Driving Chromatin Organisation through N6-methyladenosine Modification of RNA: What Do We Know and What Lies Ahead?
Abstract
:1. RNA Modifications
1.1. N6-methyladenosine in Mammals
1.2. m6A and Epigenomic Regulation
2. Transposable Elements, RNA Modifications, and Chromatin Organisation
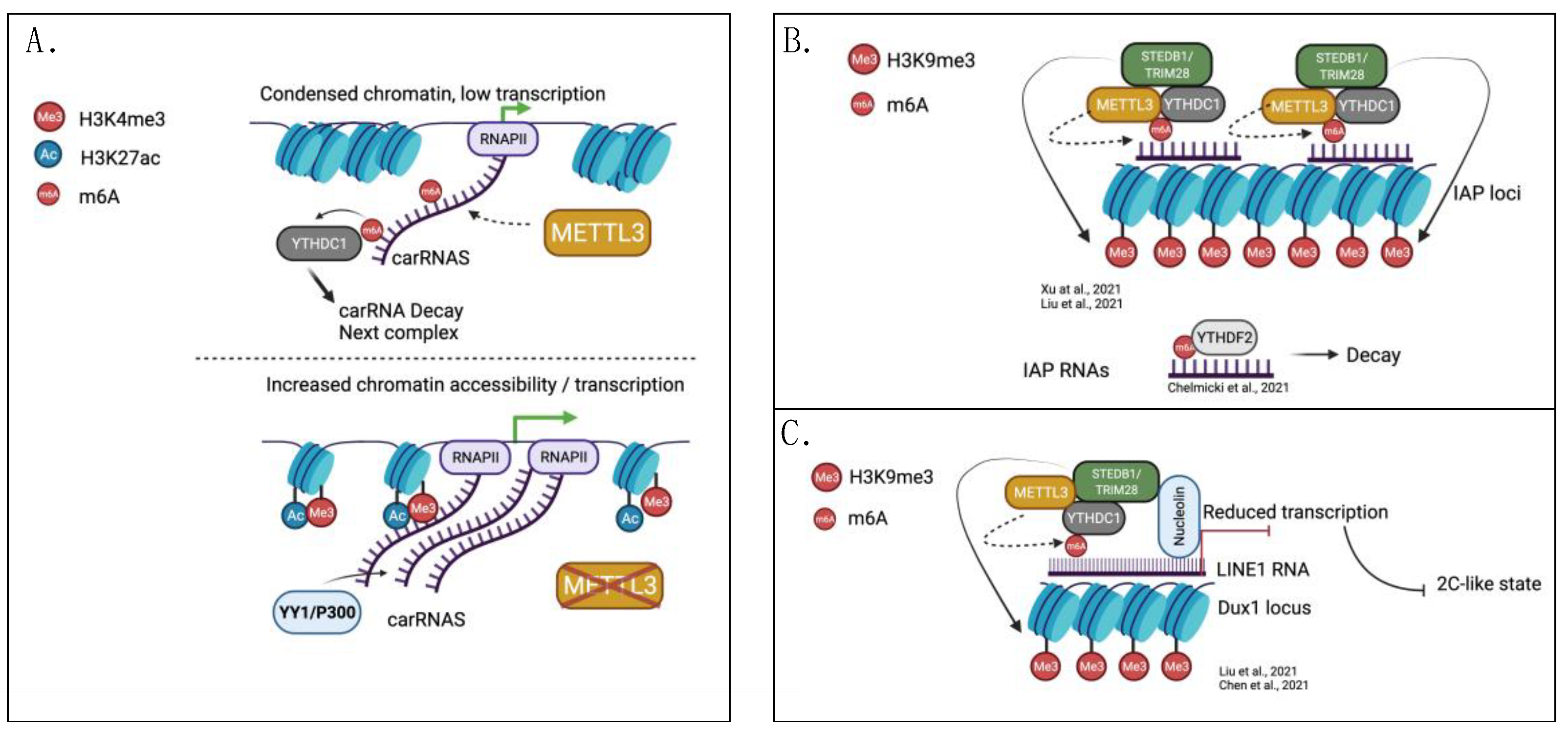
2.1. IAP and LINE Regulation by m6A and Histone Modifications in Mouse Embryonic Stem Cells
2.2. MSR Repeats Turnover and RNA:DNA Hybrid Formation by m6A
2.3. Summary of: Transposable Elements, RNA Modifications, and Chromatin Organisation
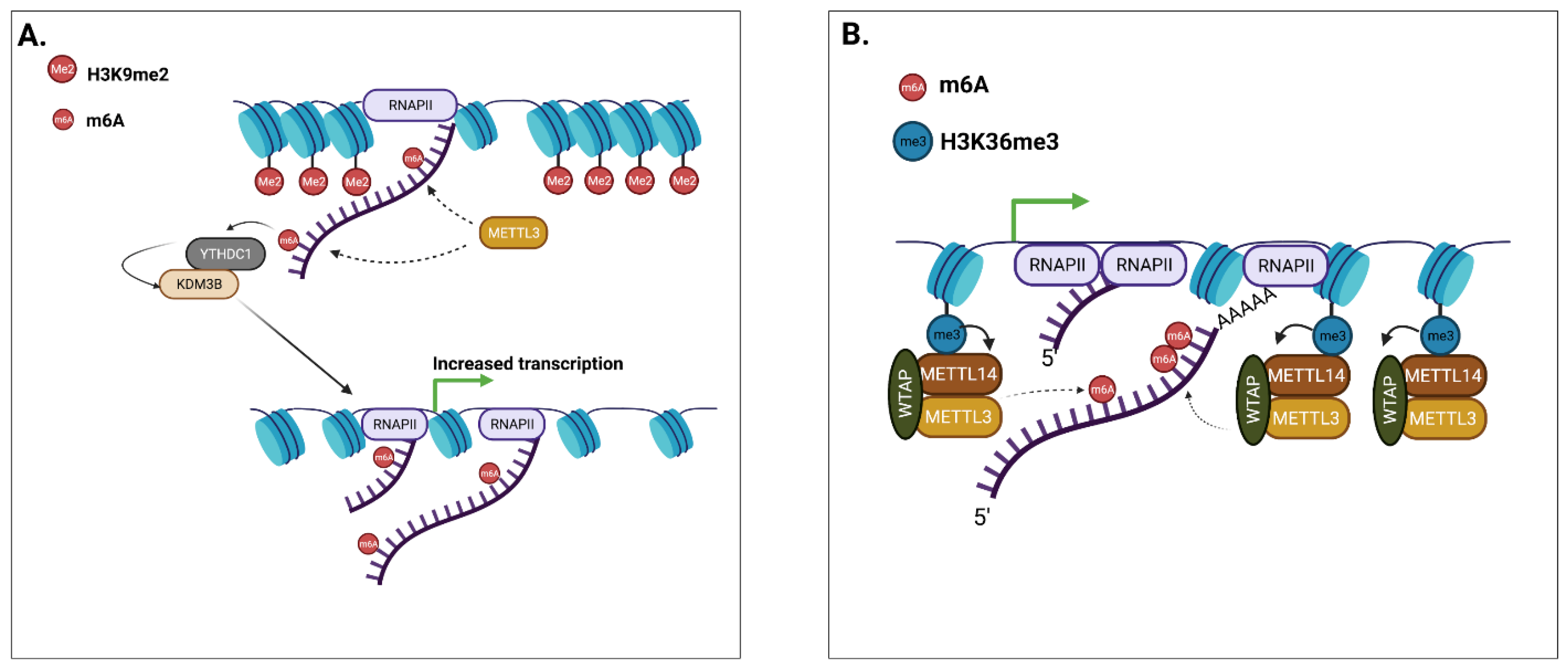
3. Chromatin Accessibility, Transcriptional Regulation, and m6A
Summary of: Chromatin Accessibility, Transcriptional Regulation, and m6A
4. m6A and Liquid–Liquid Phase Separation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piątkowski, P.; Baginski, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [Green Version]
- Wiener, D.; Schwartz, S. The epitranscriptome beyond m6A. Nat. Rev. Genet. 2021, 22, 119–131. [Google Scholar] [CrossRef]
- Khoddami, V.; Yerra, A.; Mosbruger, T.L.; Fleming, A.M.; Burrows, C.J.; Cairns, B.R. Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc. Natl. Acad. Sci. USA 2019, 116, 6784–6789. [Google Scholar] [CrossRef] [Green Version]
- Jonkhout, N.; Tran, J.; Smith, M.; Schonrock, N.; Mattick, J.; Novoa, E.M. The RNA modification landscape in human disease. RNA 2017, 23, 1754–1769. [Google Scholar] [CrossRef] [Green Version]
- Barbieri, I.; Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef]
- Perry, R.P.; Kelley, D.E.; Friderici, K.; Rottman, F. The methylated constituents of L cell messenger RNA: Evidence for an unusual cluster at the 5′ terminus. Cell 1975, 4, 387–394. [Google Scholar] [CrossRef]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [Green Version]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ utrs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [Green Version]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Slobodin, B.; Han, R.; Calderone, V.; Vrielink, J.A.F.O.; Loayza-Puch, F.; Elkon, R.; Agami, R. Transcription impacts the effi-ciency of MRNA translation via co-transcriptional N6-adenosine methylation. Cell 2017, 169, 326–337.e12. [Google Scholar] [CrossRef] [Green Version]
- Knuckles, P.; Carl, S.H.; Musheev, M.; Niehrs, C.; Wenger, A.; Bühler, M. RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat. Struct. Mol. Biol. 2017, 24, 561–569. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Hsu, P.; Xing, X.; Fang, J.; Lu, Z.; Zou, Q.; Zhang, K.-J.; Zhang, X.; Zhou, Y.; Zhang, T.; et al. Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis. Cell Res. 2017, 27, 1216–1230. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.; Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef] [Green Version]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4. [Google Scholar] [CrossRef] [Green Version]
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villaseñor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the MRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018, 32, 415–429. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 2018, 69, 1028–1038.e6. [Google Scholar] [CrossRef] [Green Version]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. M6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [Green Version]
