Tissue-Specific Expression of the Terpene Synthase Family Genes in Rosa chinensis and Effect of Abiotic Stress Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Identification of TPS Genes in R. chinensis
2.3. Chromosomal Distribution, Duplication Event of RcTPS Genes
2.4. Gene Structure, Conserved Motif and Phylogenetic Analysis
2.5. Analysis of Cis-Acting Elements in the Promoter Region of RcTPS Genes
2.6. Gene Expression Analysis Based on Transcriptome Data
2.7. qRT-PCR Analysis of RcTPS Genes under Different Treatment
3. Results
3.1. Identification, Classification and Properties of RcTPS Genes
3.2. Chromosomal Distribution, Duplication Event of RcTPS Genes
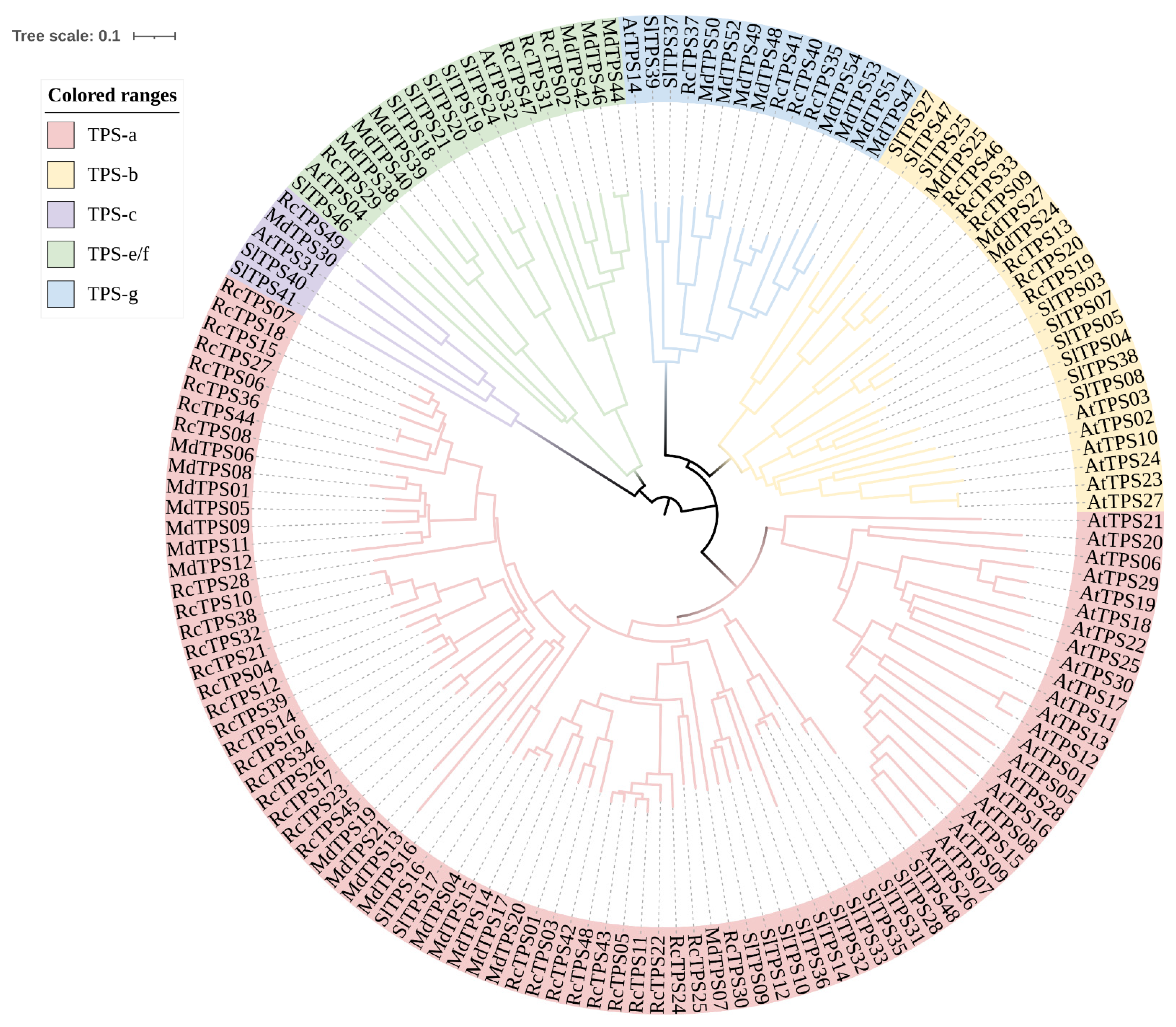
3.3. Phylogenetic Analysis of RcTPS Genes in R. chinensis
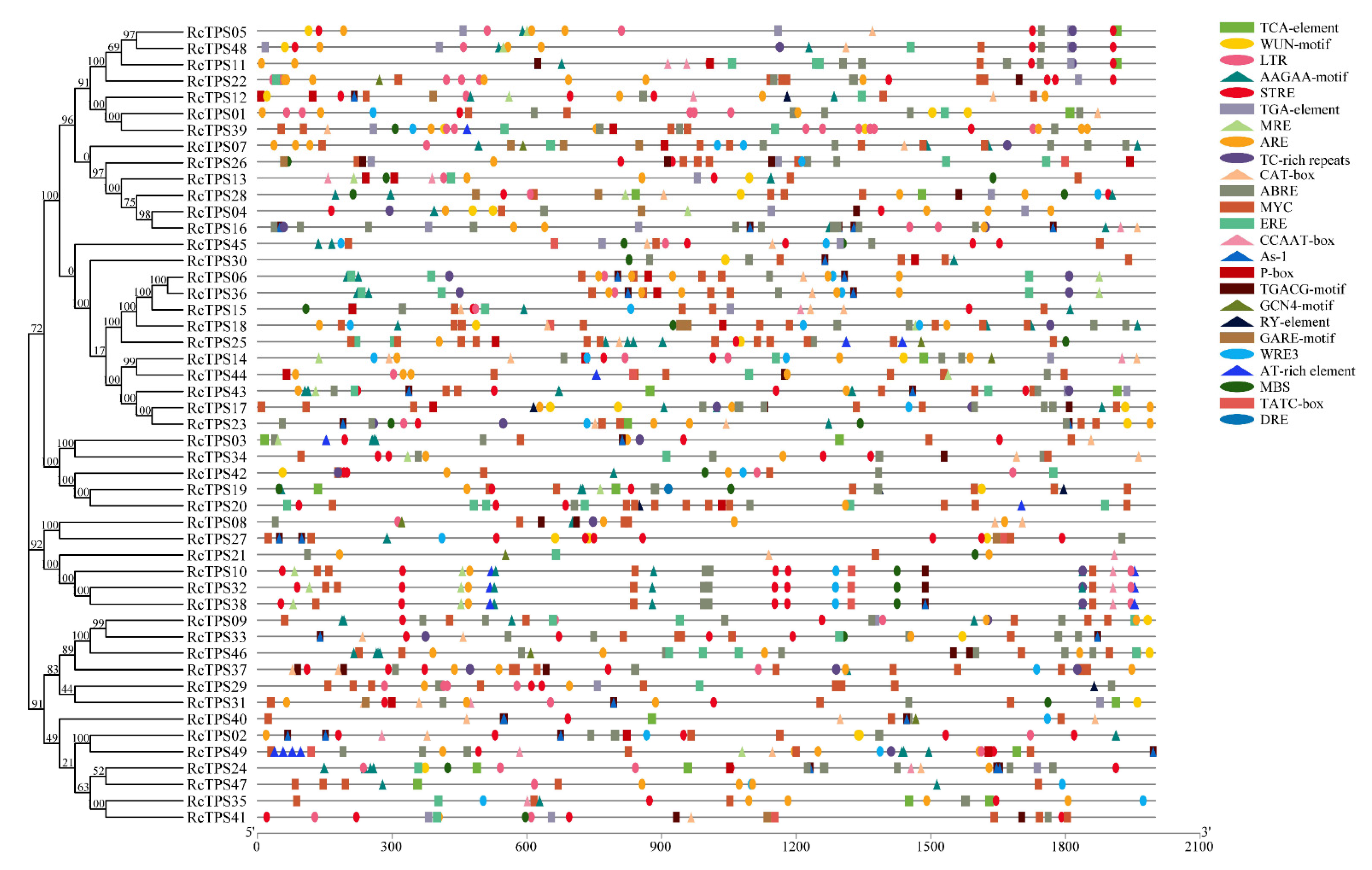
3.4. Gene Structure and Conserved Motif Analysis
3.5. Analysis of Cis-Acting Elements in the Promoter Region of RcTPS Genes
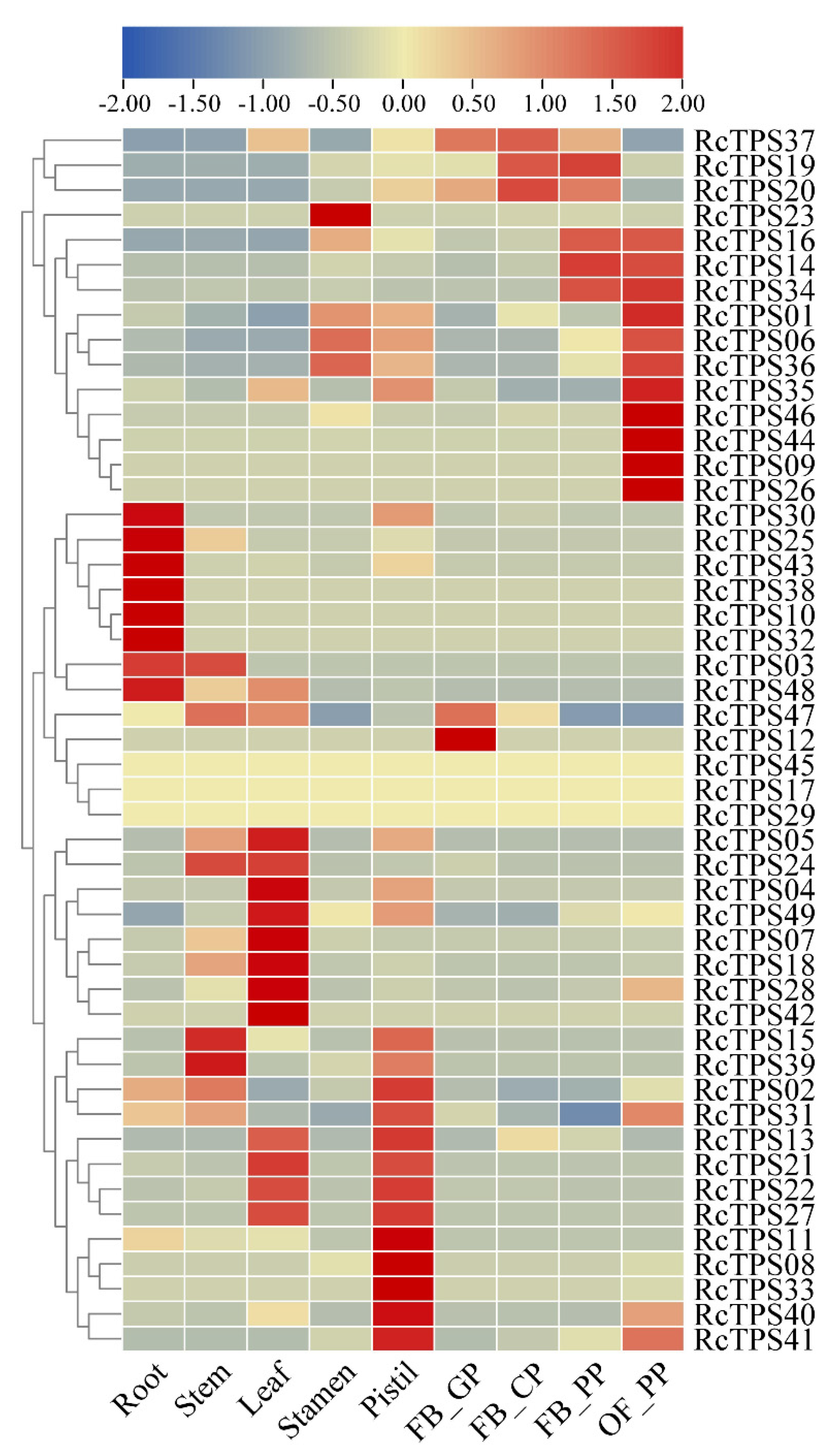
3.6. Expression Patterns of RcTPS Genes in Different Tissues of R. chinensis
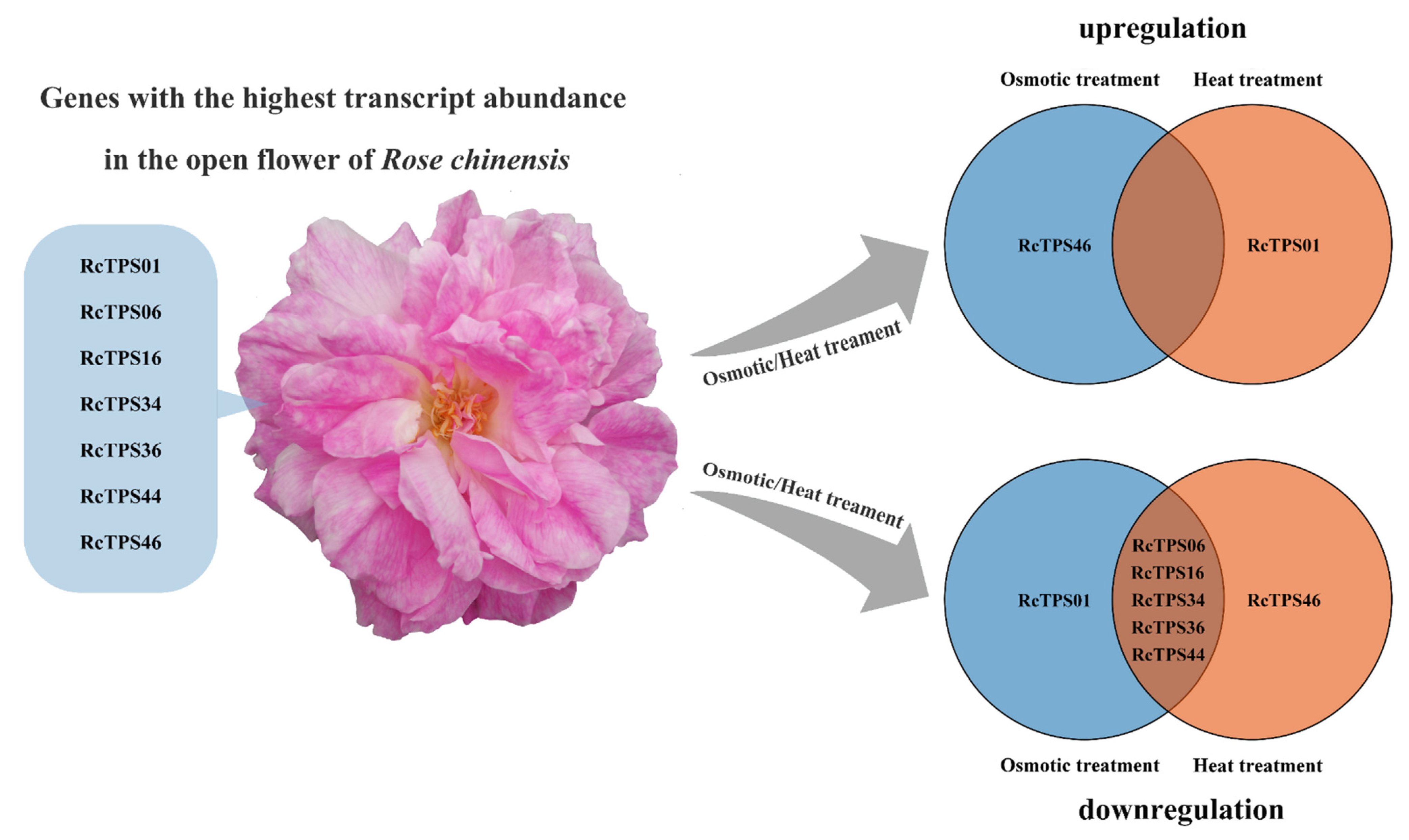
3.7. Expression Patterns of RcTPS Genes under Abiotic Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tholl, D. Biosynthesis and Biological Functions of Terpenoids in Plants. In Advances in Biochemical Engineering-Biotechnology; Schrader, J., Bohlmann, J., Eds.; Annual Reviews: San Mateo, CA, USA, 2015; Volume 148, pp. 63–106. [Google Scholar]
