Latent Membrane Protein 1 (LMP1) from Epstein–Barr Virus (EBV) Strains M81 and B95.8 Modulate miRNA Expression When Expressed in Immortalized Human Nasopharyngeal Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. LMP1 Constructs and Cell Transfections
2.3. Luciferase Assay and Cell Migration In Vitro
2.4. MicroRNA Expression Analysis
2.5. MiRNA’s Targets Prediction and Pathway Enrichment Analysis
3. Results
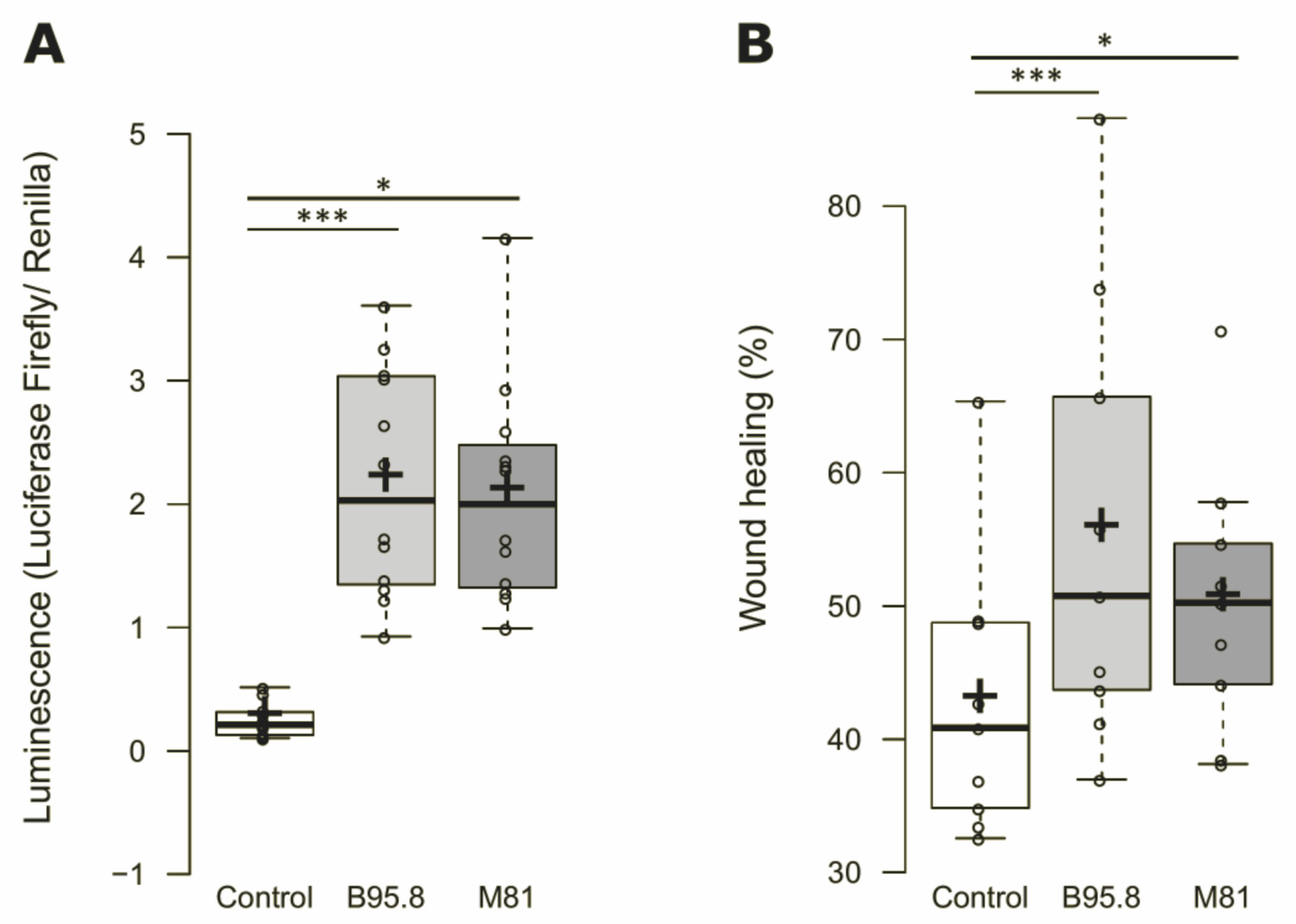
3.1. Cells Expressing EBV LMP1 from Viral Strains B95.8 and M81 Behaves Similarly in Terms of NF-κB Activation and Cell Migration Rates In Vitro
3.2. EBV LMP1 Modulate miRNA Expression in NP69SV40T Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Committee
References
- Global Burden of Disease Cancer Collaboration. The Global Burden of Cancer 2013. JAMA Oncol. 2015, 1, 505–527. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.-K.; Weiderpass, E. Infection and Cancer: Global Distribution and Burden of Diseases. Ann. Glob. Health 2014, 80, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Schottenfeld, D.; Beebe-Dimmer, J. The Cancer Burden Attributable to Biologic Agents. Ann. Epidemiol. 2015, 25, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Stewart, B.W.; Wild, C.P. World Cancer Report 2014. 2014. Available online: http://www.iahm.org/journal/vol_15/num_3/text/vol15n3p40.pdf (accessed on 20 May 2020).
