Abstract
The identification of mutants through forward genetic screens is the backbone of Drosophila genetics research, yet many mutants identified through these screens have yet to be mapped to the Drosophila genome. This is especially true of mutants that have been identified as mutagen-sensitive (mus), but have not yet been mapped to their associated molecular locus. Our study addressed the need for additional mus gene identification by determining the locus and exploring the function of the X-linked mutagen-sensitive gene mus109 using three available mutant alleles: mus109D1, mus109D2, and mus109lS. After first confirming that all three mus109 alleles were sensitive to methyl methanesulfonate (MMS) using complementation analysis, we used deletion mapping to narrow the candidate genes for mus109. Through DNA sequencing, we were able to determine that mus109 is the uncharacterized gene CG2990, which encodes the Drosophila ortholog of the highly conserved DNA2 protein that is important for DNA replication and repair. We further used the sequence and structure of DNA2 to predict the impact of the mus109 allele mutations on the final gene product. Together, these results provide a tool for researchers to further investigate the role of DNA2 in DNA repair processes in Drosophila.
1. Introduction
The development of gene mapping techniques has a long and storied history in the Drosophila melanogaster model system (reviewed in [1]), beginning with Alfred Sturtevant’s fundamental publication of the first genetic map in 1913 [2]. In this work, Sturtevant showed that genes are arranged in a linear order along chromosomes and that the recombination frequency between two genes could be used as a measure of the distance between them. This discovery created the foundation for other key advances in Drosophila gene mapping, including the generation of detailed polytene chromosome cytogenetic maps [3,4], the development of deletion kits covering the genome [5,6,7], and the sequencing of the D. melanogaster genome [8].
However, despite these advances, the current D. melanogaster genome annotation includes 14,184 genes that have not yet been mapped to the molecular genome (FlyBase R6.43; [9]), including many genes that were discovered in forward genetic screens. In these cases, alleles have been discovered that produce a phenotype of interest, but the molecular locus responsible for this phenotype remains unknown. For example, several forward genetic screens have been conducted to identify D. melanogaster mutants with defects in DNA repair (e.g., [10,11,12,13,14,15]). In these screens, flies that showed reduced survival in the presence of a mutagen—usually the alkylating agent methyl methanesulfonate (MMS)—were identified as probable DNA repair mutants. To date, 58 of these mutagen-sensitive (mus) stocks have been generated, yet the gene responsible for the mus phenotype is known for only 15 of these stocks [16,17]. Importantly, each mapped mus gene has encoded an ortholog of a human DNA repair protein [17], including proteins implicated in disorders such as Bloom syndrome [18], Fanconi anemia [16], and xeroderma pigmentosum [19].
The knowledge derived from studies of these 15 mapped mus genes demonstrates the utility of mapping mus genes to facilitate DNA repair research in Drosophila. With this in mind, we sought to map mus109, an X-linked essential gene with three extant alleles: mus109D1 [13] and mus109D2 [14] are homozygous viable hypomorphic alleles, whereas mus109lS is a homozygous lethal null allele [20]. mus109 mutants are characterized by chromosomal instability in the absence of mutagen treatment [20,21,22,23], with the majority of chromosome breaks occurring in heterochromatin [22]. Further, mus109 mutants are sensitive to MMS, 4-nitroquinoline-1-oxide (4NQO), and γ irradiation [13,14,24,25], which are mutagens that create DNA adducts (MMS and 4NQO; [26,27]) and oxidative damage (γ irradiation; [28]). In this manuscript, we present detailed mapping data obtained through complementation analysis, deletion crosses, and DNA sequence alignment showing that mus109 is the uncharacterized Drosophila gene CG2990 (human DNA2). We further discuss the potential functionality of the mus109 mutant alleles by comparing the mutations to conserved catalytic regions in DNA2.
2. Materials and Methods
2.1. Drosophila Stocks and Maintenance
D. melanogaster stocks were maintained at 25 °C in bottles containing Nutri-Fly Bloomington Formulation media (Genesee Scientific) with a 12h day/night cycle. Experimental crosses were conducted in narrow vials containing corn syrup/soy media (Archon Scientific). The following fly stocks were obtained from the Bloomington Drosophila Stock Center (BDSC): mus109D1 (BDSC# 2320), mus109D2 (BDSC# 2307), mus109lS (BDSC# 4168), Df(1)ED6991 (BDSC# 37535), Df(1)ED6989 (BDSC# 9056), Df(1)BSC539 (BDSC# 25067), Df(1)BSC754 (BDSC# 26852), DGRP-59 (wild-type; BDSC# 28129), and FM7c, P{GAL4-Kr.C}DC1, P{UAS-GFP.S65T}DC5, sn+ (BDSC# 5193).
