RNA-Seq Reveals Differentially Expressed Genes Associated with High Fiber Quality in Abaca (Musa textilis Nee)
Abstract
:1. Introduction
2. Materials and Methods
2.1. RNA Extraction
2.2. Bioinformatics Pipeline for Differential Expression
2.2.1. Reads Pre-Processing
2.2.2. Mapping
2.2.3. Read Count Quantification
2.2.4. Differential Expression (DE)
2.2.5. Gene Ontology (GO) Enrichment Analysis
2.3. Ethical Standards on the Use of Plant Materials
2.4. Availability of Data and Materials
3. Results and Discussion
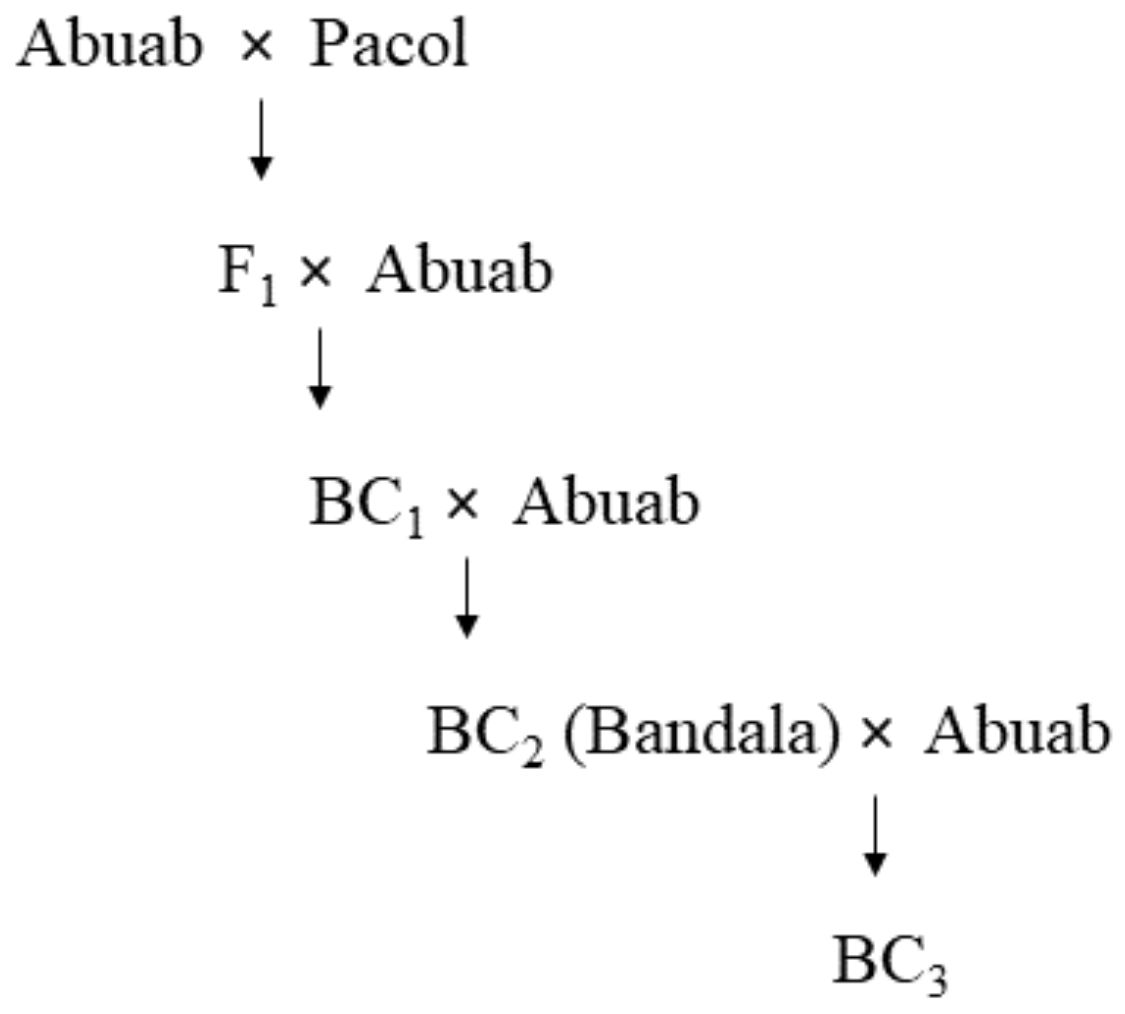
| Varieties | Resistance/Susceptibility vs. AbBTV | Fiber Quality | References |
|---|---|---|---|
| Abuab (M. textilis) | S | High | [38] |
| Inosa (M. textilis) | S | High | [38,39] |
| Tangongon (M. textilis) | S | High | [40] |