- Patil, D.P.; Chen, C.-K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. M6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Yang, D.; Qiao, J.; Wang, G.; Lan, Y.; Li, G.; Guo, X.; Xi, J.; Ye, D.; Zhu, S.; Chen, W.; et al. N6-methyladenosine modification of LincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018, 46, 3906–3920. [Google Scholar] [CrossRef] [Green Version]
- Warda, A.S.; Kretschmer, J.; Hackert, P.; Lenz, C.; Urlaub, H.; Höbartner, C.; Sloan, K.E.; Bohnsack, M.T. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-MRNAs and various non-coding RNAs. EMBO Rep. 2017, 18, 2004–2014. [Google Scholar] [CrossRef]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m 6 A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 2017, 169, 824–835.e14. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.A.; Kinzig, C.G.; DeGregorio, S.J.; Steitz, J.A. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl. Acad. Sci. USA 2016, 113, 14013–14018. [Google Scholar] [CrossRef] [Green Version]
- Ruszkowska, A. METTL16, methyltransferase-like protein 16: Current insights into structure and function. Int. J. Mol. Sci. 2021, 22, 2176. [Google Scholar] [CrossRef]
- Mendel, M.; Chen, K.-M.; Homolka, D.; Gos, P.; Pandey, R.R.; McCarthy, A.A.; Pillai, R.S. Methylation of structured RNA by the m6A writer METTL16 is essential for mouse embryonic development. Mol. Cell 2018, 71, 986–1000.e11. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Wang, X.; Liu, K.; Roundtree, I.A.; Tempel, W.; Li, Y.; Lu, Z.; He, C.; Min, J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014, 10, 927–929. [Google Scholar] [CrossRef]
- Zhang, Z.; Theler, D.; Kaminska, K.H.; Hiller, M.; de la Grange, P.; Pudimat, R.; Rafalska, I.; Heinrich, B.; Bujnicki, J.M.; Allain, F.H.-T.; et al. The YTH domain is a novel RNA binding domain. J. Biol. Chem. 2010, 285, 14701–14710. [Google Scholar] [CrossRef] [Green Version]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.-S.; Hao, Y.-J.; Sun, B.-F.; Sun, H.-Y.; Li, A.; Ping, X.-L.; Lai, W.-Y.; et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [Green Version]
- Roundtree, I.A.; Luo, G.-Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Wojtas, M.N.; Pandey, R.R.; Mendel, M.; Homolka, D.; Sachidanandam, R.; Pillai, R.S. Regulation of m6A transcripts by the 3′→5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell 2017, 68, 374–387.e12. [Google Scholar] [CrossRef] [Green Version]
- Kasowitz, S.; Ma, J.; Anderson, S.J.; Leu, N.A.; Xu, Y.; Gregory, B.D.; Schultz, R.M.; Wang, P.J. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018, 14, e1007412. [Google Scholar] [CrossRef]
- Hsu, P.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef] [Green Version]
- Lasman, L.; Krupalnik, V.; Viukov, S.; Mor, N.; Aguilera-Castrejon, A.; Schneir, D.; Bayerl, J.; Mizrahi, O.; Peles, S.; Tawil, S.; et al. Context-dependent functional compensation between Ythdf m6A reader proteins. Genes Dev. 2020, 34, 1373–1391. [Google Scholar] [CrossRef]