2.2. Complementation Analysis
Five mus109 heterozygous females—carrying either the mus109D1, mus109D2, or mus109lS chromosome over an X chromosome balancer marked with the dominant Bar eye phenotype—were crossed to five hemizygous mus109D1 or mus109D2 males per vial to establish Brood 1 (day 0). On day 3, the flies were flipped into new vials to establish Brood 2. On day 4, Brood 1 vials were mock treated with 250 µL water. On day 5, the adult flies were discarded from Brood 2 vials, and on day 6, Brood 2 vials were treated with 250 µL 0.05% methyl methanesulfonate (MMS; Sigma-Aldrich). Adult offspring were frozen on day 18 (Brood 1) or day 21 (Brood 2) and were subsequently scored for sex and eye phenotype. For each vial, relative survival was calculated as the ratio of mus109 mutant to non-mutant flies in Brood 2, normalized to the same ratio in the corresponding Brood 1 vial. Vials with fewer than 15 progeny in either Brood 1 or 2 were excluded from analysis, as in [29]. Statistical significance was determined by one-way ANOVA with Tukey’s correction for multiple comparisons. Statistical analysis and graphing were performed using GraphPad Prism 7.05.
2.3. Deletion Mapping
Four deletions covering the area predicted by Mason et al. [14] to contain mus109 were selected: Df(1)ED6991, Df(1)ED6989, Df(1)BSC539, and Df(1)BSC754. Five heterozygous females—carrying one of the deletions over an X chromosome balancer—were crossed to five mus109D2 males per vial to establish Brood 1 (day 0). The remainder of the MMS sensitivity assay proceeded as in the complementation analysis crosses.
2.4. DNA Sequencing
For the mus109D1 and mus109D2 alleles, DNA was extracted from single adult hemizygous males using the protocol described in [30]. For mus109lS, flies were balanced with the FM7c, P{GAL4-Kr.C}DC1, P{UAS-GFP.S65T}DC5, sn+ chromosome and homozygous third-instar larvae were identified by the absence of green fluorescence. DNA was then extracted from single homozygous third-instar larvae using the same protocol described in [30]. From these extracts, the CG2990 coding region was amplified, purified with a GeneJet Gel Extraction Kit (Thermo Scientific), and sequenced (Eurofins Genomics). The primers used in PCR and sequencing are shown in Table S1. Sequences were aligned to the FlyBase [31] CG2990 reference sequence and identified mutations were confirmed on a second DNA sample.
