KASP Markers Specific for the Fertility Restorer Locus Rf1 and Application for Genetic Purity Testing in Sunflowers (Helianthus annuus L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sunflower Plant Material
2.2. EMS-Treatment of Sunflower Seeds
2.3. DNA Extraction
2.4. SNP Analyses Using Amplicon Targeted Sequences
2.5. KASP Primer Design
2.6. KASP-Marker Assay
2.7. Data Analysis
3. Results
3.1. Development of KASP Markers Based on SNPs Associated with Fertility Restoration
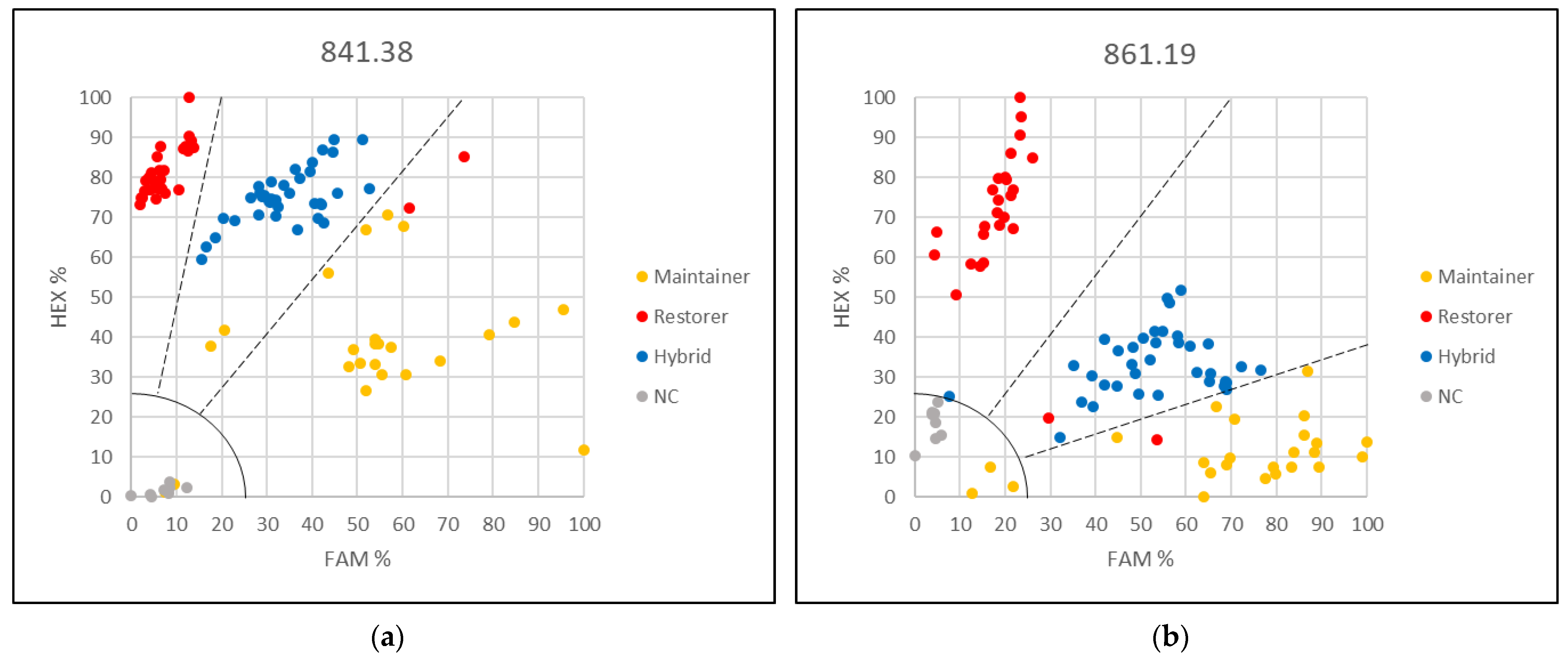
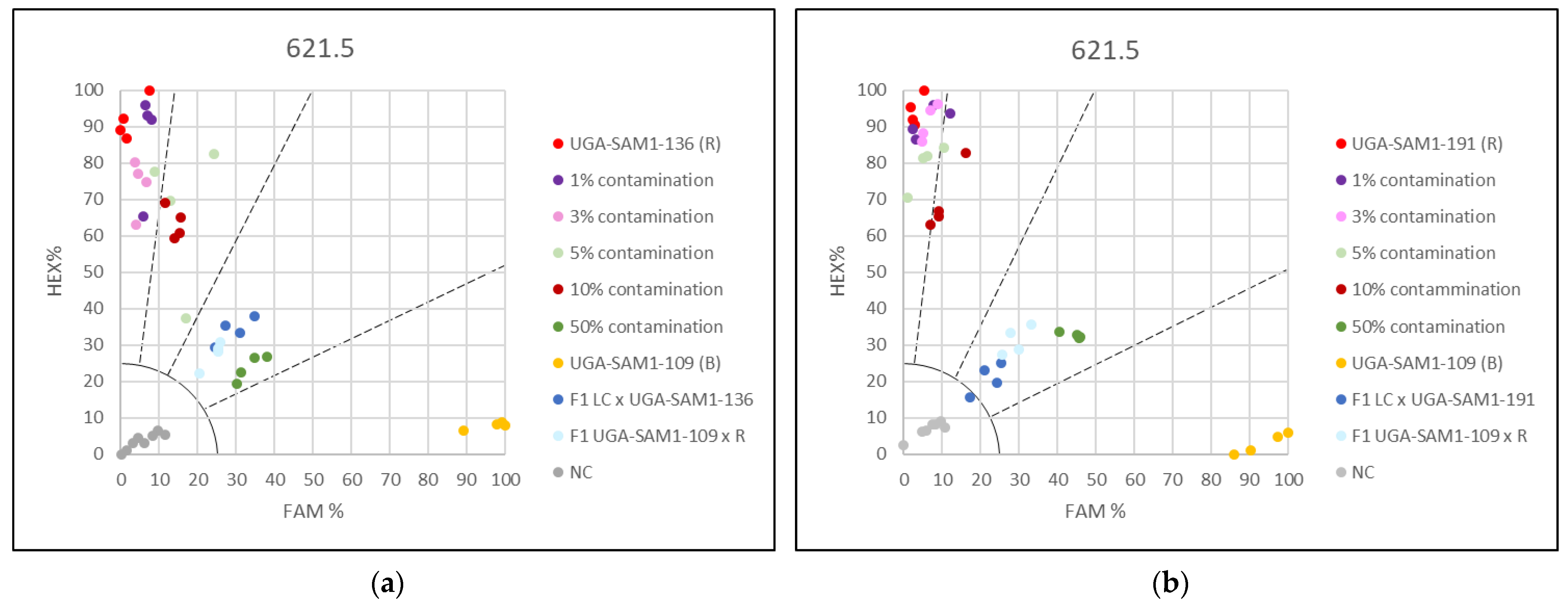
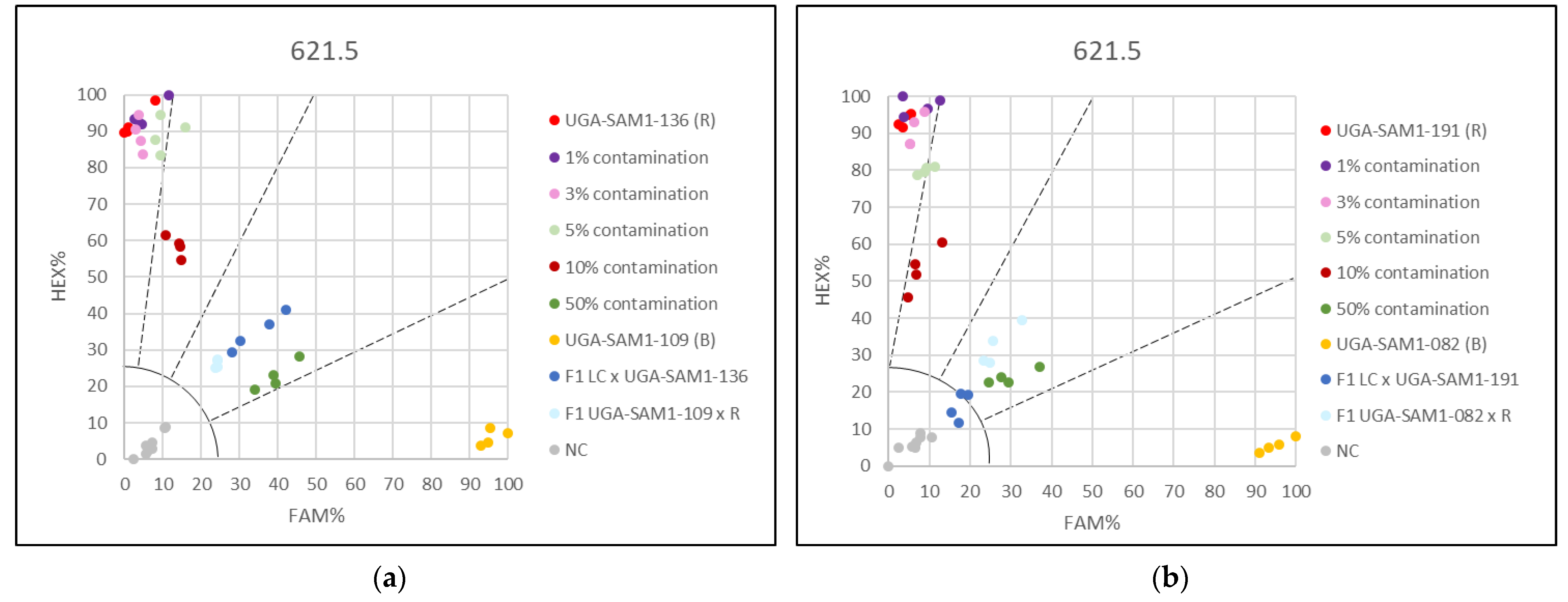
3.2. Applying the KASP Marker 621.5 for Genetic Purity Testing
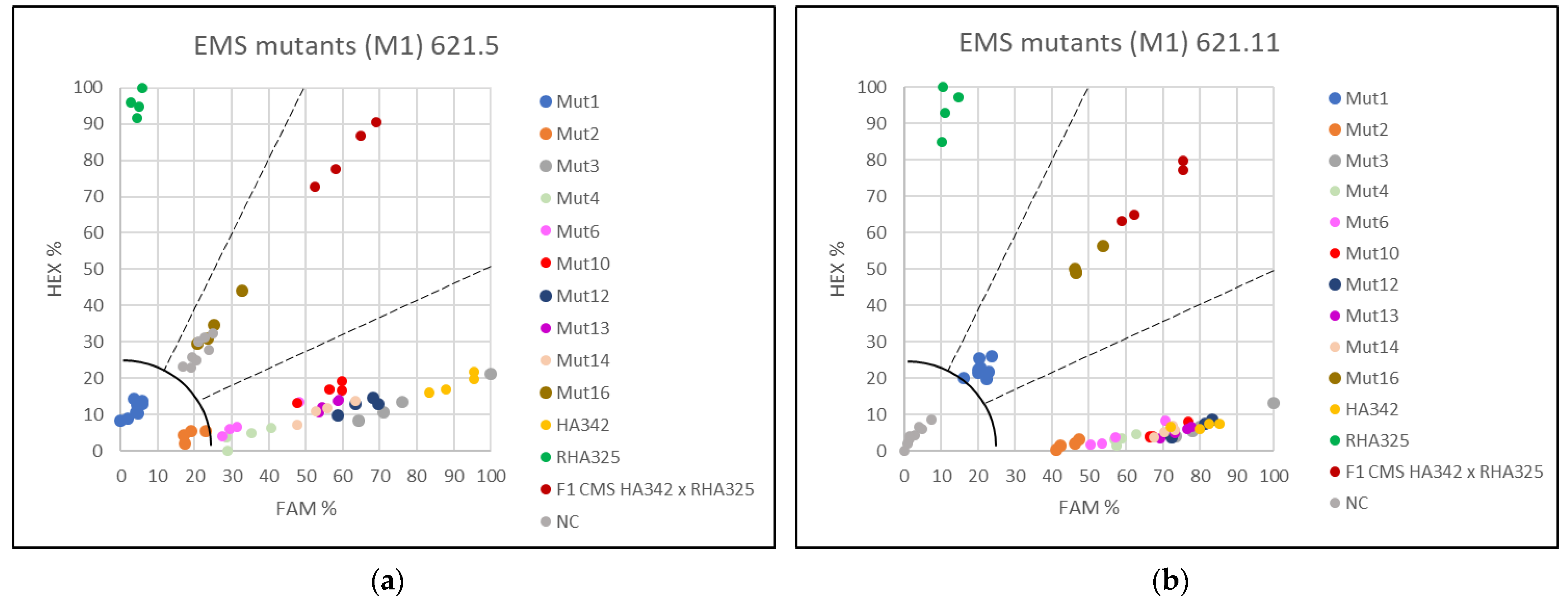
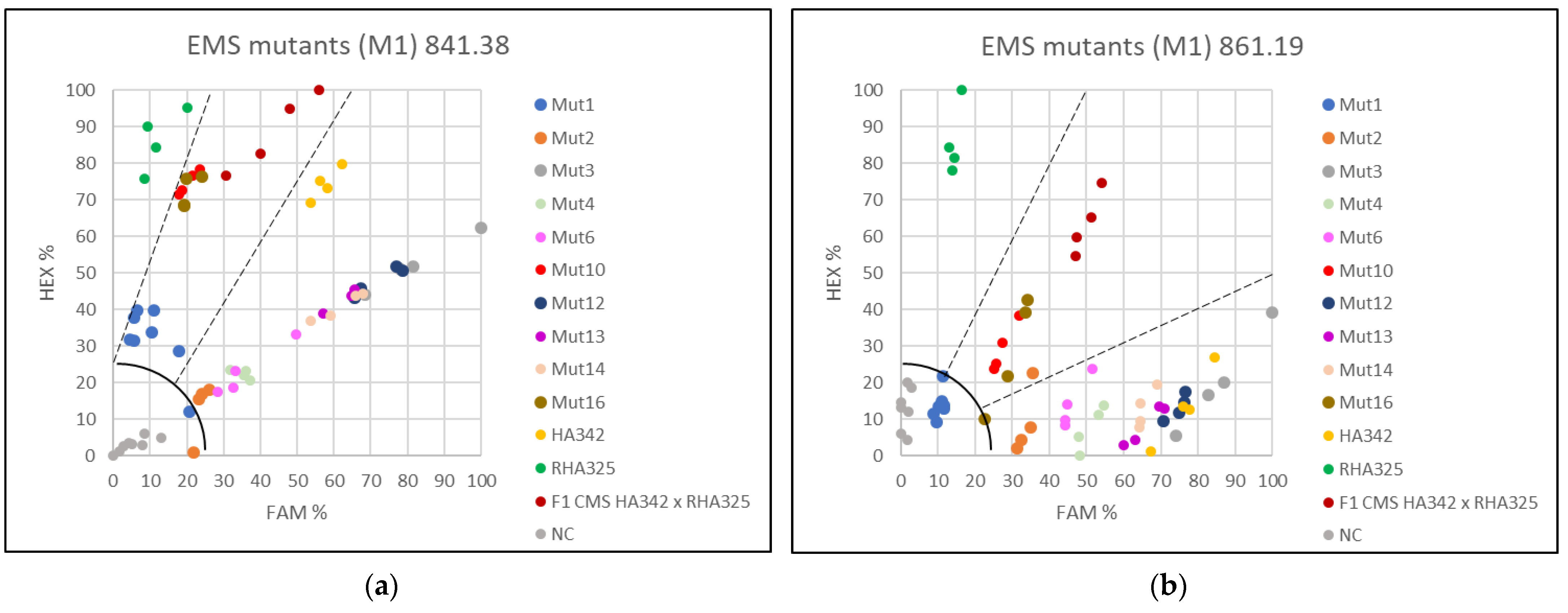
3.3. Applying the KASP Markers for Single Plant Testing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vear, F. Changes in sunflower breeding over the last fifty years. OCL 2016, 23, D202. [Google Scholar] [CrossRef] [Green Version]
- Köhler, R.H.; Horn, R.; Lossl, A.; Zetsche, K. Cytoplasmic male sterility in sunflower is correlated with the co-transcription of a new open reading frame with the atpA gene. Mol. Gen. Genet. 1991, 227, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Hanson, M.R.; Bentolila, S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 2004, 16, S154–S169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korell, M.; Mösges, G.; Friedt, W. Construction of a sunflower pedigree map. Helia 1992, 15, 7–16. [Google Scholar]