| BC2 (87.5% Abuab; 12.5% Pacol) | R | High | [15,38,41,42] |
| BC3 (93.75% Abuab; 6.25% Pacol) * | R | High | |
| Pacol (M. balbisiana) (wild banana) | R | Low | [38] |
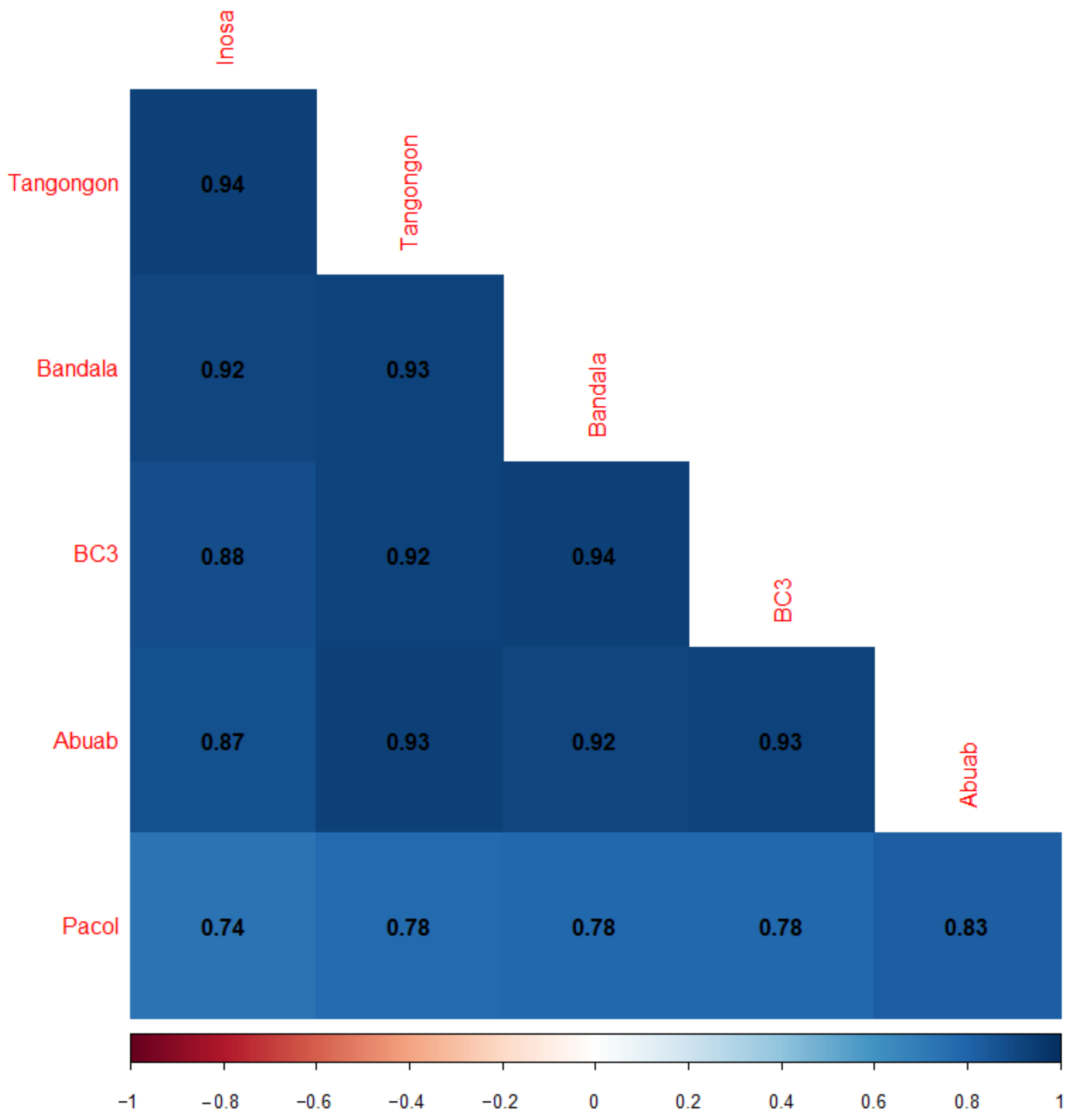
3.1. RNA-Seq Data Information
3.2. DE between and among the Varieties
3.2.1. Pairwise Differential Expression (PDE)
3.2.2. Genotypic Differential Expression between Musa Groups
3.2.3. Non-Differential Expression (NDE) Model across Abaca Varieties
3.2.4. GDE between Resistant and Susceptible Varieties
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- PHILFIDA. Philippine Fiber Industry Development Authority. Available online: http://www.philfida.da.gov.ph/index.php/archived-articles/19-philippine-abaca-helps-in-global-environment-conservation (accessed on 14 May 2020).