- Zaccara, S.; Jaffrey, S.R. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 2020, 181, 1582–1595.e18. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, K.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N 6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2020, 20, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Sanchez de Groot, N.; Armaos, A.; Graña-Montes, R.; Alriquet, M.; Calloni, G.; Vabulas, R.M.; Tartaglia, G.G. RNA structure drives interaction with proteins. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Roost, C.; Lynch, S.R.; Batista, P.J.; Qu, K.; Chang, H.Y.; Kool, E.T. Structure and thermodynamics of N6-methyladenosine in RNA: A spring-loaded base modification. J. Am. Chem. Soc. 2015, 137, 2107–2115. [Google Scholar] [CrossRef] [Green Version]
- Edupuganti, R.R.; Geiger, S.; Lindeboom, R.G.H.; Shi, H.; Hsu, P.J.; Lu, Z.; Wang, S.-Y.; Baltissen, M.P.A.; Jansen, P.W.T.C.; Rossa, M.; et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 2017, 24, 870–878. [Google Scholar] [CrossRef]
- Fedeles, B.I.; Singh, V.; Delaney, J.C.; Li, D.; Essigmann, J.M. The AlkB family of Fe(II)/α-Ketoglutarate-dependent dioxygenases: Repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 2015, 290, 20734–20742. [Google Scholar] [CrossRef] [Green Version]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Samanta, D.; Lu, H.; Bullen, J.W.; Zhang, H.; Chen, I.; He, X.; Semenza, G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E2047–E2056. [Google Scholar] [CrossRef] [Green Version]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Mauer, J.; Sindelar, M.; Despic, V.; Guez, T.; Hawley, B.R.; Vasseur, J.-J.; Rentmeister, A.; Gross, S.S.; Pellizzoni, L.; Debart, F.; et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 2019, 15, 340–347. [Google Scholar] [CrossRef]
- Bartosovic, M.; Molares, H.C.; Gregorova, P.; Hrossova, D.; Kudla, G.; Vanacova, S. N6-methyladenosine demethylase FTO targets pre-MRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 2017, 45, 11356–11370. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommer, S.; Lavi, U.; Darnell, J.E. The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol. 1978, 124, 487–499. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. M6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zhao, B.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5′ UTR m(6)A promotes cap-independent translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Yang, Y.; Sun, B.-F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.-J.; Ping, X.-L.; Chen, Y.-S.; Wang, W.-J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef]
- Ivanova, I.; Much, C.; di Giacomo, M.; Azzi, C.; Morgan, M.; Moreira, P.N.; Monahan, J.; Carrieri, C.; Enright, A.J.; O’Carroll, D. The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell 2017, 67, 1059–1067.e4. [Google Scholar] [CrossRef] [Green Version]