2.5. Protein Alignment
Clustal Omega [32] was used to conduct amino acid sequence alignment between DNA2 orthologs in wild-type D. melanogaster (NCBI Accession# NP_727386), Mus musculus (NCBI Accession# NP_796346.2), Homo sapiens (NCBI Accession# NP_001073918.2), and Caenorhabditis elegans (NCBI Accession# NP_496516.1). Alignment was visualized using Jalview version 2.11.1.4 [33]. Domains were mapped and analyzed according to the domain map devised by Zhou et al. [34] based on the structure of M. musculus DNA2.
3. Results and Discussion
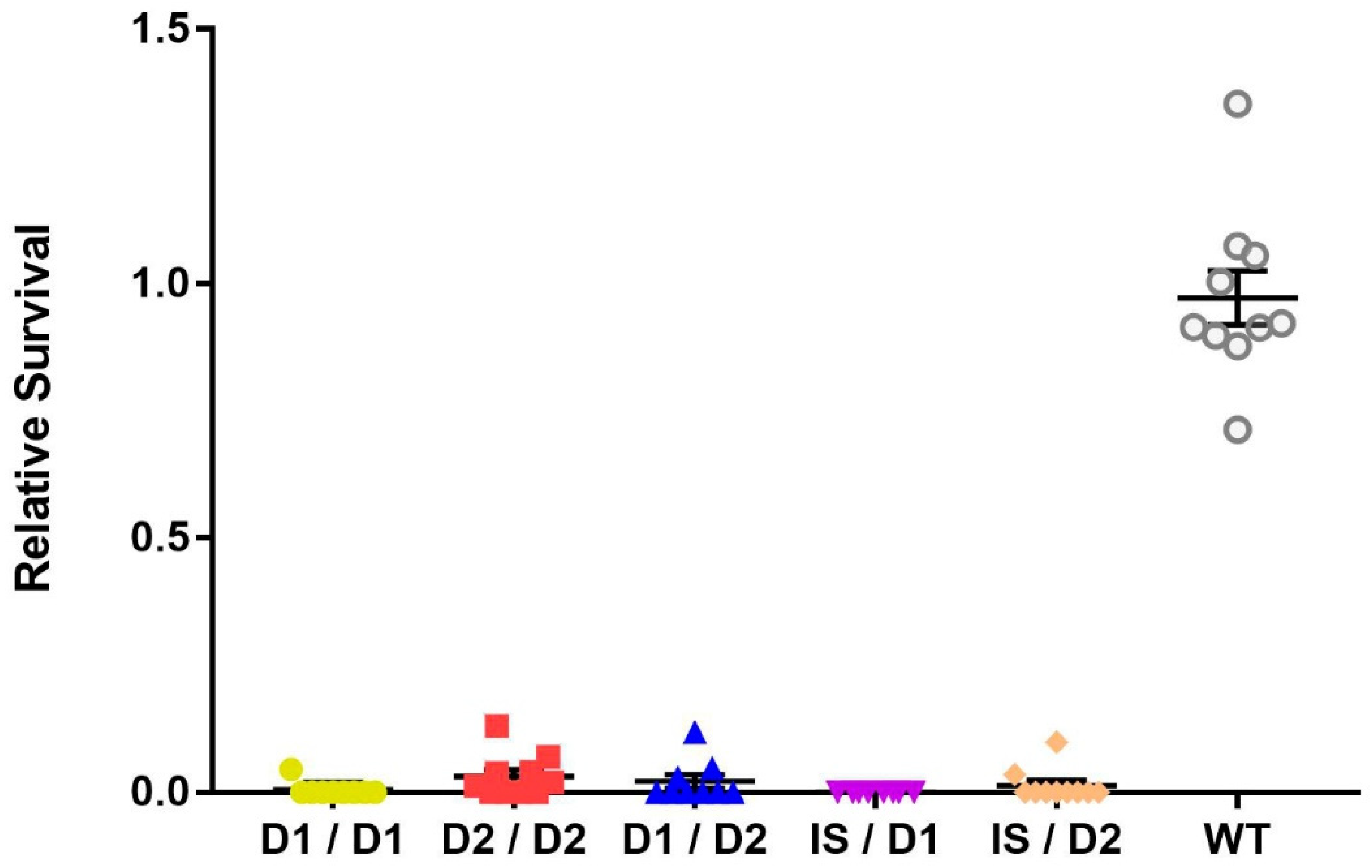
Since the three mus109 alleles—mus109D1, mus109D2, and mus109lS—were identified in the early 1980s [14,20,24], we first used complementation analysis to confirm that the fly stocks were still mutagen-sensitive. All possible mus109 allelic combinations showed sensitivity to MMS with significantly lower relative survival values compared to wild-type (one-way ANOVA, F(5,50) = 255.7, p < 0.0001; Figure 1). Although the relative survival values were not significantly different between the mus109 allele combinations (p = 0.221), the relative survival values were lower in genotypes containing mus109lS than in combinations without mus109lS (Figure S1), consistent with previous suggestions that mus109lS is amorphic [20] whereas mus109D1 and mus109D2 are hypomorphic [21].
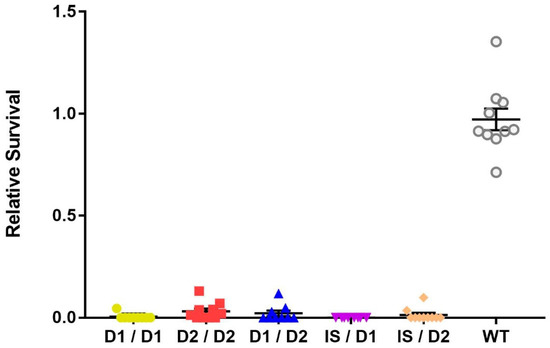
Figure 1.
Relative survival of flies exposed to 0.05% methyl methanesulfonate for the indicated mus109 allelic combinations and wild-type (WT). mus109lS/mus109lS could not be tested because the mus109lS allele is homozygous lethal. Each point represents one vial containing between 16 and 134 progeny (average = 55 progeny across all Brood 2 vials of all genotypes). The large horizontal line is the mean, while the upper and lower lines show the standard deviation.
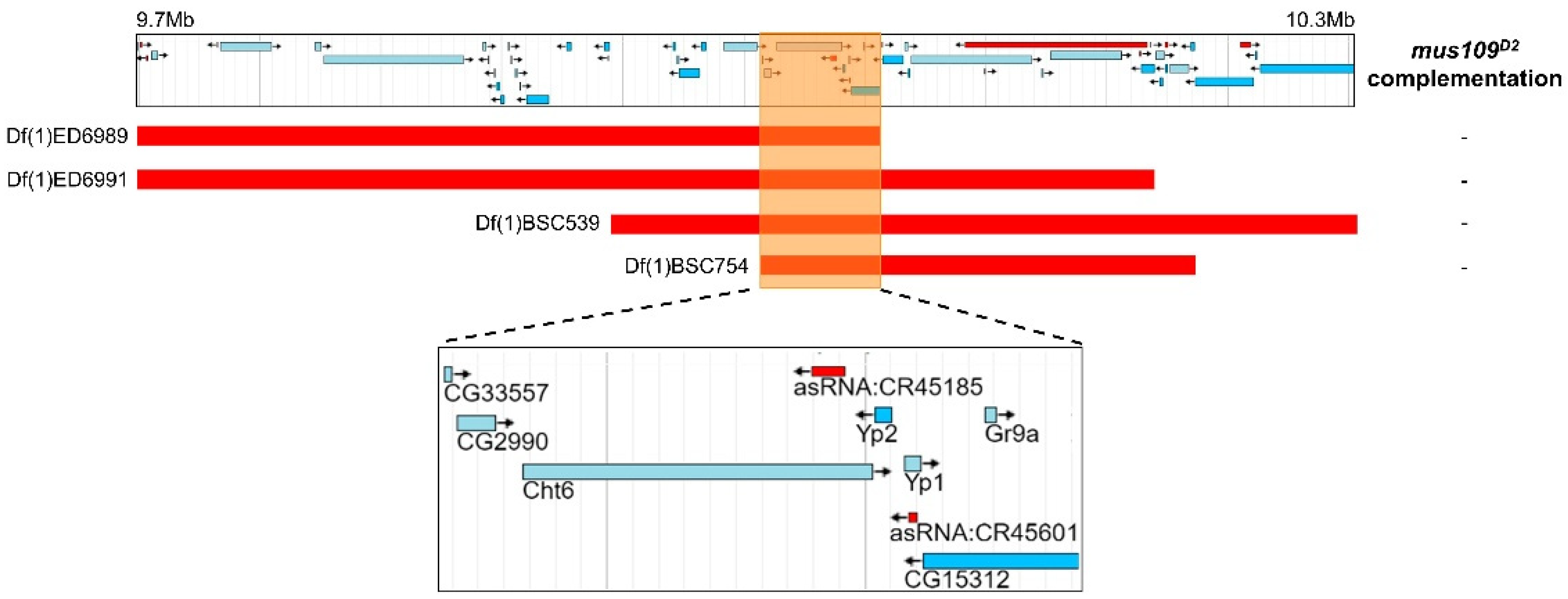