- De Martel, C.; Ferlay, J.; Franceschi, S.; Vignat, J.; Bray, F.; Forman, D.; Plummer, M. Global Burden of Cancers Attributable to Infections in 2008: A Review and Synthetic Analysis. Lancet Oncol. 2012, 13, 607–615. [Google Scholar] [CrossRef]
- Vineis, P.; Wild, C.P. Global Cancer Patterns: Causes and Prevention. Lancet 2014, 383, 549–557. [Google Scholar] [CrossRef]
- Dunmire, S.K.; Hogquist, K.A.; Balfour, H.H. Infectious Mononucleosis. In Epstein Barr Virus Volume 1; Münz, C., Ed.; Springer International Publishing: Cham, Switzerland, 2015; Volume 390, pp. 211–240. ISBN 978-3-319-22821-1. [Google Scholar]
- Elgui de Oliveira, D. DNA Viruses in Human Cancer: An Integrated Overview on Fundamental Mechanisms of Viral Carcinogenesis. Cancer Lett. 2007, 247, 182–196. [Google Scholar] [CrossRef]
- Tang, L.-L.; Chen, W.-Q.; Xue, W.-Q.; He, Y.-Q.; Zheng, R.-S.; Zeng, Y.-X.; Jia, W.-H. Global Trends in Incidence and Mortality of Nasopharyngeal Carcinoma. Cancer Lett. 2016, 374, 22–30. [Google Scholar] [CrossRef]
- Colditz, G.A.; Wolin, K.Y.; Gehlert, S. Applying What We Know to Accelerate Cancer Prevention. Sci. Transl. Med. 2012, 4, 127rv4. [Google Scholar] [CrossRef] [Green Version]
- Brousset, P.; Keryer, C.; Ooca, T.; Corbex, M. EBV-Associated Nasopharyngeal Carcinomas: From Epidemiology to Virus-Targeting Strategies. Trends Microbiol. 2004, 12, 353–356. [Google Scholar] [CrossRef]
- The Enigmatic Epidemiology of Nasopharyngeal Carcinoma|Cancer Epidemiology, Biomarkers & Prevention. Available online: http://cebp.aacrjournals.org/content/15/10/1765.long (accessed on 5 July 2018).
