Open Sci. 2019, 6, 190387. [Google Scholar] [CrossRef] [Green Version]
- Upham, N.S.; Esselstyn, J.A.; Jetz, W. Inferring the mammal tree: Species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 2019, 17, e3000494. [Google Scholar] [CrossRef]
- Honeycutt, R.L. Rodents (Rodentia). In The Timetree of Life; Hedges, S.B., Kumar, S., Eds.; Oxford University Press: Oxford, UK, 2009; pp. 490–494. [Google Scholar]
- Kumar, V.; Hallström, B.M.; Janke, A. Coalescent-based genome analyses resolve the early branches of the euarchontoglires. PLoS ONE 2013, 8, e60019. [Google Scholar] [CrossRef] [Green Version]
- Flynn, L. Rodent Suborders. Foss. Impr. 2019, 75, 292–298. [Google Scholar] [CrossRef]
- Douzery, E.J.; Huchon, D. Rabbits, if anything, are likely Glires. Mol. Phylogenet. Evol. 2004, 33, 922–935. [Google Scholar] [CrossRef]
- Misawa, K.; Janke, A. Revisiting the Glires concept—phylogenetic analysis of nuclear sequences. Mol. Phylogenet. Evol. 2003, 28, 320–327. [Google Scholar] [CrossRef]
- Kriegs, J.O.; Churakov, G.; Jurka, J.; Brosius, J.; Schmitz, J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007, 23, 158–161. [Google Scholar] [CrossRef]
- Rokas, A.; Williams, B.L.; King, N.; Carroll, S.B. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 2003, 425, 798–804. [Google Scholar] [CrossRef]
- Patel, S.; Gupta, R.S. A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: Proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 406–438. [Google Scholar] [CrossRef]
- Baldauf, S.L.; Palmer, J.D. Animals and fungi are each other’s closest relatives: Congruent evidence from multiple proteins. Proc. Natl. Acad. Sci. USA 1993, 90, 11558–11562. [Google Scholar] [CrossRef] [Green Version]
- Springer, M.S.; Stanhope, M.J.; Madsen, O.; De Jong, W.W. Molecules consolidate the placental mammal tree. Trends Ecol. Evol. 2004, 19, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Gupta, R.S. Novel molecular synapomorphies demarcate different main groups/subgroups of Plasmodium and Piroplasmida species clarifying their evolutionary relationships. Genes 2019, 10, 490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, R.S. Molecular signatures that are distinctive characteristics of the vertebrates and chordates and supporting a grouping of vertebrates with the tunicates. Mol. Phylogenet. Evol. 2016, 94, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S. Identification of conserved indels that are useful for classification and evolutionary studies. Methods Microbiol. 2014, 41, 153–182. [Google Scholar]
- Rokas, A.; Holland, P.W. Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 2000, 15, 454–459. [Google Scholar] [CrossRef]
- Gupta, R.S. Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 1998, 62, 1435–1491. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.S. Impact of genomics on the understanding of microbial evolution and classification: The importance of Darwin’s views on classification. FEMS Microbiol. Rev. 2016, 40, 520–553. [Google Scholar] [CrossRef]
- Baldauf, S.L. Phylogeny for the faint of heart: A tutorial. Trends Genet. 2003, 19, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Inferring Phylogenies; Sinauer Associates, Inc.: Sunderland, MA, USA, 2004. [Google Scholar]
- Rivera, M.C.; Lake, J.A. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science 1992, 257, 74–76. [Google Scholar] [CrossRef]
- Khadka, B.; Chatterjee, T.; Gupta, B.P.; Gupta, R.S. Genomic analyses identify novel molecular signatures specific for the caenorhabditis and other nematode taxa providing novel means for genetic and biochemical studies. Genes 2019, 10, 739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khadka, B.; Gupta, R.S. Conserved molecular signatures in the spike protein provide evidence indicating the origin of SARS-CoV-2 and a Pangolin-CoV (MP789) by recombination(s) between specific lineages of Sarbecoviruses. PeerJ 2021, 9, e12434. [Google Scholar] [CrossRef]