- Edens, B.M.; Vissers, C.; Su, J.; Arumugam, S.; Xu, Z.; Shi, H.; Miller, N.; Ringeling, F.R.; Ming, G.-L.; He, C.; et al. FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep. 2019, 28, 845–854.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, Y.; Yue, M.; Wang, J.; Kumar, S.; Wechsler-Reya, R.J.; Zhang, Z.; Ogawa, Y.; Kellis, M.; Duester, G.; et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 2018, 21, 195–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Zhao, X.; Wang, W.; Shi, H.; Pan, Q.; Lu, Z.; Perez, S.P.; Suganthan, R.; He, C.; Bjørås, M.; et al. Ythdf2-mediated m6A mRNA clearance modulates neural development in mice. Genome Biol. 2018, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fustin, J.-M.; Doi, M.; Yamaguchi, Y.; Hida, H.; Nishimura, S.; Yoshida, M.; Isagawa, T.; Morioka, M.S.; Kakeya, H.; Manabe, I.; et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013, 155, 793–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, K.-W.; Zealy, R.W.; Davila, S.; Fomin, M.; Cummings, J.C.; Makowsky, D.; Mcdowell, C.H.; Thigpen, H.; Hafner, M.; Kwon, S.-H.; et al. Profiling of m6A RNA modifications identified an age-associated regulation of AGO2 mRNA stability. Aging Cell 2018, 17, e12753. [Google Scholar] [CrossRef]
- Liu, J.; Eckert, M.A.; Harada, B.T.; Liu, S.-M.; Lu, Z.; Yu, K.; Tienda, S.M.; Chryplewicz, A.; Zhu, A.C.; Yang, Y.; et al. M6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018, 20, 1074–1083. [Google Scholar] [CrossRef]
- Barbieri, I.; Tzelepis, K.; Pandolfini, L.; Namshik, H.; Millan-Zambrano, G.; Robson, S.C.; Aspris, D.; Migliori, V.; Bannister, A.J.; Hannes, P.; et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017, 552, 126–131. [Google Scholar] [CrossRef]
- Lin, X.; Chai, G.; Wu, Y.; Li, J.; Chen, F.; Liu, J.; Luo, G.; Tauler, J.; Du, J.; Lin, S.; et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of snail. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Wei, J.; Cui, Y.-H.; Park, G.; Shah, P.; Deng, Y.; Aplin, A.E.; Lu, Z.; Hwang, S.; He, C.; et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019, 10, 2782. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Chen, J. M6A modification in coding and non-coding RNAs: Roles and therapeutic implications in cancer. Cancer Cell 2020, 37, 270–288. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; Mackay, M.; et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Liu, J.; Chen, C.; Dong, L.; Liu, Y.; Chang, R.; Huang, X.; Liu, Y.; Wang, J.; Dougherty, U.; et al. Anti-tumour immunity controlled through MRNA m6A methylation and YTHDF1 in dendritic cells. Nature 2019, 566, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Song, J.; Cheng, M.; Zheng, M.; Wang, T.; Tian, S.; Flavell, R.A.; Zhu, S.; Li, H.-B.; Ding, C.; et al. METTL3-mediated m6A RNA methylation promotes the anti-tumour immunity of natural killer cells. Nat. Commun. 2021, 12, 5522. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M. The regulation of RNA modification systems: The next frontier in epitranscriptomics? Genes 2021, 12, 345. [Google Scholar] [CrossRef]
- Porman, A.M.; Roberts, J.T.; Duncan, E.D.; Chrupcala, M.L.; Levine, A.A.; Kennedy, M.A.; Williams, M.M.; Richer, J.K.; Johnson, A.M. A single N6-methyladenosine site in LncRNA HOTAIR regulates its function in breast cancer cells. bioRxiv 2021. Available online: https://www.biorxiv.org/content/10.1101/2020.06.08.140954v3 (accessed on 29 December 2021). [CrossRef]