- Tsao, S.-W.; Tsang, C.M.; To, K.-F.; Lo, K.-W. The Role of Epstein–Barr Virus in Epithelial Malignancies: Role of EBV in Epithelial Malignancies. J. Pathol. 2015, 235, 323–333. [Google Scholar] [CrossRef]
- De Oliveira, D.E.; Müller-Coan, B.G.; Pagano, J.S. Viral Carcinogenesis Beyond Malignant Transformation: EBV in the Progression of Human Cancers. Trends Microbiol. 2016, 24, 649–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinkmann, M.M. Regulation of Intracellular Signalling by the Terminal Membrane Proteins of Members of the Gammaherpesvirinae. J. Gen. Virol. 2006, 87, 1047–1074. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Tang, M.; Tao, Y.; Li, L.; Liu, S.; Guo, L.; Li, Z.; Ma, X.; Xu, J.; Cao, Y. Epstein–Barr Virus-Encoded LMP1 Triggers Regulation of the ERK-Mediated Op18/Stathmin Signaling Pathway in Association with Cell Cycle. Cancer Sci. 2012, 103, 993–999. [Google Scholar] [CrossRef]
- Masoud, G.N.; Li, W. HIF-1α Pathway: Role, Regulation and Intervention for Cancer Therapy. Acta Pharm. Sin. B 2015, 5, 378–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakisaka, N.; Kondo, S.; Yoshizaki, T.; Murono, S.; Furukawa, M.; Pagano, J.S. Epstein–Barr Virus Latent Membrane Protein 1 Induces Synthesis of Hypoxia-Inducible Factor 1. Mol. Cell. Biol. 2004, 24, 5223–5234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, M.-H.; Lin, X.; Shumilov, A.; Bernhardt, K.; Feederle, R.; Poirey, R.; Kopp-Schneider, A.; Pereira, B.; Almeida, R.; Delecluse, H.-J. The Biological Properties of Different Epstein–Barr Virus Strains Explain Their Association with Various Types of Cancers. Oncotarget 2016, 8, 10238–10254. [Google Scholar] [CrossRef] [Green Version]
- Tsai, M.-H.; Raykova, A.; Klinke, O.; Bernhardt, K.; Gärtner, K.; Leung, C.S.; Geletneky, K.; Sertel, S.; Münz, C.; Feederle, R.; et al. Spontaneous Lytic Replication and Epitheliotropism Define an Epstein–Barr Virus Strain Found in Carcinomas. Cell Rep. 2013, 5, 458–470. [Google Scholar] [CrossRef] [Green Version]
- Abba, M.; Patil, N.; Allgayer, H. MicroRNAs in the Regulation of MMPs and Metastasis. Cancers 2014, 6, 625–645. [Google Scholar] [CrossRef] [Green Version]
- Acunzo, M.; Romano, G.; Wernicke, D.; Croce, C.M. MicroRNA and Cancer—A Brief Overview. Adv. Biol. Regul. 2015, 57, 1–9. [Google Scholar] [CrossRef]
- Abba, M.L.; Patil, N.; Leupold, J.H.; Allgayer, H. MicroRNA Regulation of Epithelial to Mesenchymal Transition. J. Clin. Med. 2016, 5, 8. [Google Scholar] [CrossRef]
- Chen, H.-C.; Chen, G.-H.; Chen, Y.-H.; Liao, W.-L.; Liu, C.-Y.; Chang, K.-P.; Chang, Y.-S.; Chen, S.-J. MicroRNA Deregulation and Pathway Alterations in Nasopharyngeal Carcinoma. Br. J. Cancer 2009, 100, 1002–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, G.-D.; Huang, T.-J.; Peng, L.-X.; Yang, C.-F.; Liu, R.-Y.; Huang, H.-B.; Chu, Q.-Q.; Yang, H.-J.; Huang, J.-L.; Zhu, Z.