Next, deletion mapping was used to narrow the genomic location of mus109. Four deletions spanning the approximately 630 kb region predicted to contain mus109 [14] were each crossed to mus109D2 and assayed for sensitivity to MMS. With relative survival values of 0 in each case (Table S2), all four deletions failed to complement mus109D2. Thus, the location of mus109 was narrowed to the approximately 62kb region shared by all deletions (Figure 2). The FlyBase entries for the nine genes within this region were reviewed to identify genes involved in DNA metabolism (Table 1), a characteristic of all mapped mus genes. Notably, one of these genes, CG2990, is orthologous to the well-characterized DNA repair gene DNA2 [17]. Similar to Drosophila mus109, DNA2 is essential in yeast and mice [35,36], its downregulation causes genome instability in yeast and human cells [37,38], and yeast Dna2 mutants are sensitive to MMS [39]. Collectively, these observations suggested that CG2990 is an ideal mus109 candidate gene. To test our hypothesis that mus109 was CG2990, we sequenced the CG2990 coding region in wild-type flies and in each of the three mus109 alleles. In comparing these sequences, we identified mutations resulting in premature stop codons in all three mus109 alleles as well as eight missense mutations in mus109D1 (Figure 3A), all of which likely affect the functionality of the mus109 gene product.
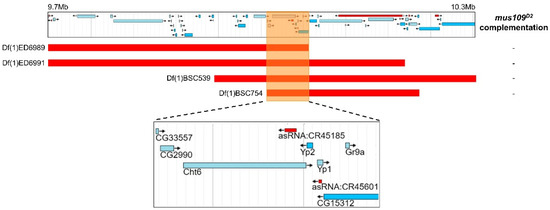
Figure 2.
Results of deletion mapping assay where four deletions were crossed to mus109D2. Each deletion is shown as a red bar aligned with its genomic location on the Drosophila melanogaster X chromosome in the jBrowse [40] screenshot above. The orange box highlights the region of overlap between the four deletions, and the jBrowse area within this box is enlarged in the inset below. This insert shows the nine genes contained in the overlapping region. “-” indicates non-complementation of a deletion with mus109D2.

Table 1.
Predicted function of genes within mus109D2 and the non-complementing region.
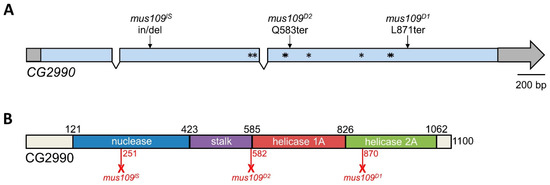
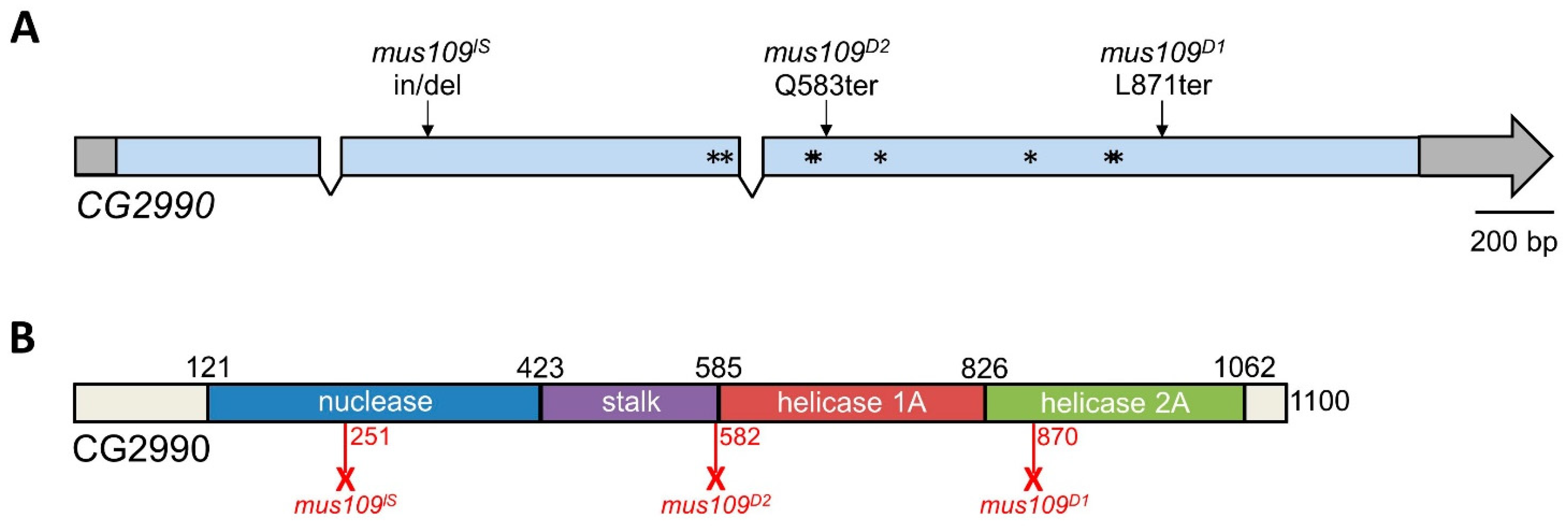
Figure 3.