- Kan, R.L.; Chen, J.; Sallam, T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 2021, 38, 182–193. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, Y.; Jin, M.; Wang, J. The crosstalk between m6A RNA methylation and other epigenetic regulators: A novel perspective in epigenetic remodeling. Theranostics 2021, 11, 4549–4566. [Google Scholar] [CrossRef]
- Liu, J.; Dou, X.; Chen, C.; Chen, C.; Liu, C.; Xu, M.M.; Zhao, S.; Shen, B.; Gao, Y.; Han, D.; et al. N 6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 2020, 367, 580–586. [Google Scholar] [CrossRef]
- Liu, J.; Gao, M.; He, J.; Wu, K.; Lin, S.; Jin, L.; Chen, Y.; Liu, H.; Shi, J.; Wang, X.; et al. The RNA m6A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature 2021, 591, 322–326. [Google Scholar] [CrossRef]
- Chen, C.; Liu, W.; Guo, J.; Liu, Y.; Liu, X.; Liu, J.; Dou, X.; Le, R.; Huang, Y.; Li, C.; et al. Nuclear m6A reader YTHDC1 regulates the scaffold function of LINE1 RNA in mouse ESCs and early embryos. Protein Cell 2021, 12, 455–474. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Li, J.; He, C.; Wen, J.; Ma, H.; Rong, B.; Diao, J.; Wang, L.; Wang, J.; Wu, F.; et al. METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature 2021, 591, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Chelmicki, T.; Roger, E.; Teissandier, A.; Dura, M.; Bonneville, L.; Rucli, S.; Dossin, F.; Fouassier, C.; Lameiras, S.; Bourc’His, D. M6A RNA methylation regulates the fate of endogenous retroviruses. Nature 2021, 591, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Duda, K.J.; Ching, R.W.; Jerabek, L.; Shukeir, N.; Erikson, G.; Engist, B.; Onishi-Seebacher, M.; Perrera, V.; Richter, F.; Mittler, G.; et al. M6A RNA methylation of major satellite repeat transcripts facilitates chromatin association and RNA:DNA hybrid formation in mouse heterochromatin. Nucleic Acids Res. 2021, 49, 5568–5587. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xia, L.; Tan, K.; Ye, X.; Zuo, Z.; Li, M.; Xiao, R.; Wang, Z.; Liu, X.; Deng, M.; et al. N6-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat. Genet. 2020, 52, 870–877. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Zhou, K.; Wu, T.; Zhao, B.S.; Sun, M.; Chen, Z.; Deng, X.; Xiao, G.; Auer, F.; et al. Histone H3 tri-methylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 2019, 567, 414–419. [Google Scholar] [CrossRef]
- Wu, C.; Chen, W.; He, J.; Jin, S.; Liu, Y.; Yi, Y.; Gao, Z.; Yang, J.; Yang, J.; Cui, J.; et al. Interplay of m6A and H3K27 trimethylation restrains inflammation during bacterial infection. Sci. Adv. 2020, 6, eaba0647. [Google Scholar] [CrossRef]
- Padeken, J.; Zeller, P.; Gasser, S.M. Repeat DNA in genome organization and stability. Curr. Opin. Genet. Dev. 2015, 31, 12–19. [Google Scholar] [CrossRef]
- Sassaman, D.M.; Dombroski, B.A.; Moran, J.V.; Kimberland, M.L.; Naas, T.P.; DeBerardinis, R.J.; Gabriel, A.; Swergold, G.D.; Kazazian, H.H., Jr. Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997, 16, 37–43. [Google Scholar] [CrossRef]
- Platt, R.N.; Vandewege, M.W.; Ray, D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosom. Res. 2018, 26, 25–43. [Google Scholar] [CrossRef] [Green Version]