-Y.; et al. Epstein–Barr Virus_Encoded LMP1 Upregulates MicroRNA-21 to Promote the Resistance of Nasopharyngeal Carcinoma Cells to Cisplatin-Induced Apoptosis by Suppressing PDCD4 and Fas-L. PLoS ONE 2013, 8, e78355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlmann, S.; Zhang, J.D.; Schwäger, A.; Mannsperger, H.; Riazalhosseini, Y.; Burmester, S.; Ward, A.; Korf, U.; Wiemann, S.; Sahin, O. MiR-200bc/429 Cluster Targets PLCgamma1 and Differentially Regulates Proliferation and EGF-Driven Invasion than MiR-200a/141 in Breast Cancer. Oncogene 2010, 29, 4297–4306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, X.; Li, J.-Y.; Guo, J.; Li, P.-S.; Zhang, W.-H. Influence of MiR-451 on Drug Resistances of Paclitaxel-Resistant Breast Cancer Cell Line. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2015, 21, 3291–3297. [Google Scholar] [CrossRef] [Green Version]
- Raza, U.; Zhang, J.D.; Sahin, O. MicroRNAs: Master Regulators of Drug Resistance, Stemness, and Metastasis. J. Mol. Med. Berl. Ger. 2014, 92, 321–336. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent Deletions and Down-Regulation of Micro- RNA Genes MiR15 and MiR16 at 13q14 in Chronic Lymphocytic Leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [Green Version]
- Graham, F.L.; Smiley, J.; Russell, W.C.; Nairn, R. Characteristics of a Human Cell Line Transformed by DNA from Human Adenovirus Type 5. J. Gen. Virol. 1977, 36, 59–72. [Google Scholar] [CrossRef]
- Tsao, S.W.; Wang, X.; Liu, Y.; Cheung, Y.C.; Feng, H.; Zheng, Z.; Wong, N.; Yuen, P.W.; Lo, A.K.; Wong, Y.C.; et al. Establishment of Two Immortalized Nasopharyngeal Epithelial Cell Lines Using SV40 Large T and HPV16E6/E7 Viral Oncogenes. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2002, 1590, 150–158. [Google Scholar] [CrossRef] [Green Version]
- Delecluse, H.-J.; Hilsendegen, T.; Pich, D.; Zeidler, R.; Hammerschmidt, W. Propagation and Recovery of Intact, Infectious Epstein–Barr Virus from Prokaryotic to Human Cells. Proc. Natl. Acad. Sci. USA 1998, 95, 8245–8250. [Google Scholar] [CrossRef] [Green Version]
- Feederle, R.; Bartlett, E.J.; Delecluse, H.-J. Epstein–Barr Virus Genetics: Talking about the BAC Generation. Herpesviridae 2010, 1, 6. [Google Scholar] [CrossRef] [Green Version]
- Keller, S.A.; Hernandez-Hopkins, D.; Vider, J.; Ponomarev, V.; Hyjek, E.; Schattner, E.J.; Cesarman, E. NF-ΚB Is Essential for the Progression of KSHV- and EBV-Infected Lymphomas in Vivo. Blood 2006, 107, 3295–3302. [Google Scholar] [CrossRef] [PubMed]
- Gebäck, T.; Shultz, M.M.P.; Koumoutsakos, P.; Detmar, M. TScratch: A Novel and Simple Software Tool for Automated Analysis of Monolayer Wound Healing Assays. BioTechniques 2009, 46, 265–274. [Google Scholar] [CrossRef]
- Marí-Alexandre, J.; Barceló-Molina, M.; Belmonte-López, E.; García-Oms, J.; Estellés, A.; Braza-Boïls, A.; Gilabert-Estellés, J. Micro-RNA Profile and Proteins in Peritoneal Fluid from Women with Endometriosis: Their Relationship with Sterility. Fertil. Steril. 2018, 109, 675–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrante, S.C.; Nadler, E.P.; Pillai, D.K.; Hubal, M.J.; Wang, Z.; Wang, J.M.; Gordish-Dressman, H.; Koeck, E.; Sevilla, S.; Wiles, A.A.; et al. Adipocyte-Derived Exosomal MiRNAs: A Novel Mechanism for Obesity-Related Disease. Pediatr. Res. 2015, 77, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.-C.; Tsay, M.; Lu, R.; Jurisica, I. MirDIP 4.1—Integrative Database of Human MicroRNA Target Predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef] [PubMed]