CG2990 gene and protein structure. (A) CG2990 contains three exons and two introns. Untranslated regions are shown in gray. Asterisks represent the missense mutations found in mus109D1 mutants: E500D, L512F, S571G, E572Q, I633V, C755S, E825A, and K826E. mus109D1 and mus109D2 are truncated by nonsense mutations (L871ter and Q583ter, respectively), while mus109lS is truncated by a stop codon created by a 40 nucleotide deletion and four nucleotide (GAGG) insertion after amino acid 251. (B) CG2990 protein structure. The location of the nuclease, stalk, helicase 1A, and helicase 2A domains as identified in mouse DNA2 by Zhou et al. [34] are shown. Numbers above the bar denote the amino acid position of domain boundaries. Red numbers below the bar represent the predicted length of the CG2990 protein generated in each mus109 mutant. Full-length CG2990 is 1100 amino acids long.
The DNA2 protein is an essential and conserved nuclease-helicase with roles in several pathways that are crucial for maintaining genome integrity (reviewed in [41]). These pathways include long-track end resection during homologous recombination [42], Okazaki fragment processing [43], the recovery of stalled replication forks [44], and the maintenance of mitochondrial DNA [45]. Underscoring the importance of this protein, human DNA2 mutations have been implicated in mitochondrial myopathy [46], microcephalic primordial dwarfism [47], and some cancers [48]. DNA2 consists of a structure-specific nuclease and helicase/DNA-dependent ATPase connected by a β-barrel stalk [34]. While the nuclease activity is most critical to DNA2 repair functions [49,50,51], the helicase domains contribute to the narrow tunnel-like structure of DNA2 that allows single-stranded DNA access to the nuclease [34].
To explore the possible impact of the nonsense and missense mutations on mus109 mutant allele functionality, we mapped DNA2 domains onto CG2990 using the mouse DNA2 protein structure [34] (Figure 3B). Like mouse DNA2, CG2990 contains a structure-specific nuclease domain and a helicase/DNA-dependent ATPase domain connected by a β-barrel stalk sequence, as well as two helicase motifs (1A and 2A) [34]. The two helicase motifs are common to members of the Upf1 subfamily of helicases and contain an ATPase at their cleft [52]; however, helicase and ATPase activity are considered weak and non-essential to DNA2 nuclease function [34,49].
We further compared the CG2990 and human DNA2 protein sequences. The amino acid sequence alignment of CG2990 confirmed sequence homology to human DNA2 as well as with other model species (Figure 4). CG2990 contains the highly conserved DEXXQ-box helicase motif, as well as all known active site residues as defined in Zhou et al. [34]. Similarly, CG2990 contains most of the DNA contact site residues found in mouse DNA2 [34]. The insertion/deletion mutation in mus109lS creates a premature stop codon prior to the active site residues in the nuclease domain. This mutation likely abolishes nuclease function, which is known to be essential for viability in yeast [49]. If so, this could explain the homozygous lethal phenotype of mus109lS mutants. In contrast, the nonsense mutations in the mus109D1 and mus109D2 alleles occur after the conserved nuclease domain, which may allow for functional nuclease activity. While the nonsense mutation in mus109D1 occurs in the second helicase domain, the I663V mutation in the stalk domain changes a highly conserved amino acid (Figure 4), which may impact protein folding and/or helicase functionality.

Figure 4.
DNA2 amino acid sequence alignment. Residues conserved in all four species (Homo sapiens (Hsap), Mus musculus (Mmus), D. melanogaster (Dmel), and Caenorhabditis elegans (Cele)) are highlighted. Colored underlines represent the protein domains identified by Zhou et al. [34] in mouse DNA2. Conserved nuclease active sites are indicated by blue scissors, DNA contact site residues are indicated with orange arrows, and the DEXXQ-box helicase motif is indicated in bold text, all shown above the corresponding residues. Asterisks are located above the residues mutated in mus109D1 mutants: E500D, L512F, S571G, E572Q, I633V, C755S, E825A, and K826E.