- Diehl, A.G.; Ouyang, N.; Boyle, A.P. Transposable elements contribute to cell and species-specific chromatin looping and gene regulation in mammalian genomes. Nat. Commun. 2020, 11, 1796. [Google Scholar] [CrossRef] [Green Version]
- Horváth, V.; Merenciano, M.; González, J. Revisiting the relationship between transposable elements and the eukaryotic stress response. Trends Genet. 2017, 33, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Phalke, S.; Nickel, O.; Walluscheck, D.; Hortig, F.; Onorati, M.C.; Reuter, G. Retrotransposon silencing and telomere integrity in somatic cells of drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat. Genet. 2009, 41, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Genenncher, B.; Durdevic, Z.; Hanna, K.; Zinkl, D.; Mobin, M.B.; Senturk, N.; da Silva, B.; Legrand, C.; Carré, C.; Lyko, F.; et al. Mutations in cytosine-5 tRNA methyltransferases impact mobile element expression and genome stability at specific DNA repeats. Cell Rep. 2018, 22, 1861–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jachowicz, J.; Bing, X.; Pontabry, J.; Bošković, A.; Rando, O.J.; Torres-Padilla, M.-E. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 2017, 49, 1502–1510. [Google Scholar] [CrossRef]
- Percharde, M.; Lin, C.J.; Yin, Y.; Guan, J.; Peixoto, G.A.; Bulut-Karslioglu, A.; Biechele, S.; Huang, B.; Shen, X.; Ramalho-Santos, M. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 2018, 174, 391–405.e19. [Google Scholar] [CrossRef] [Green Version]
- Walter, M.; Teissandier, A.; Pérez-Palacios, R.; Bourc’his, D. An epigenetic switch ensures transposon repression upon dy-namic loss of DNA methylation in embryonic stem cells. eLife 2016, 5, e11418. [Google Scholar] [CrossRef]
- Hutnick, L.K.; Huang, X.; Loo, T.-C.; Ma, Z.; Fan, G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J. Biol. Chem. 2010, 285, 21082–21091. [Google Scholar] [CrossRef] [Green Version]
- Rowe, H.; Jakobsson, J.; Mesnard, D.; Rougemont, J.; Reynard, S.; Aktas, T.; Maillard, P.V.; Layard-Liesching, H.; Verp, S.; Marquis, J.; et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010, 463, 237–240. [Google Scholar] [CrossRef]
- Janssen, A.; Colmenares, S.U.; Karpen, G.H. Heterochromatin: Guardian of the genome. Annu. Rev. Cell Dev. Biol. 2018, 34, 265–288. [Google Scholar] [CrossRef] [Green Version]
- Johnson, W.L.; Yewdell, W.T.; Bell, J.C.; McNulty, S.M.; Duda, Z.; O’Neill, R.J.; A Sullivan, B.; Straight, A.F. RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. eLife 2017, 6, 25299. [Google Scholar] [CrossRef]
- Camacho, O.V.; Galan, C.; Swist-Rosowska, K.; Ching, R.; Gamalinda, M.; Karabiber, F.; de la Rosa-Velazquez, I.; Engist, B.; Koschorz, B.; Shukeir, N.; et al. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA: DNA hybrid formation. eLife 2017, 6, e25293. [Google Scholar] [CrossRef] [PubMed]
- Skourti-Stathaki, K.; Proudfoot, N.J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014, 28, 1384–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Chen, L.; Peng, D.; Jiang, A.; He, Y.; Zeng, Y.; Xie, C.; Zhou, H.; Luo, X.; Liu, H.; et al. METTL3 and N6-methyladenosine promote homologous recombination-mediated repair of DSBs by modulating DNA-RNA hybrid accumulation. Mol. Cell 2020, 79, 425–442.e7. [Google Scholar] [CrossRef] [PubMed]