- Shirdel, E.A.; Xie, W.; Mak, T.W.; Jurisica, I. NAViGaTing the Micronome—Using Multiple MicroRNA Prediction Databases to Identify Signalling Pathway-Associated MicroRNAs. PLoS ONE 2011, 6, e17429. [Google Scholar] [CrossRef] [Green Version]
- Croft, D.; Mundo, A.F.; Haw, R.; Milacic, M.; Weiser, J.; Wu, G.; Caudy, M.; Garapati, P.; Gillespie, M.; Kamdar, M.R.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2014, 42, D472–D477. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wasil, L.R.; Shair, K.H.Y. Epstein–Barr Virus LMP1 Induces Focal Adhesions and Epithelial Cell Migration through Effects on Integrin-A5 and N-Cadherin. Oncogenesis 2015, 4, e171. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Tian, Z.; Qin, H.; Li, N.; Zhou, X.; Li, J.; Ni, B.; Ruan, Z. High Expression of Sphingosine Kinase 1 Is Associated with Poor Prognosis in Nasopharyngeal Carcinoma. Biochem. Biophys. Res. Commun. 2015, 460, 341–347. [Google Scholar] [CrossRef]
- Lee, H.M.; Lo, K.-W.; Wei, W.; Tsao, S.W.; Chung, G.T.Y.; Ibrahim, M.H.; Dawson, C.W.; Murray, P.G.; Paterson, I.C.; Yap, L.F. Oncogenic S1P Signalling in EBV-Associated Nasopharyngeal Carcinoma Activates AKT and Promotes Cell Migration through S1P Receptor 3. J. Pathol. 2017, 242, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-H.; Chang, H.-C.; Hung, W.-C. Transcriptional Repression of Tissue Inhibitor of Metalloproteinase-3 by Epstein–Barr Virus Latent Membrane Protein 1 Enhances Invasiveness of Nasopharyngeal Carcinoma Cells. Oral Oncol. 2008, 44, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Aga, M.; Bentz, G.L.; Raffa, S.; Torrisi, M.R.; Kondo, S.; Wakisaka, N.; Yoshizaki, T.; Shackelford, J. Exosomal HIF1α Supports Invasive Potential of Nasopharyngeal Carcinoma-Associated LMP1-Positive Exosomes. Oncogene 2014, 33, 4613–4622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.-C.; Tang, K.-F. Clinical Value of Integrated-Signature MiRNAs in Esophageal Cancer. Cancer Med. 2017, 6, 1893–1903. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Yu, S.; Zhou, L.; Shi, M.; Hu, Y.; Xu, X.; Shen, B.; Liu, S.; Yan, D.; Feng, J. Cisplatin-Resistant Lung Cancer Cell–Derived Exosomes Increase Cisplatin Resistance of Recipient Cells in Exosomal MiR-100–5p-Dependent Manner. Int. J. Nanomed. 2017, 12, 3721–3733. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Sun, Y.; Yuan, Y.; Han, Z.; Zhang, P.; Zhang, J.; You, M.J.; Teruya-Feldstein, J.; Wang, M.; Gupta, S.; et al. MiR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion. PLoS Genet. 2014, 10, e1004177. [Google Scholar] [CrossRef]
- Nabavi, N.; Saidy, N.R.N.; Venalainen, E.; Haegert, A.; Parolia, A.; Xue, H.; Wang, Y.; Wu, R.; Dong, X.; Collins, C.; et al. MiR-100-5p Inhibition Induces Apoptosis in Dormant Prostate Cancer Cells and Prevents the Emergence of Castration-Resistant Prostate Cancer. Sci. Rep. 2017, 7, 4079. [Google Scholar] [CrossRef] [Green Version]