- Ocampo, K.R. Demand Rises for Philippine Abaca as Raw Material for PPE. Inquirer. Available online: https://newsinfo.inquirer.net/ (accessed on 10 May 2020).
- Delicano, J.A. A review on abaca fiber reinforced composites. Compos. Interfaces 2018, 25, 1039–1066. [Google Scholar] [CrossRef]
- Barba, B.J.; Madrid, J.F.; Penaloza, D.P. A review of abaca fiber-reinforced polymer composites: Different modes of preparation and their applications. J. Chil. Chem. Soc. 2020, 65, 4919–4924. [Google Scholar] [CrossRef]
- GBIF. Global Biodiversity Information Facility. Available online: https://www.gbif.org/species/113660435 (accessed on 15 May 2020).
- Halos, S.C. The Abaca; Department of Agriculture-Biotechnology Program Office: Quezon City, Philippines, 2008. [Google Scholar]
- D’Hont, A.; Denoeud, F.; Aury, J.M.; Baurens, F.-C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suthanthiram, B.; Uma, S.; Saraswathi, M.; Saravanakumar, A.; Arumugam, C. Transcriptome analysis of banana (Musa balbisiana) based on next-generation sequencing technology. Turk. J. Agric. For. 2015, 39, 705–717. [Google Scholar] [CrossRef]
- Galvez, L.C.; Koh, R.B.L.; Barbosa, C.F.C.; Asunto, J.C.; Catalla, J.L.; Atienza, R.G.; Costales, K.T.; Aquino, V.M.; Zhang, D. Sequencing and de Novo Assembly of Abaca (Musa textilis Née) var. Abuab Genome. Genes 2021, 12, 1202. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.E. Abaca and the Philippines. Econ. Geogr. 1951, 27, 95–106. [Google Scholar] [CrossRef]
- Lasalita-Zapico, F.C.; Aguilar, C.H.M.; Aujero, J.M. Phenotypic variability of Manila hemp (Musa textilis L. Nee) genotypes in southern Mindanao Island, Philippines. J. Trop. Agric. 2010, 48, 68–70. [Google Scholar]
- Bhattacharyya, D.; Subasinghe, A.; Kim, N.K. Chapter 4—Natural fibers: Their composites and flammability characterizations. In Multifunctionality of Polymer Composites; Friedrich, K., Breuer, U., Eds.; William Andrew Publishing: Norwich, NY, USA, 2015; pp. 102–143. [Google Scholar] [CrossRef]
- Armecin, R.; Gabon, F. Biomass, organic carbon and mineral matter contents of abaca (Musa textilis Nee) at different stages of growth. Ind. Crop. Prod. 2008, 28, 340–345. [Google Scholar] [CrossRef]
- Kumar, P.L.; Selvarajan, R.; Iskra-Caruana, M.L.; Chabannes, M.; Hanna, R. Biology, Etiology, and Control of Virus Diseases of Banana and Plantain. Adv. Virus Res. 2014, 91, 229–269. [Google Scholar]
- Lalusin, A.G.; Villavicencio, M.L.H. Abaca (Musa textilis Nee) Breeding in the Philippines. In Industrial Crops; Breeding for Bioenergy and Bioproducts; Springer: New York, NY, USA, 2015; pp. 265–289. [Google Scholar]
- PHILFIDA. 2017. Available online: http://www.philfida.da.gov.ph/index.php/news-articles/116-3-abaca-varities-now-regsitered-with-nsic (accessed on 31 August 2021).