- Talukder, Z.I.; Ma, G.; Hulke, B.S.; Jan, C.C.; Qi, L. Linkage mapping and genome-wide association studies of the Rf gene cluster in sunflower (Helianthus annuus L.) and their distribution in world sunflower collections. Front. Genet. 2019, 14, 216. [Google Scholar] [CrossRef] [Green Version]
- Kinman, M.L. New developments in the USDA and state experiment station sunflower breeding programs. In Proceedings of the 4th International Sunflower Conference, Memphis, TN, USA, 23 June 1970; pp. 181–183. [Google Scholar]
- Kusterer, B.; Horn, R.; Friedt, W. Molecular mapping of the fertility restoration locus Rf1 in sunflower and development of diagnostic markers for the restorer gene. Euphytica 2005, 143, 35–43. [Google Scholar] [CrossRef]
- Horn, R.; Kusterer, B.; Lazarescu, E.; Prüfe, M.; Friedt, W. Molecular mapping of the Rf1 gene restoring pollen fertility in PET1-based F1-hybrids in sunflower (Helianthus annuus L.). Theor. Appl. Genet. 2003, 106, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Vick, B.A.; Cai, X.; Hu, J. Genetic mapping for the Rf1 (fertility restoration) gene in sunflower (Helianthus annuus L.) by SSR and TRAP markers. Plant Breed. 2010, 129, 24–28. [Google Scholar] [CrossRef]
- Markin, N.; Usatov, A.; Makarenko, M.; Azarin, K.; Gorbachenko, O.; Kolokolova, N.; Usatenko, T.; Markina, O.; Gavrilova, V. Study of informative DNA markers of the Rf1 gene in sunflower for breeding practice. Czech J. Plant Breed. 2017, 53, 69–75. [Google Scholar] [CrossRef] [Green Version]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef]