- Ajawatanawong, P.; Baldauf, S.L. Evolution of protein indels in plants, animals and fungi. BMC Evol. Biol. 2013, 13, 140. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.; Gupta, R.S. Conserved inserts in the Hsp60 (GroEL) and Hsp70 (DnaK) proteins are essential for cellular growth. Mol. Genet. Genom. 2009, 281, 361–373. [Google Scholar] [CrossRef]
- Khadka, B.; Gupta, R.S. Identification of a conserved 8 aa insert in the PIP5K protein in the Saccharomycetaceae family of fungi and the molecular dynamics simulations and structural analysis to investigate its potential functional role. Proteins 2017, 85, 1454–1467. [Google Scholar] [CrossRef]
- Akiva, E.; Itzhaki, Z.; Margalit, H. Built-in loops allow versatility in domain-domain interactions: Lessons from self-interacting domains. Proc. Natl. Acad. Sci. USA 2008, 105, 13292–13297. [Google Scholar] [CrossRef] [Green Version]
- Chatterji, M.; Unniraman, S.; Maxwell, A.; Nagaraja, V. The additional 165 amino acids in the B protein of Escherichia coli DNA gyrase have an important role in DNA binding. J. Biol. Chem. 2000, 275, 22888–22894. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, K.; Panchenko, A.R. Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states. Proc. Natl. Acad. Sci. USA 2010, 107, 20352–20357. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Sun, F.; Xu, S.; Yang, G.; Li, M. The position of tree shrews in the mammalian tree: Comparing multi-gene analyses with phylogenomic results leaves monophyly of Euarchonta doubtful. Integr. Zool. 2015, 10, 186–198. [Google Scholar] [CrossRef]
- Jeanmougin, F.; Thompson, J.D.; Gouy, M.; Higgins, D.G.; Gibson, T.J. Multiple sequence alignment with Clustal x. Trends Biochem. Sci. 1998, 23, 403–405. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. CABIOS 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Gao, B.; Adeolu, M.; Khadka, B.; Gupta, R.S. Phylogenomic analyses and comparative studies on genomes of the Bifidobacteriales: Identification of molecular signatures specific for the order Bifidobacteriales and its different subclades. Front. Microbiol. 2016, 7, 978. [Google Scholar] [CrossRef]
- Bhandari, V.; Naushad, H.S.; Gupta, R.S. Protein Based Molecular Markers Provide Reliable Means to Understand Prokaryotic Phylogeny and Support a Predominantly Darwinian Mode of Evolution. Front. Cell. Infect. Microbiol. 2012, 2, 98. [Google Scholar] [CrossRef] [Green Version]
- Puigbo, P.; Wolf, Y.I.; Koonin, E.V. Seeing the Tree of Life behind the phylogenetic forest. BMC Biol. 2013, 11, 46. [Google Scholar] [CrossRef] [Green Version]
- McCormack, J.E.; Faircloth, B.C.; Crawford, N.G.; Gowaty, P.A.; Brumfield, R.T.; Glenn, T.C. Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis. Genome Res. 2012, 22, 746–754. [Google Scholar] [CrossRef] [Green Version]
- Akashi, M.; Higashi, T.; Masuda, S.; Komori, T.; Furuse, M. A coronary artery disease-associated gene product, JCAD/KIAA1462, is a novel component of endothelial cell–cell junctions. Biochem. Biophys. Res. Commun. 2011, 413, 224–229. [Google Scholar] [CrossRef]
- Osawa, T.; Mizuno, Y.; Fujita, Y.; Takatama, M.; Nakazato, Y.; Okamoto, K. Optineurin in neurodegenerative diseases. Neuropathology 2011, 31, 569–574. [Google Scholar] [CrossRef]
- Sparwel, M.; Doronina, L.; Churakov, G.; Stegemann, A.; Brosius, J.; Robinson, T.J.; Schmitz, J. The volcano rabbit in the phylogenetic network of Lagomorphs. Genome Biol. Evol. 2019, 11, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Bronsert, P.; Kohler, I.; Timme, S.; Kiefer, S.; Werner, M.; Schilling, O.; Vashist, Y.; Makowiec, F.; Brabletz, T.; Hopt, U.T.; et al. Prognostic significance of Zinc finger E-box binding homeobox 1 (ZEB1) expression in cancer cells and cancer-associated fibroblasts in pancreatic head cancer. Surgery 2014, 156, 97–108. [Google Scholar] [CrossRef]