- Biswas, S.; Agrawal, Y.N.; Mucyn, T.S.; Dang, J.L.; Jones, C.D. Biological Averaging in RNA-Seq. arXiv 2013, arXiv:1309.0670. [Google Scholar]
- Assefa, A.T.; Vandesompele, J.; Thas, O. On the utility of RNA sample pooling to optimize cost and statistical power in RNA sequencing experiments. BMC Genom. 2020, 21, 312. [Google Scholar] [CrossRef]
- Miao, G.; Qin, Y.; Guo, J.; Zhang, Q.; Bao, Y. Transcriptome characterization and expression profile of Coix lacryma-jobi L. in response to drought. PLoS ONE 2021, 16, e0256875. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, T.; Rao, P.; Gao, K.; Yang, X.; Chen, Z.; An, X. Transcriptome profiling of Populus tomentosa under cold stress. Ind. Crop. Prod. 2019, 135, 283–293. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 15 September 2021).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Wang, Z.; Miao, H.; Liu, J.; Xu, B.; Yao, X.; Xu, C.; Zhao, S.; Fang, X.; Jia, C.; Wang, J.; et al. Musa balbisiana genome reveals subgenome evolution and functional divergence. Nat. Plants 2019, 5, 810–821. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup, the Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. Feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Ballereau, S.; Couturier, D.L.; Dunning, M.; Edwards, A.; Sawle, A. RNA-seq Analysis in R: Counting Reads with SubRead. 2019. Available online: https://bioinformatics-core-shared-training.github.io/ (accessed on 7 September 2021).
- Hardcastle, T.J. BaySeq: Empirical Bayesian Analysis of Patterns of Differential Expression in Count Data. 2021. Available online: https://bioconductor.org/packages/release/bioc/html/baySeq.html (accessed on 9 September 2021).
- Hardcastle, T.J.; Kelly, K.A. BaySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinform. 2010, 11, 422. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Bell, G.D.; Kane, N.C.; Rieseberg, L.H.; Adams, K.L. RNA-Seq Analysis of Allele-Specific Expression, Hybrid Effects, and Regulatory Divergence in Hybrids Compared with Their Parents from Natural Populations. Genome Biol. Evol. 2013, 5, 1309–1323. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Ng, D.W.-K.; Zhang, C.; Comai, L.; Ye, W.; Chen, Z.J. Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat. Commun. 2012, 3, 950. [Google Scholar] [CrossRef]
- BioBam Bioinformatics. OmicsBox—Bioinformatics Made Easy (Version 2.0.29). 2019. Available online: https://www.biobam.com/omicsbox (accessed on 12 September 2019).
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [Green Version]
- Labrador, D.A.; Lalusin, A.G.; Mendoza, M.R.R.; dela Viña, C.B. Morphological Characterization and Karyotype Analysis of Abaca (Musa textilis Nee) and Its Hybrids with Musa balbisiana Colla. Philipp. Agric. Sci. 2020, 103, 13–28. [Google Scholar]
- Parac, E.P.; Lalusin, A.G.; Pangga, I.B.; Sta Cruz, F.C. Characteristics of Selected Hybrids of Abaca (Musa textilis Nee) with Resistance to Bunchy Top. Philipp. Agric. Sci. 2020, 103, 1–12. [Google Scholar]
- Boguero, A.P.B.; Parducho, M.A.L.; Mendoza, M.R.; Abustan, M.A.; Lalusin, A.G. Molecular Screening of Abaca (Musa textilis Nee). Philipp. J. Crop Sci. 2016, 41, 13–19. [Google Scholar]
- CFC; UNIDO; FAO; FIDA. Abaca Improvement of Fiber Extraction and Identification of Higher Yielding Varieties. Final Technical Report CFC/FIGHF/09. Activities in the Philippines. 2004. Available online: https://www.yumpu.com/en/document/view/27575439/abaca-activities-in-the-philippines-unido (accessed on 1 September 2021).