- Goryunov, D.V.; Anisimova, I.N.; Gavrilova, V.A.; Chernova, A.I.; Sotnikova, E.A.; Martynova, E.U.; Boldyrev, S.V.; Ayupova, A.F.; Gubaev, R.F.; Mazin, P.V.; et al. Association mapping of fertility restorer gene for CMS PET1 in sunflower. Agronomy 2019, 9, 49. [Google Scholar] [CrossRef] [Green Version]
- Owens, G.L.; Baute, G.J.; Hubner, S.; Rieseberg, L.H. Genomic sequence and copy number evolution during hybrid crop development in sunflowers. Evol. Appl. 2019, 12, 54–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, R.; Radanovic, A.; Fuhrmann, L.; Sprycha, Y.; Hamrit, S.; Jockovic, M.; Miladinovic, D.; Jansen, C. Development and validation of markers for the fertility restorer gene Rf1 in sunflower. Int. J. Mol. Sci. 2019, 20, 1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polivanova, O.B.; Sivolapova, A.B.; Goryunov, D.V.; Fedorova, A.V.; Sotnikova, E.A.; Chebanova, Y.V.; Karabitsina, Y.U.; Benko, N.I.; Demurin, Y.N.; Goryunova, S.V. Structural diversity of sunflower (Helianthus annuus L.) candidate Rf1 loci based on gene-specific PCR. Res. Crops 2021, 22, 40–46. [Google Scholar] [CrossRef]
- Seiler, G.J.; Qi, L.L.; Marek, L.F. Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. 2017, 57, 1083–1101. [Google Scholar] [CrossRef] [Green Version]
- Pallavi, H.M.; Gowda, R.; Vishwanath, K.; Shadakshari, Y.G.; Bhanuprakash, K. Identification of SSR markers for hybridity and seed genetic purity testing in sunflower (Helianthus annuus L.). Seed Sci. Technol. 