Considering our deletion mapping data and our identification of deleterious mutations in CG2990, we conclude that mus109 is CG2990, the Drosophila ortholog of DNA2 [17]. This knowledge will be immediately useful to the DNA repair community, as there are no existing non-transgenic alleles of CG2990. With the identification of three (two hypomorphic and one amorphic) alleles of CG2990, future genetic studies on the functions of DmDNA2 in DNA repair can be conducted. For example, comparisons between the mus109D1 and mus109D2 alleles exposed to mutagens that impact DNA replication could be used to dissect the function of the DmDNA2 helicase 1A domain, which is present in mus109D1 but not mus109D2. Likewise, investigations of the mus109D1 allele may further uncover the importance of the DmDNA2 helicase 2A domain, as this domain is not predicted to contribute to the tunnel structure needed for the nucleolytic activity of DNA2. Both of these genetic studies would also benefit from complementary biochemical analyses of the truncated DmDNA2 proteins produced in mus109D1 and mus109D2 mutants. Further, because DNA2 has been shown to act as a tumor suppressor (reviewed in [41]), the nuclease domain mutant allele may serve as a model with which to study DNA2-deficient cancer processes. Future studies may also aim to investigate genetic interactions with DmDNA2 by creating flies mutant in both DNA2 and a critical gene in a redundant double-strand break repair pathway, such as tosca (Exo1). These and other experimental possibilities will greatly contribute to the growing body of work on DNA repair mechanisms and strengthen the use of Drosophila as a model for biomedical research.
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/article/10.3390/genes13020312/s1, Figure S1: Relative survival of flies exposed to 0.05% methyl methanesulfonate; Table S1: Primers used in this study. Table S2: Relative survival of deletions crossed to mus109D2.
Author Contributions
Conceptualization, E.B. and K.P.K.; methodology, E.B. and K.P.K.; investigation, C.M., V.B., J.D., E.N., E.B., and K.P.K.; writing—original draft preparation, E.B. and K.P.K.; writing—review and editing, C.M., V.B., J.D., E.N., E.B., and K.P.K.; supervision, E.B. and K.P.K.; funding acquisition, C.M., V.B., E.B., and K.P.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by grant P20GM103499 (SC INBRE) and grant T34-GM105549 (Maximizing Access to Research Careers (MARC)) from the National Institute of General Medical Sciences of the National Institutes of Health and through the Ciliate Genomics Consortium funded by the National Science Foundation IUSE grant DUE1431837. C.M. was supported in part by Winthrop University’s Ronald E. McNair Postbaccalaureate Achievement Program funded by the U.S. Department of Education TRiO grant P217A130111.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
The authors would also like to thank Eric Stoffregen and Christina Swanson for their comments, suggestions, and proofreading. Additionally, thank you to all of the Kohl Lab members for their help with experiments and in maintaining the fly stocks.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Adryan, B.; Russell, S. Genome Mapping and Genomics in Drosophila. In Genome Mapping and Genomics in Laboratory Animals; Denny, P., Kole, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 31–86. [Google Scholar]
- Sturtevant, A.H. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Zool. 1913, 14, 43–59. [Google Scholar] [CrossRef]
- Painter, T.S. A New Method for the Study of Chromosome Aberrations and the Plotting of Chromosome Maps in Drosophila Melanogaster. Genetics 1934, 19, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Bridges, C.B. Salivary Chromosome Maps: With a Key to the Banding of the Chromosomes of Drosophila Melanogaster. J. Hered. 1935, 26, 60–64. [Google Scholar] [CrossRef]
- Lindsley, D.L.; Sandler, L.; Baker, B.S.; Carpenter, A.T.; Denell, R.E.; Hall, J.C.; Jacobs, P.A.; Miklos, G.L.; Davis, B.K.; Gethmann, R.C.; et al. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics 1972, 71, 157–184. [Google Scholar] [CrossRef]
- Parks, A.L.; Cook, K.R.; Belvin, M.; Dompe, N.A.; Fawcett, R.; Huppert, K.; Tan, L.R.; Winter, C.G.; Bogart, K.P.; Deal, J.E.; et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 2004, 36, 288–292. [Google Scholar] [CrossRef]