- Lalusin, A.G. Revitalizing the Abaca Industry through S & T Interventions for Higher Crop Productivity Using High-Yielding and Bunchy Top-Resistant Abaca Hybrids; Terminal Report 2016–2020; Institute of Plant Breeding, College of Agriculture and Food Science, University of the Philippines: Laguna, Philippines, 2020. [Google Scholar]
- Parducho, M.A.L.; Rama RA, B.; Lalusin, A.G. Stability Analysis of BC2 Abaca (Musa textilis Nee) Hybrids across Different Locations in the Philippines. Philipp. J. Crop Sci. PJCS 2020, 45, 44–49. [Google Scholar]
- Zhou, Y.; Zhu, J.; Tong, T.; Wang, J.; Lin, B.; Zhang, J. A statistical normalization method and differential expression analysis for RNA-seq data between different species. BMC Bioinform. 2019, 20, 163. [Google Scholar] [CrossRef]
- Galvez, L.C.; Catalla, J.L.; Borromeo, T.H.; Altoveros, N.C. Abaca Germplasm Conservation; Philippine Fiber Industry Development Authority: Quezon City, Philippines, 2018.
- Bao, Y.; Hu, G.; Grover, C.E.; Conover, J.; Yuan, D.; Wendel, J.F. Unraveling cis and trans regulatory evolution during cotton domestication. Nat. Commun. 2019, 10, 5399. [Google Scholar] [CrossRef] [Green Version]
- Lovell, J.T.; Schwartz, S.; Lowry, D.B.; Shakirov, E.V.; Bonnette, J.E.; Weng, X.; Wang, M.; Johnson, J.; Sreedasyam, A.; Plott, C.; et al. Drought responsive gene expression regulatory divergence between upland and lowland ecotypes of a perennial C4 grass. Genome Res. 2016, 26, 510–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ereful, N.C.; Laurena, A.; Liu, L.-Y.; Kao, S.-M.; Tsai, E.; Greenland, A.; Powell, W.; Mackay, I.; Leung, H. Unraveling regulatory divergence, heterotic malleability, and allelic imbalance switching in rice due to drought stress. Sci. Rep. 2021, 11, 13489. [Google Scholar] [CrossRef] [PubMed]
- Saragih, S.W.; Wirjosentono, B.; Eddyanto, M.Y. Extraction and Characterization of Cellulose from Abaca Pseudo Stem (Musa textilis). J. Phys. Conf. Ser. 2019, 1232, 012018. [Google Scholar] [CrossRef]
- Sinha, A.K.; Bhattacharya, S.; Narang, H.K. Abaca fibre reinforced polymer composites: A review. J. Mater. Sci. 2021, 56, 4569–4587. [Google Scholar] [CrossRef]
- Uniprot. Available online: https://www.uniprot.org/ (accessed on 14 September 2021).
- Wang, G.F.; He, Y.; Strauch, R.; Olukolu, B.A.; Nielsen, D.; Li, X.; Balint-Kurti, P.J. Maize Homologs of Hydroxycinnamoyltransferase, a Key Enzyme in Lignin Biosynthesis, Bind the Nucleotide Binding Leucine-Rich Repeat Rp1 Proteins to Modulate the Defense Response. Plant Physiol. 2015, 169, 2230–2243. [Google Scholar] [PubMed]
- Serrani-Yarce, J.C.; Escamilla-Trevino, L.; Barros, J.; Gallego-Giraldo, L.; Pu, Y.; Ragauskas, A.; Dixon, R.A. Targeting hydroxycinnamoyl CoA: Shikimate hydroxycinnamoyl transferase for lignin modification in Brachypodium distachyon. Biotechnol. Biofuels 2021, 14, 50. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, G.V.; Da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Li, S.; Sun, T.; Ren, H. The functions of the cytoskeleton and associated proteins during mitosis and cytokinesis in plant cells. Front. Plant Sci. 2015, 6, 282. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Zhao, J.; Fu, G.; Pei, X.; Pan, Z.; Li, H.; Ahmed, H.; He, S.; DU, X. Identification and expression analysis of Tubulin gene family in upland cotton. J. Cotton Res. 2021, 4, 20. [Google Scholar] [CrossRef]
- Pydiura, N.; Pirko, Y.; Galinousky, D.; Postovoitova, A.; Yemets, A.; Kilchevsky, A.; Blume, Y. Genome-wide identification. Phylogenetic classification. And exon-intron structure characterization of the tubulin and actin genes in flax (Linum usitatissimum). Cell Biol. Int. 2019, 43, 1010–1019. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 12 September 2021).