2011, 34, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Miklič, V.; Dušanić, N.; Jocić, S. Sunflower seed production. In Seed Production; Milošević, M., Kobiljski, B., Eds.; Institute of Field and Vegetable Crops: Novi Sad, Serbia, 2011; Volume 2, pp. 196–264. (In Serbian) [Google Scholar]
- Gongshe, L.; Jiesheng, M.; Jie, L.; Baoqi, S. Use of RAPD markers to screen hybrids of oilseed sunflower. In Proceedings of the 15th International Sunflower Conference, Toulouse, France, 12–16 June 2000; pp. M-16–M-19. [Google Scholar]
- Anusha, S.; Dhammaprakash, W.; Yamini, K.; Kumar, C.A.; Kumar, V.D. Assessment of genetic purity of two sunflower (Helianthus annuus L.) hybrids using sequence characterized amplified region (SCAR) markers. Indian Soc. Oilseeds Res. 2020, 15, 60–61. [Google Scholar]
- Solodenko, A.E.; Sanalatij, A.V.; Sivolap, Y.M. Sunflower genotypes identification by means of microsatellite markers. Cytol. Genet. 2003, 38, 84–90. (In Russian) [Google Scholar]
- Antonova, T.S.; Guchetl, S.Z.; Tchelustnikova, T.A.; Ramasanova, S.A. Development of marker system for identification and certification of sunflower lines and hybrids on the basis of SSR-analysis. Helia 2006, 29, 63–72. [Google Scholar] [CrossRef]
- Ayalew, H.; Tsang, P.W.; Chu, C.; Wang, J.; Liu, S.; Chen, C.; Ma, X.-F. Comparison of TaqMan, KASP and rhAmp SNP genotyping platforms in hexaploid wheat. PLoS ONE 2019, 14, e0217222. [Google Scholar] [CrossRef] [Green Version]