- Ryder, E.; Ashburner, M.; Bautista-Llacer, R.; Drummond, J.; Webster, J.; Johnson, G.; Morley, T.; Chan, Y.S.; Blows, F.; Coulson, D.; et al. The DrosDel deletion collection: A Drosophila genomewide chromosomal deficiency resource. Genetics 2007, 177, 615–629. [Google Scholar] [CrossRef]
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F.; et al. The genome sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, R.A.; Carlson, J.W.; Wan, K.H.; Park, S.; Mendez, I.; Galle, S.E.; Booth, B.W.; Pfeiffer, B.D.; George, R.A.; Svirskas, R.; et al. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 2015, 25, 445–458. [Google Scholar] [CrossRef]
- Smith, P.D. Mutagen sensitivity of Drosophila melanogaster. I. Isolation and preliminary characterization of a methyl methanesulphonate-sensitive strain. Mutat. Res. 1973, 20, 215–220. [Google Scholar] [CrossRef]
- Boyd, J.B.; Golino, M.D.; Nguyen, T.D.; Green, M.M. Isolation and characterization of X-linked mutants of Drosophila melanogaster which are sensitive to mutagens. Genetics 1976, 84, 485–506. [Google Scholar] [CrossRef]
- Boyd, J.B.; Golino, M.D.; Shaw, K.E.; Osgood, C.J.; Green, M.M. Third-chromosome mutagen-sensitive mutants of Drosophila melanogaster. Genetics 1981, 97, 607–623. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.D.; Green, M.M.; Boyd, J.B. Isolation of two X-linked mutants in Drosophila melanogaster which are sensitive to γ-rays. Mutat. Res. 1978, 49, 139–143. [Google Scholar] [CrossRef]
- Mason, J.M.; Green, M.M.; Shaw, K.E.; Boyd, J.B. Genetic analysis of X-linked mutagen-sensitive mutants of Drosophila melanogaster. Mutat. Res. 1981, 81, 329–343. [Google Scholar] [CrossRef]
- Laurencon, A.; Orme, C.M.; Peters, H.K.; Boulton, C.L.; Vladar, E.K.; Langley, S.A.; Bakis, E.P.; Harris, D.T.; Harris, N.J.; Wayson, S.M.; et al. A large-scale screen for mutagen-sensitive loci in Drosophila. Genetics 2004, 167, 217–231. [Google Scholar] [CrossRef]
- Carvajal-Garcia, J.; Gales, E.R.; Ramsden, D.A.; Sekelsky, J. The Drosophila melanogaster Ortholog of RFWD3 Functions Independently of RAD51 During DNA Repair. G3 Genes Genomes Genet. 2020, 10, 999–1004. [Google Scholar] [CrossRef]
- Sekelsky, J. DNA Repair in Drosophila: Mutagens, Models, and Missing Genes. Genetics 2017, 205, 471–490. [Google Scholar] [CrossRef]
- Kusano, K.; Johnson-Schlitz, D.M.; Engels, W.R. Sterility of Drosophila with mutations in the Bloom syndrome gene--complementation by Ku70. Science 2001, 291, 2600–2602. [Google Scholar] [CrossRef]
- Sekelsky, J.J.; McKim, K.S.; Chin, G.M.; Hawley, R.S. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 1995, 141, 619–627. [Google Scholar] [CrossRef]
- Baker, B.S.; Smith, D.A.; Gatti, M. Region-specific effects on chromosome integrity of mutations at essential loci in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1982, 79, 1205–1209. [Google Scholar] [CrossRef]
- Baker, B.S.; Smith, D.A. The effects of mutagen-sensitive mutants of Drosophila melanogaster in nonmutagenized cells. Genetics 1979, 92, 833–847. [Google Scholar] [CrossRef]
- Gatti, M. Genetic control of chromosome breakage and rejoining in Drosophila melanogaster: Spontaneous chromosome aberrations in X-linked mutants defective in DNA metabolism. Proc. Natl. Acad. Sci. USA 1979, 76, 1377–1381. [Google Scholar] [CrossRef] [PubMed]
- Gatti, M.; Baker, B.S. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 1989, 3, 438–453. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.D. Mutagen sensitivity of Drosophila melanogaster. III. X-linked loci governing sensitivity to methyl methanesulfonate. Mol. Gen. Genet. 1976, 149, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.D.; Boyd, J.B.; Green, M.M. Sensitivity of drosophila mutants to chemical carcinogens. Mutat. Res. 1979, 63, 67–77. [Google Scholar] [CrossRef]
- Beranek, D.T. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 1990, 231, 11–30. [Google Scholar] [CrossRef]
- Ikenaga, M.; Ichikawa-Ryo, H.; Kondo, S. The major cause of inactivation and mutation by 4-nitroquinoline 1-oixde in Escherichia coli: Excisable 4NQO-purine adducts. J. Mol. Biol. 1975, 92, 341–356. [Google Scholar] [CrossRef]
- Azzam, E.I.; Jay-Gerin, J.P.; Pain, D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012, 327, 48–60. [Google Scholar] [CrossRef]
- Romero, N.E.; Matson, S.W.; Sekelsky, J. Biochemical Activities and Genetic Functions of the Drosophila melanogaster Fancm Helicase in DNA Repair. Genetics 2016, 204, 531–541. [Google Scholar] [CrossRef][Green Version]