- Torriglia, A.; Martin, E.; Jaadane, I. The hidden side of SERPINB1/Leukocyte Elastase Inhibitor. Semin. Cell Dev. Biol. 2017, 62, 178–186. [Google Scholar] [CrossRef]
- Lehnart, S.E.; Mongillo, M.; Bellinger, A.; Lindegger, N.; Chen, B.-X.; Hsueh, W.; Reiken, S.; Wronska, A.; Drew, L.J.; Ward, C.W.; et al. Leaky Ca2+ release channel/ryanodine receptor 2 causes seizures and sudden cardiac death in mice. J. Clin. Investig. 2008, 118, 2230–2245. [Google Scholar] [CrossRef] [Green Version]
- Fang, C.-L.; Uen, Y.-H.; Chen, H.-K.; Hseu, Y.-C.; Lin, C.-C.; Hung, S.-T.; Sun, D.-P.; Lin, K.-Y. Loss of cyclin-dependent kinase-like 2 predicts poor prognosis in gastric cancer, and its overexpression suppresses cells growth and invasion. Cancer Med. 2018, 7, 2993–3002. [Google Scholar] [CrossRef]
- Yersin, A. La peste bubonique à Hong-Kong. Ann. L’institut Pasteur 1894, 8, 662–667. [Google Scholar]
- Alnajar, S.; Khadka, B.; Gupta, R.S. Ribonucleotide reductases from Bifidobacteria contain multiple conserved indels distinguishing them from all other organisms: In silico analysis of the possible role of a 43 aa Bifidobacteria-specific insert in the Class III RNR homolog. Front. Microbiol. 2017, 8, 1409. [Google Scholar] [CrossRef] [Green Version]
- Ahmod, N.Z.; Gupta, R.S.; Shah, H.N. Identification of a Bacillus anthracis specific indel in the yeaC gene and development of a rapid pyrosequencing assay for distinguishing B. anthracis from the B. cereus group. J. Microbiol. Methods 2011, 87, 278–285. [Google Scholar] [CrossRef]
- Wong, S.Y.; Paschos, A.; Gupta, R.S.; Schellhorn, H.E. Insertion/deletion-based approach for the detection of Escherichia coli O157:H7 in freshwater environments. Environ. Sci. Technol. 2014, 48, 11462–11470. [Google Scholar] [CrossRef]


| Protein Name | Accession No. | Figure Number | Indel Size | Indel Location | Specificity |
|---|---|---|---|---|---|
| junctional protein associated with coronary artery disease | BAD90447 | Figure 2A Figure S1 | 1aa Ins | 792–822 | Glires |
| adenylyl cyclase-associated protein 2 | EDL41025 | Figure 2B Figure S2 | 1aa Del | 280–313 | Glires and Scandentia |
| activity-dependent neuroprotector homeobox protein 2 | XP_036017036 | Figure 3A Figure S3 | 28aa Ins | 696–747 | Rodentia |
| optineurin | NP_001343416 | Figure 3B Figure S4 | 3aa Del | 153–178 | Lagomorpha |
| U3 small nucleolar RNA-associated protein 6 homolog | 74146777 | Figure S5 | 1aa Ins | 192–227 | |
| ankyrin repeat and KH domain-containing protein 1 | NP_780584 | Figure S6 | 1aa Ins | 1893–1927 | |
| prickle-like protein 1 | NP_001028389 | Figure S7 | 3aa Del | 553–586 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, R.S.; Suggett, C. Conserved Signatures in Protein Sequences Reliably Demarcate Different Clades of Rodents/Glires Species and Consolidate Their Evolutionary Relationships. Genes 2022, 13, 288. https://doi.org/10.3390/genes13020288
Gupta RS, Suggett C. Conserved Signatures in Protein Sequences Reliably Demarcate Different Clades of Rodents/Glires Species and Consolidate Their Evolutionary Relationships. Genes. 2022; 13(2):288. https://doi.org/10.3390/genes13020288
Chicago/Turabian StyleGupta, Radhey S., and Carson Suggett. 2022. "Conserved Signatures in Protein Sequences Reliably Demarcate Different Clades of Rodents/Glires Species and Consolidate Their Evolutionary Relationships" Genes 13, no. 2: 288. https://doi.org/10.3390/genes13020288
APA StyleGupta, R. S., & Suggett, C. (2022). Conserved Signatures in Protein Sequences Reliably Demarcate Different Clades of Rodents/Glires Species and Consolidate Their Evolutionary Relationships. Genes, 13(2), 288. https://doi.org/10.3390/genes13020288