- Kante, M.; Rattunde, H.F.W.; Nébié, B.; Weltzien, E.; Haussmann, B.I.; Leiser, W.L. QTL mapping and validation of fertility restoration in West African sorghum A 1 cytoplasm and identification of a potential causative mutation for Rf2. Theor. Appl. Genet. 2018, 131, 2397–2412. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Bosland, P.W.; Zhang, Z.; Wang, Y.; Zhang, G.; Wang, L.; Yu, J. A predicted NEDD8 conjugating enzyme gene identified as a Capsicum candidate Rf gene using bulk segregant RNA sequencing. Hortic. Res. 2020, 7, 210. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Wang, L.; Song, J.; Ma, G.; Wang, J. Development and utilization of the functional co-dominant KASP marker for thermo-sensitive genic male sterility in rice Oryza sativa L. Genet. Resour. Crop Evol. 2022, 69, 635–643. [Google Scholar] [CrossRef]
- Rasheed, A.; Wen, W.; Gao, F.; Zhai, S.; Jin, H.; Liu, J.; Guo, Q.; Zhang, Y.; Dreisigacker, S.; Xia, X.; et al. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 2016, 129, 1843–1860. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Patterson, E.L.; Fleming, M.B.; Kessler, K.C.; Nissen, S.J.; Gaines, T.A. A KASP genotyping method to identify Northern watermilfoil, Eurasian watermilfoil, and their interspecific hybrids. Front. Plant Sci. 2017, 8, 752. [Google Scholar] [CrossRef]
- Oliveira, M.C.; Gaines, T.A.; Patterson, E.L.; Jhala, A.J.; Irmak, S.; Amundsen, K.; Knezevic, S.Z. Interspecific and intraspecific transference of metabolism-based mesotrione resistance in dioecious weedy Amaranthus. Plant J. 2018, 96, 1051–1063. [Google Scholar] [CrossRef] [Green Version]