- Yan-Chun, L.; Hong-Mei, Y.; Zhi-Hong, C.; Qing, H.; Yan-Hong, Z.; Ji-Fang, W. MicroRNA-192-5p Promote the Proliferation and Metastasis of Hepatocellular Carcinoma Cell by Targeting SEMA3A. Appl. Immunohistochem. Mol. Morphol. 2015, 25, 251–260. [Google Scholar] [CrossRef]
- Kim, Y.S.; Park, S.J.; Lee, Y.S.; Kong, H.K.; Park, J.H. MiRNAs Involved in LY6K and Estrogen Receptor α Contibute to Tamoxifen-Suceptibility in Breast Cancer. Oncotarget 2016, 5, 42261–42273. [Google Scholar]
- Su, Y.; Ni, Z.; Wang, G.; Cui, J.; Wei, C.; Wang, J.; Yang, Q.; Xu, Y.; Li, F. Aberrant Expression of MicroRNAs in Gastric Cancer and Biological Significance of MiR-574-3p. Int. Immunopharmacol. 2012, 13, 468–475. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Bai, J.; Zhuan, Z.; Li, B.; Zhang, Z.; Wu, X.; Luo, X.; Yang, L. EBV-LMP1 Is Involved in Vasculogenic Mimicry Formation via VEGFA/VEGFR1 Signaling in Nasopharyngeal Carcinoma. Oncol. Rep. 2018, 40, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Tworkoski, K.; Raab-Traub, N. LMP1 Promotes Expression of Insulin-Like Growth Factor 1 (IGF1) To Selectively Activate IGF1 Receptor and Drive Cell Proliferation. J. Virol. 2014, 89, 2590–2602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawson, C.W.; Laverick, L.; Morris, M.A.; Tramoutanis, G.; Young, L.S. Epstein–Barr Virus-Encoded LMP1 Regulates Epithelial Cell Motility and Invasion via the ERK-MAPK Pathway. J. Virol. 2008, 82, 3654–3664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuda, M.; Kurosaki, W.; Yanagihara, K.; Kuratsune, H.; Sairenji, T. A Mechanism in Epstein–Barr Virus Oncogenesis: Inhibition of Transforming Growth Factor-Β1-Mediated Induction of MAPK/P21 by LMP1. Virology 2002, 302, 310–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- QingLing, Z.; LiNa, Y.; Liu, L.; Shuang, W.; YuFang, Y.; Yi, D.; Divakaran, J.; Xin, L.; YanQing, D. LMP1 Antagonizes WNT/β-Catenin Signalling through Inhibition of WTX and Promotes Nasopharyngeal Dysplasia but Not Tumourigenesis in LMP1B95−8 Transgenic Mice. J. Pathol. 2011, 223, 574–583. [Google Scholar] [CrossRef]
- Zeng, Z.-Y.; Zhou, Y.-H.; Zhang, W.-L.; Xiong, W.; Fan, S.-Q.; Li, X.-L.; Luo, X.-M.; Wu, M.-H.; Yang, Y.-X.; Huang, C.; et al. Gene Expression Profiling of Nasopharyngeal Carcinoma Reveals the Abnormally Regulated Wnt Signaling Pathway. Hum. Pathol. 2007, 38, 120–133. [Google Scholar] [CrossRef]
- Leonard, S.; Wei, W.; Anderton, J.; Vockerodt, M.; Rowe, M.; Murray, P.G.; Woodman, C.B. Epigenetic and Transcriptional Changes Which Follow Epstein–Barr Virus Infection of Germinal Center B Cells and Their Relevance to the Pathogenesis of Hodgkin’s Lymphoma. J. Virol. 2011, 85, 9568–9577. [Google Scholar] [CrossRef] [Green Version]
- Seo, S.Y.; Kim, E.-O.; Jang, K.L. Epstein–Barr Virus Latent Membrane Protein 1 Suppresses the Growth-Inhibitory Effect of Retinoic Acid by Inhibiting Retinoic Acid Receptor-Β2 Expression via DNA Methylation. Cancer Lett. 2008, 270, 66–76. [Google Scholar] [CrossRef]