- Gloor, G.B.; Preston, C.R.; Johnson-Schlitz, D.M.; Nassif, N.A.; Phillis, R.W.; Benz, W.K.; Robertson, H.M.; Engels, W.R. Type I repressors of P element mobility. Genetics 1993, 135, 81–95. [Google Scholar] [CrossRef]
- Larkin, A.; Marygold, S.J.; Antonazzo, G.; Attrill, H.; Dos Santos, G.; Garapati, P.V.; Goodman, J.L.; Gramates, L.S.; Millburn, G.; Strelets, V.B.; et al. FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021, 49, D899–D907. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Pourmal, S.; Pavletich, N.P. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Budd, M.E.; Campbell, J.L. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. USA 1995, 92, 7642–7646. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Sampathi, S.; Dai, H.; Liu, C.; Zhou, M.; Hu, J.; Huang, Q.; Campbell, J.; Shin-Ya, K.; Zheng, L.; et al. Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J. 2013, 32, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Cheng, E.; Vaisica, J.A.; Ou, J.; Baryshnikova, A.; Lu, Y.; Roth, F.P.; Brown, G.W. Genome rearrangements caused by depletion of essential DNA replication proteins in Saccharomyces cerevisiae. Genetics 2012, 192, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Duxin, J.P.; Dao, B.; Martinsson, P.; Rajala, N.; Guittat, L.; Campbell, J.L.; Spelbrink, J.N.; Stewart, S.A. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell Biol. 2009, 29, 4274–4282. [Google Scholar] [CrossRef] [PubMed]
- Formosa, T.; Nittis, T. Dna2 mutants reveal interactions with DNA polymerase α and Ctf4, a Pol α accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics 1999, 151, 1459–1470. [Google Scholar] [CrossRef]
- Buels, R.; Yao, E.; Diesh, C.M.; Hayes, R.D.; Munoz-Torres, M.; Helt, G.; Goodstein, D.M.; Elsik, C.G.; Lewis, S.E.; Stein, L.; et al. JBrowse: A dynamic web platform for genome visualization and analysis. Genome Biol. 2016, 17, 66. [Google Scholar] [CrossRef]
- Zheng, L.; Meng, Y.; Campbell, J.L.; Shen, B. Multiple roles of DNA2 nuclease/helicase in DNA metabolism, genome stability and human diseases. Nucleic Acids Res. 2020, 48, 16–35. [Google Scholar] [CrossRef]
- Zhu, Z.; Chung, W.H.; Shim, E.Y.; Lee, S.E.; Ira, G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 2008, 134, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Levikova, M.; Cejka, P. The Saccharomyces cerevisiae Dna2 can function as a sole nuclease in the processing of Okazaki fragments in DNA replication. Nucleic Acids Res. 2015, 43, 7888–7897. [Google Scholar] [CrossRef] [PubMed]
- Thangavel, S.; Berti, M.; Levikova, M.; Pinto, C.; Gomathinayagam, S.; Vujanovic, M.; Zellweger, R.; Moore, H.; Lee, E.H.; Hendrickson, E.A.; et al. DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 2015, 208, 545–562. [Google Scholar] [CrossRef]
- Zheng, L.; Zhou, M.; Guo, Z.; Lu, H.; Qian, L.; Dai, H.; Qiu, J.; Yakubovskaya, E.; Bogenhagen, D.F.; Demple, B.; et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell 2008, 32, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Ronchi, D.; Di Fonzo, A.; Lin, W.; Bordoni, A.; Liu, C.; Fassone, E.; Pagliarani, S.; Rizzuti, M.; Zheng, L.; Filosto, M.; et al. Mutations in DNA2 link progressive myopathy to mitochondrial DNA instability. Am. J. Hum. Genet. 2013, 92, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Tarnauskaite, Z.; Bicknell, L.S.; Marsh, J.A.; Murray, J.E.; Parry, D.A.; Logan, C.V.; Bober, M.B.; de Silva, D.C.; Duker, A.L.; Sillence, D.; et al. Biallelic variants in DNA2 cause microcephalic primordial dwarfism. Hum. Mutat. 2019, 40, 1063–1070. [Google Scholar] [CrossRef]
- Strauss, C.; Kornowski, M.; Benvenisty, A.; Shahar, A.; Masury, H.; Ben-Porath, I.; Ravid, T.; Arbel-Eden, A.; Goldberg, M. The DNA2 nuclease/helicase is an estrogen-dependent gene mutated in breast and ovarian cancers. Oncotarget 2014, 5, 9396–9409. [Google Scholar] [CrossRef]
- Budd, M.E.; Choe, W.; Campbell, J.L. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 2000, 275, 16518–16529. [Google Scholar] [CrossRef]
- Lee, K.H.; Kim, D.W.; Bae, S.H.; Kim, J.A.; Ryu, G.H.; Kwon, Y.N.; Kim, K.A.; Koo, H.S.; Seo, Y.S. The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res. 2000, 28, 2873–2881. [Google Scholar] [CrossRef]
- Pawlowska, E.; Szczepanska, J.; Blasiak, J. DNA2-An Important Player in DNA Damage Response or Just Another DNA Maintenance Protein? Int. J. Mol. Sci. 2017, 18, 1562. [Google Scholar] [CrossRef]
- Fairman-Williams, M.E.; Guenther, U.P.; Jankowsky, E. SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 2010, 20, 313–324. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).