- Horn, R.; Friedt, W. Fertility restoration of new CMS sources in sunflower (Helianthus annuus L.). Plant Breed. 1997, 116, 317–322. [Google Scholar] [CrossRef]
- Li, H.; Bariana, H.; Singh, D.; Zhang, L.; Dillon, S.; Whan, A.; Bansal, U.; Ayliffe, M. A durum wheat adult plant stripe rust resistance QTL and its relationship with the bread wheat Yr80 locus. Theor. Appl. Genet. 2020, 133, 3049–3066. [Google Scholar] [CrossRef]
- Grewal, S.; Hubbart-Edwards, S.; Yang, C.; Devi, U.; Baker, L.; Heath, J.; Ashling, S.; Scholefield, D.; Howells, C.; Yarde, J.; et al. Rapid identification of homozygosity and site of wild relative introgressions in wheat through chromosome-specific KASP genotyping assays. Plant Biotechnol. J. 2020, 18, 743–755. [Google Scholar] [CrossRef] [Green Version]
- Sajer, O.; Schirmak, U.; Hamrit, S.; Horn, R. Mapping of the new fertility restorer gene Rf-PET2 close to Rf1 on linkage group 13 in sunflower (Helianthus annuus L.). Genes 2020, 11, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brusa, A.; Patterson, E.L.; Gaines, T.A.; Dorn, K.; Westra, P.; Sparks, C.D.; Wyse, D. A needle in a seedstack: An improved method for detection of rare alleles in bulk seed testing through KASP. Pest Manag. Sci. 2021, 77, 2477–2484. [Google Scholar] [CrossRef] [PubMed]
- OECD Seed Schemes Rules and Regulations 2021, OECD Schemes for the Varietal Certification or the Control of Seed Moving in International Trade. Available online: https://www.oecd.org/agriculture/seeds/documents/oecd-seed-schemes-rules-and-regulations.pdf (accessed on 13 January 2022).