- Vanholme, R.; Cesarino, I.; Rataj, K. Caffeoyl Shikimate Esterase (CSE) Is an Enzyme in the Lignin Biosynthetic Pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [Green Version]
- Del Río, J.C.; Prinsen, P.; Cadena, E.M.; Martínez, Á.T.; Gutiérrez, A.; Rencoret, J. Lignin-carbohydrate complexes from sisal (Agave sisalana) and abaca (Musa textilis): Chemical composition and structural modifications during the isolation process. Planta 2016, 243, 1143–1158. [Google Scholar] [CrossRef]
- Del Río, J.C.; Gutiérrez, A. Chemical composition of abaca (Musa textilis) leaf fibers used for manufacturing of high quality paper pulps. J. Agric. Food. Chem. 2006, 54, 13, 4600–4610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging Insights into the Functions of Pathogenesis-Related Protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef]







| Sample | Sample Quality Metrics | Pre-Library Prep Info | ||||
|---|---|---|---|---|---|---|
| Conc. (ng/μL) | Volume (μL) | Total Amount (μg) | Conc. (ng/μL) | Conc. (nM) | Result | |
| Abuab | 462.418 | 21 | 9.711 | 25.1 | 105 | Pass |
| BC2 (Bandala) | 137.261 | 21 | 2.882 | 4.64 | 19.9 | Pass |
| BC3 | 339.573 | 21 | 7.131 | 9.52 | 40.7 | Pass |
| Inosa | 150.325 | 21 | 3.157 | 2.65 | 10.9 | Pass |
| Tangongon | 366.006 | 21 | 7.686 | 7.07 | 29.8 | Pass |
| Pacol | 577.261 | 21 | 12.122 | 11.8 | 51.3 | Pass |
| Sample | Total Read Bases (bp) | Total Reads | Q30 (%) |
|---|---|---|---|
| Abuab | 3,487,334,262 | 34,528,062 | 95.28 |
| BC3 | 4,327,484,582 | 42,846,382 | 95.49 |
| BC2 (Bandala) | 3,225,244,514 | 31,933,114 | 95.36 |
| Inosa | 4,690,066,098 | 46,436,298 | 95.3 |
| Pacol | 3,186,258,918 | 31,547,118 | 95.76 |
| Tangongon | 3,097,216,712 | 30,665,512 | 94.93 |
| Variety | Uniquely Mapped Reads (%) | Reads Mapped to Multiple Loci (%) | Reads Unmapped (%) |
|---|---|---|---|
| Abuab | 89.12 | 6.59 | 3.68 |
| Tangongon | 87.00 | 2.57 | 10.35 |
| Inosa | 83.11 | 3.43 | 13.35 |
| BC2 | 59.96 | 3.03 | 35.87 |
| BC3 | 65.37 | 2.66 | 31.40 |
| Pacol | 43.10 | 32.80 | 9.14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ereful, N.C.; Lalusin, A.G.; Laurena, A.C. RNA-Seq Reveals Differentially Expressed Genes Associated with High Fiber Quality in Abaca (Musa textilis Nee). Genes 2022, 13, 519. https://doi.org/10.3390/genes13030519
Ereful NC, Lalusin AG, Laurena AC. RNA-Seq Reveals Differentially Expressed Genes Associated with High Fiber Quality in Abaca (Musa textilis Nee). Genes. 2022; 13(3):519. https://doi.org/10.3390/genes13030519
Chicago/Turabian StyleEreful, Nelzo C., Antonio G. Lalusin, and Antonio C. Laurena. 2022. "RNA-Seq Reveals Differentially Expressed Genes Associated with High Fiber Quality in Abaca (Musa textilis Nee)" Genes 13, no. 3: 519. https://doi.org/10.3390/genes13030519
APA StyleEreful, N. C., Lalusin, A. G., & Laurena, A. C. (2022). RNA-Seq Reveals Differentially Expressed Genes Associated with High Fiber Quality in Abaca (Musa textilis Nee). Genes, 13(3), 519. https://doi.org/10.3390/genes13030519