- Liao, J.; Karnik, R.; Gu, H.; Ziller, M.J.; Clement, K.; Tsankov, A.M.; Akopian, V.; Gifford, C.A.; Donaghey, J.; Galonska, C.; et al. Targeted Disruption of DNMT1, DNMT3A and DNMT3B in Human Embryonic Stem Cells. Nat. Genet. 2015, 47, 469–478. [Google Scholar] [CrossRef] [Green Version]
- Perez-Balaguer, A.; Ortiz-Martínez, F.; García-Martínez, A.; Pomares-Navarro, C.; Lerma, E.; Peiró, G. FOXA2 MRNA Expression Is Associated with Relapse in Patients with Triple-Negative/Basal-like Breast Carcinoma. Breast Cancer Res. Treat. 2015, 153, 465–474. [Google Scholar] [CrossRef]
- Yao, H.S.; Wang, J.; Zhang, X.P.; Wang, L.Z.; Wang, Y.; Li, X.X.; Jin, K.Z.; Hu, Z.Q.; Wang, W.J. Hepatocyte Nuclear Factor 4α Suppresses the Aggravation of Colon Carcinoma. Mol. Carcinog. 2016, 55, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Adams, A.; Roberts, B.; O’Neil, M.; Vittal, A.; Schmitt, T.; Kumer, S.; Cox, J.; Li, Z.; Weinman, S.A.; et al. Protein Arginine Methyl Transferase 1– and Jumonji C Domain-Containing Protein 6–Dependent Arginine Methylation Regulate Hepatocyte Nuclear Factor 4 α Expression and Hepatocyte Proliferation in Mice. Hepatology 2018, 67, 1109–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clorennec, C.L.; Ouk, T.-S.; Youlyouz-Marfak, I.; Panteix, S.; Martin, C.-C.; Rastelli, J.; Adriaenssens, E.; Zimber-Strobl, U.; Coll, J.; Feuillard, J.; et al. Molecular Basis of Cytotoxicity of Epstein–Barr Virus (EBV) Latent Membrane Protein 1 (LMP1) in EBV Latency III B Cells: LMP1 Induces Type II Ligand-Independent Autoactivation of CD95/Fas with Caspase 8-Mediated Apoptosis. J. Virol. 2008, 82, 6721–6733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clorennec, C.L.; Youlyouz-Marfak, I.; Adriaenssens, E.; Coll, J.; Bornkamm, G.W.; Feuillard, J. EBV Latency III Immortalization Program Sensitizes B Cells to Induction of CD95-Mediated Apoptosis via LMP1: Role of NF-ΚB, STAT1, and P53. Blood 2006, 107, 2070–2078. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Uthaisang, W.; Hu, L.; Ernberg, I.T.; Fadeel, B. Epstein–Barr Virus-Encoded Latent Membrane Protein 1 Promotes Stress-Induced Apoptosis Upstream of Caspase-2-Dependent Mitochondrial Perturbation. Int. J. Cancer 2005, 113, 397–405. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller Coan, B.G.; Cesarman, E.; Acencio, M.L.; Elgui de Oliveira, D. Latent Membrane Protein 1 (LMP1) from Epstein–Barr Virus (EBV) Strains M81 and B95.8 Modulate miRNA Expression When Expressed in Immortalized Human Nasopharyngeal Cells. Genes 2022, 13, 353. https://doi.org/10.3390/genes13020353
Müller Coan BG, Cesarman E, Acencio ML, Elgui de Oliveira D. Latent Membrane Protein 1 (LMP1) from Epstein–Barr Virus (EBV) Strains M81 and B95.8 Modulate miRNA Expression When Expressed in Immortalized Human Nasopharyngeal Cells. Genes. 2022; 13(2):353. https://doi.org/10.3390/genes13020353
Chicago/Turabian StyleMüller Coan, Barbara G., Ethel Cesarman, Marcio Luis Acencio, and Deilson Elgui de Oliveira. 2022. "Latent Membrane Protein 1 (LMP1) from Epstein–Barr Virus (EBV) Strains M81 and B95.8 Modulate miRNA Expression When Expressed in Immortalized Human Nasopharyngeal Cells" Genes 13, no. 2: 353. https://doi.org/10.3390/genes13020353