- Chen, Z.; Tang, D.; Ni, J.; Li, P.; Wang, L.; Zhou, J.; Li, C.; Lan, H.; Li, L.; Liu, J. Development of genic KASP SNP markers from RNA-Seq data for map-based cloning and marker-assisted selection in maize. BMC Plant Biol. 2021, 21, 157. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Wen, Z.; Gu, C.; Wang, D. Introduction of high throughput and cost effective SNP genotyping platforms in soybean. Plant. Genet. Genome Biotech. 2014, 2, 90–94. [Google Scholar] [CrossRef]
- Josia, C.; Mashingaidze, K.; Amelework, A.B.; Kondwakwenda, A.; Musvosvi, C.; Sibiya, J. SNP-based assessment of genetic purity and diversity in maize hybrid breeding. PLoS ONE 2021, 16, e0249505. [Google Scholar] [CrossRef]
- Satya Srii, V.; Nethra, N.; Umarani, K.; Lohithaswa, H.C.; Shadakshari, Y.G.; Rajendra Prasad, S. SNP genotyping of maize (Zea mays) hybrids and parental inbred lines for genetic purity testing using double digest restriction site-associated DNA sequencing. Seed Sci. Technol. 2021, 49, 193–206. [Google Scholar] [CrossRef]
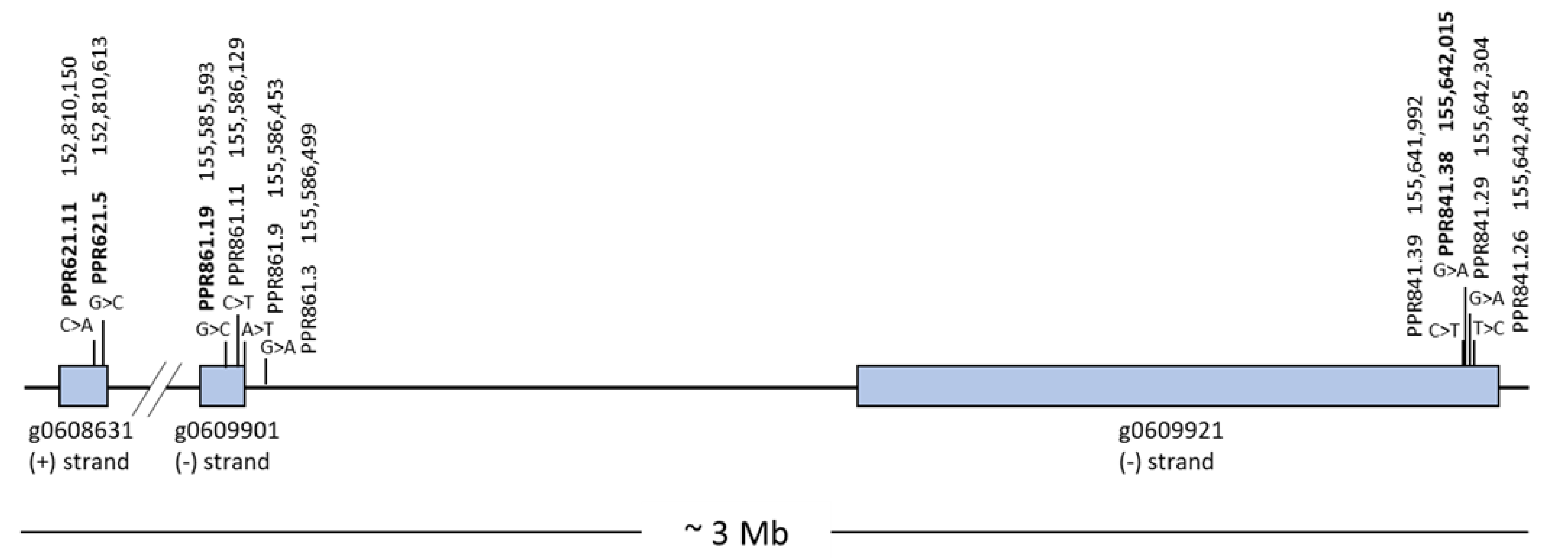


| Primer Name | Primer Sequences (5′-3′) 1 |
|---|---|
| PPR621.5 (G/C) | |
| 621.5 F1_FAM | GAAGGTGACCAAGTTCATGCTACCAGTAATCTCCACATGAACATTG |
| 621.5 F2_HEX | GAAGGTCGGAGTCAACGGATTACCAGTAATCTCCACATGAACATTC |
| 621.5 R1 | GCGATAAAGAAGCGGGAGATTA |
| PPR621.11 (C/A) | |
| 621.11 F3_FAM | GAAGGTGACCAAGTTCATGCTGCGGACGCTTGTATGTTC |
| 621.11 F4_HEX | GAAGGTCGGAGTCAACGGATTGCGGACGCTTGTATGTTA |
| 621.11 R3 | TACGGGTGGACCCACAT |
| PPR841.38 (G/A) | |
| 841.38 F3_FAM | GAAGGTGACCAAGTTCATGCTGCAAAGCACTTGTTTCGTAG |
| 841.38 F4_HEX | GAAGGTCGGAGTCAACGGATTGCAAAGCACTTGTTTCGTAA |
| 841.38 R3 | ATCCCTGGAGAAGAACATTGT |
| PPR861.19 (G/C) | |
| 861.19 F3_FAM | GAAGGTGACCAAGTTCATGCTAAAAGAAATGGAGGAGGATG |
| 861.19 F4_HEX | GAAGGTCGGAGTCAACGGATTAAAAGAAATGGAGGAGGATC |
| 861.19 R3 | CTTCATGCACCTTACCTTCC |
| Common Name | Sequence-Based SNP Analyses | KASP Marker 1 | ||||||
|---|---|---|---|---|---|---|---|---|
| PPR621.5 | PPR621.11 | PPR841.38 | PPR861.19 | 621.5 | 621.11 | 841.38 | 861.19 | |
| Armavirsky 3497 | GG | CC | GG | GG | GG | CC | GG | GG |
| Arrowhead | GG | CC | GG | GG | GG | CC | GG | GG |
| CM259 | GG | CC | GG | GG | GG | CC | GG | GG |
| CM63 | GG | CC | GG | GG | GG | CC | - | - |
| Krasnodaret | GG | CC | GG | GG | GG | CC | GG | GG |
| No. 2 | GG | CC | GG | GG | GG | CC | GG | GG |