- Abakir, A.; Giles, T.C.; Cristini, A.; Foster, J.M.; Dai, N.; Starczak, M.; Rubio-Roldan, A.; Li, M.; Eleftheriou, M.; Crutchley, J.; et al. N6-methyladenosine regulates the stability of RNA: DNA hybrids in human cells. Nat. Genet. 2019, 52, 48–55. [Google Scholar] [CrossRef]
- Cheng, J.X.; Chen, L.; Li, Y.; Cloe, A.; Yue, M.; Wei, J.; Watanabe, K.A.; Shammo, J.M.; Anastasi, J.; Shen, Q.J.; et al. RNA Cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia. Nat. Commun. 2018, 9, 1163. [Google Scholar] [CrossRef]
- Aguilo, F.; Li, S.; Balasubramaniyan, N.; Sancho, A.; Benko, S.; Zhang, F.; Vashisht, A.; Rengasamy, M.; Andino, B.; Chen, C.-H.; et al. Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1α. Cell Rep. 2016, 14, 479–492. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Li, X.; Tang, H.; Jiang, B.; Dou, Y.; Gorospe, M.; Wang, W. NSUN2-mediated M5C methylation and METTL3/ METTL14-mediated m6A methylation cooperatively enhance P21 translation. J. Cell. Biochem. 2017, 118, 2587–2598. [Google Scholar] [CrossRef]
- Van Tran, N.; Ernst, F.G.M.; Hawley, B.R.; Zorbas, C.; Ulryck, N.; Hackert, P.; Bohnsack, K.E.; Bohnsack, M.T.; Jaffrey, S.R.; Graille, M.; et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019, 47, 7719–7733. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Wang, X.; Cai, J.; Dai, Q.; Natchiar, S.K.; Lv, R.; Chen, K.; Lu, Z.; Chen, H.; Shi, Y.G.; et al. N6-methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 2018, 15, 88–94. [Google Scholar] [CrossRef]
- Bertero, A.; Brown, S.; Madrigal, P.; Osnato, A.; Ortmann, D.; Yiangou, L.; Kadiwala, J.; Hubner, N.C.; de los Mozos, I.R.; Sadée, C.; et al. The SMAD2/3 interactome reveals that TGFβ controls m6A MRNA methylation in pluripotency. Nature 2018, 555, 256–259. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, Y.; Tang, Q.; Wei, L.; Zhang, X.; Jia, G. An elongation and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N6-methyladenosine modifications. Angew. Chem. 2018, 57, 15995–16000. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, B.; Chen, L.; Gou, L.-T.; Li, H.; Fu, X.-D. GRID-seq reveals the global RNA–chromatin interactome. Nat. Biotechnol. 2017, 35, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Hyman, A.A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 196–213. [Google Scholar] [CrossRef] [PubMed]
- Sabari, B.R.; Dall’Agnese, A.; Young, R.A. Biomolecular condensates in the nucleus. Trends Biochem. Sci. 2020, 45, 961–977. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Ditlev, J.A.; Hui, E.; Xing, W.; Banjade, S.; Okrut, J.; King, D.S.; Taunton, J.; Rosen, M.K.; Vale, R.D. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 2016, 352, 595–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, J.H.; Davis, E.S.; Daugird, T.A.; Zhao, S.; Quiroga, I.Y.; Uryu, H.; Li, J.; Storey, A.J.; Tsai, Y.-H.; Keeley, D.P.; et al. Phase separation drives aberrant chromatin looping and cancer development. Nature 2021, 595, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Roden, C.; Gladfelter, A.S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 2021, 22, 183–195. [Google Scholar] [CrossRef]
- Ries, R.J.; Zaccara, S.; Klein, P.; Olarerin-George, A.; Namkoong, S.; Pickering, B.F.; Patil, D.P.; Kwak, H.; Lee, J.H.; Jaffrey, S.R. M6A enhances the phase separation potential of mRNA. Nature 2019, 571, 424–428. [Google Scholar] [CrossRef]
- Fu, Y.; Zhuang, X. M6A-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol. 2020, 16, 955–963. [Google Scholar] [CrossRef]
- Gao, Y.; Pei, G.; Li, D.; Li, R.; Shao, Y.; Zhang, Q.C.; Li, P. Multivalent m6A motifs promote phase separation of YTHDF proteins. Cell Res. 2019, 29, 767–769. [Google Scholar] [CrossRef]
- Cheng, Y.; Xie, W.; Pickering, B.F.; Chu, K.L.; Savino, A.M.; Yang, X.; Luo, H.; Nguyen, D.T.; Mo, S.; Barin, E.; et al. N6-methyladenosine on MRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell 2021, 39, 958–972.e8. [Google Scholar] [CrossRef] [PubMed]
| Cells | Writer/Reader | Target RNA | Histone Signature | Effect of Writer or Reader KOon Histone Signature | Affected Genomic Loci | Biological Function of m6A | Refs. |
|---|---|---|---|---|---|---|---|
| mNSc | METTL14 | CBP, P300 mRNAs | H3K27ac H3K27me3 | Increased | Proliferation/differentiation gene sets | Regulating self-renewal and differentiation | [62] |
| mESc | METTL3, METTL14, YTHDC1 | carRNAs, LINE1 | H3K4me3 H3K27ac | Increased | eRNAs, paRNAs, repeat RNAs | Repressing transcription at intergenic regions | [79] |
| mESc | YTHDC1, METTL3 | IAPs, LINE1 | H3K9me3 | Decreased | IAP repeats, Dux1 locus | Repressing transcription of 2C-like state genes | [80,81] |
| mESc | METTL3, YTHDC1 | IAPs | H3K9me3 H3K20me3 | Decreased | IAP repeats | Maintaining heterochromatin at repeat elements | [82] |
| mESc | METTL3, METTL14, YTHDF2 | IAPs | H3K9me3 | No significant change | ERVs | Repressing transcription of ERVs | [83] |
| mESC | METTL3, METTL4 | MSR LINE1 | H3K9me3 | No significant change | MSR LINE1 | Promoting the formation of RNA:DNA hybrids | [84] |
| HEK293, mESc | METTL3, YTHDC1 | mRNAs | H3K9me2 | Decreased | Gene bodies | Promoting co-transcriptional histone de-methylation and transcriptional output | [85] |
| mESc | METTL14 | MYC ACTB | H3K36me3 | No significant change | Gene rich regions | Methylate nascent RNAs | [86] |
| THP1 | YTHDF2 | KDM6B mRNA | H3K27me3 | Decreased | IL-6, IL-12B, CCL22, ICAM1 | Restraining the expression of pro-inflammatory cytokines | [87] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selmi, T.; Lanzuolo, C. Driving Chromatin Organisation through N6-methyladenosine Modification of RNA: What Do We Know and What Lies Ahead? Genes 2022, 13, 340. https://doi.org/10.3390/genes13020340
Selmi T, Lanzuolo C. Driving Chromatin Organisation through N6-methyladenosine Modification of RNA: What Do We Know and What Lies Ahead? Genes. 2022; 13(2):340. https://doi.org/10.3390/genes13020340
Chicago/Turabian StyleSelmi, Tommaso, and Chiara Lanzuolo. 2022. "Driving Chromatin Organisation through N6-methyladenosine Modification of RNA: What Do We Know and What Lies Ahead?" Genes 13, no. 2: 340. https://doi.org/10.3390/genes13020340