| UGA-SAM1-082 | GG | CC | GG | GG | GG | CC | GG | GG |
| UGA-SAM1-109 | GG | CC | GG | GG | GG | CC | GC | GG |
| UGA-SAM1-156 | GG | CC | GG | GG | GG | CC | GG | GG |
| UGA-SAM1-185 | GG | CC | GG | GG | GG | CC | GG | GG |
| HA342 | - | - | - | - | GG | CC | GG | GG |
| HA383 | - | - | - | - | GG | CC | GG | GG |
| UGA-SAM1-010 | CC | AA | AA | CC | CC | AA | AA | CC |
| UGA-SAM1-024 | CC | AA | AA | CC | CC | AA | AA | CC |
| UGA-SAM1-100 | CC | AA | AA | CC | CC | AA | AA | CC |
| UGA-SAM1-101 | CC | AA | AA | CC | CC | AA | AA | CC |
| UGA-SAM1-121 | CC | AA | AA | CC | CC | AA | AA | CC |
| UGA-SAM1-136 | CC | AA | AA | CC | CC | AA | AA | CC |
| UGA-SAM1-161 | CC | AA | AA | CC | CC | AA | AA | CC |
| UGA-SAM1-169 | CC | AA | AA | CC | CC | AA | AA | CC |
| UGA-SAM1-191 | CC | AA | AA | CC | CC | AA | AA | CC |
| UGA-SAM1-204 | CC | AA | AA | CC | CC | AA | AA | CC |
| RHA325 | - | - | - | - | CC | AA | AA | CC |
| RHA265 | - | - | - | - | CC | AA | AA | CC |
| IH-51 | - | - | - | - | GG | CC | GG | GG |
| NS-H-27 | - | - | - | - | CC | AA | AA | CC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radanović, A.; Sprycha, Y.; Jocković, M.; Sundt, M.; Miladinović, D.; Jansen, C.; Horn, R. KASP Markers Specific for the Fertility Restorer Locus Rf1 and Application for Genetic Purity Testing in Sunflowers (Helianthus annuus L.). Genes 2022, 13, 465. https://doi.org/10.3390/genes13030465
Radanović A, Sprycha Y, Jocković M, Sundt M, Miladinović D, Jansen C, Horn R. KASP Markers Specific for the Fertility Restorer Locus Rf1 and Application for Genetic Purity Testing in Sunflowers (Helianthus annuus L.). Genes. 2022; 13(3):465. https://doi.org/10.3390/genes13030465
Chicago/Turabian StyleRadanović, Aleksandra, Yves Sprycha, Milan Jocković, Monja Sundt, Dragana Miladinović, Constantin Jansen, and Renate Horn. 2022. "KASP Markers Specific for the Fertility Restorer Locus Rf1 and Application for Genetic Purity Testing in Sunflowers (Helianthus annuus L.)" Genes 13, no. 3: 465. https://doi.org/10.3390/genes13030465
APA StyleRadanović, A., Sprycha, Y., Jocković, M., Sundt, M., Miladinović, D., Jansen, C., & Horn, R. (2022). KASP Markers Specific for the Fertility Restorer Locus Rf1 and Application for Genetic Purity Testing in Sunflowers (Helianthus annuus L.). Genes, 13(3), 465. https://doi.org/10.3390/genes13030465