- Boncan, D.A.T.; Tsang, S.S.K.; Li, C.; Lee, I.H.T.; Lam, H.-M.; Chan, T.-F.; Hui, J.H.L. Terpenes and Terpenoids in Plants: Interactions with Environment and Insects. Int. J. Mol. Sci. 2020, 21, 7382. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef] [PubMed]
- McGarvey, D.J.; Croteau, R. TERPENOID METABOLISM. Plant Cell 1995, 7, 1015–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichtenthaler, H.K. The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Bohlmann, J.; Meyer-Gauen, G.; Croteau, R. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 1998, 95, 4126–4133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vranova, E.; Coman, D.; Gruissem, W. Network Analysis of the MVA and MEP Pathways for Isoprenoid Synthesis. In Annual Review of Plant Biology; Merchant, S.S., Ed.; Springer: Berlin, Germany, 2013; Volume 64, pp. 665–700. [Google Scholar]
- Boutanaev, A.M.; Moses, T.; Zi, J.; Nelson, D.R.; Mugford, S.T.; Peters, R.J.; Osbourn, A. Investigation of terpene diversification across multiple sequenced plant genomes. Proc. Natl. Acad. Sci. USA 2015, 112, E81–E88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichersky, E.; Noel, J.P.; Dudareva, N. Biosynthesis of plant volatiles: Nature's diversity and ingenuity. Science 2006, 311, 808–811. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Honzatko, R.B.; Peters, R.J. Terpenoid synthase structures: A so far incomplete view of complex catalysis. Nat. Prod. Rep. 2012, 29, 1153–1175. [Google Scholar] [CrossRef] [Green Version]
- Hansen, N.L.; Heskes, A.M.; Hamberger, B.; Olsen, C.E.; Hallstrom, B.M.; Andersen-Ranberg, J.; Hamberger, B. The terpene synthase gene family in Tripterygium wilfordii harbors a labdane-type diterpene synthase among the monoterpene synthase TPS-b subfamily. Plant J. 2017, 89, 429–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christianson, D.W. Structural and Chemical Biology of Terpenoid Cyclases. Chem. Rev. 2017, 117, 11570–11648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Degenhardt, J.; Koellner, T.G.; Gershenzon, J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 2009, 70, 1621–1637. [Google Scholar] [CrossRef]
- Jiang, S.-Y.; Jin, J.; Sarojam, R.; Ramachandran, S. A Comprehensive Survey on the Terpene Synthase Gene Family Provides New Insight into Its Evolutionary Patterns. Genome Biol. Evol. 2019, 11, 2078–2098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zerbe, P.; Bohlmann, J. Plant diterpene synthases: Exploring modularity and metabolic diversity for bioengineering. Trends Biotechnol. 2015, 33, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Martin, D.; Kish, C.M.; Kolosova, N.; Gorenstein, N.; Faldt, J.; Miller, B.; Bohlmann, J. (E)-β-ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: Function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 2003, 15, 1227–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubourg, S.; Lecharny, A.; Bohlmann, J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genom. 2002, 267, 730–745. [Google Scholar] [CrossRef]
- Martin, D.M.; Aubourg, S.; Schouwey, M.B.; Daviet, L.; Schalk, M.; Toub, O.; Lund, S.T.; Bohlmann, J. Functional Annotation, Genome Organization and Phylogeny of the Grapevine (Vitis vinifera) Terpene Synthase Gene Family Based on Genome Assembly, FLcDNA Cloning, and Enzyme Assays. Bmc Plant Biol. 2010, 10. [Google Scholar] [CrossRef] [Green Version]
- Falara, V.; Akhtar, T.A.; Nguyen, T.T.H.; Spyropoulou, E.A.; Bleeker, P.M.; Schauvinhold, I.; Matsuba, Y.; Bonini, M.E.; Schilmiller, A.L.; Last, R.L.; et al. The Tomato Terpene Synthase Gene Family. Plant Physiol. 2011, 157, 770–789. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Pichersky, E. The complete functional characterisation of the terpene synthase family in tomato. New Phytol. 2020, 226, 1341–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieuwenhuizen, N.J.; Green, S.A.; Chen, X.; Bailleul, E.J.D.; Matich, A.J.; Wang, M.Y.; Atkinson, R.G. Functional Genomics Reveals That a Compact Terpene Synthase Gene Family Can Account for Terpene Volatile Production in Apple. Plant Physiol. 2013, 161, 787–804. [Google Scholar] [CrossRef] [Green Version]
- Kuelheim, C.; Padovan, A.; Hefer, C.; Krause, S.T.; Koellner, T.G.; Myburg, A.A.; Degenhardt, J.; Foley, W.J. The Eucalyptus terpene synthase gene family. Bmc Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alquezar, B.; Rodriguez, A.; de la Pena, M.; Pena, L. Genomic Analysis of Terpene Synthase Family and Functional Characterization of Seven Sesquiterpene Synthases from Citrus sinensis. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karunanithi, P.S.; Berrios, D.I.; Wang, S.; Davis, J.; Shen, T.; Fiehn, O.; Maloof, J.N.; Zerbe, P. The foxtail millet (Setaria italica) terpene synthase gene family. Plant J. 2020, 103, 781–800. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-C.; Shamala, L.F.; Yi, X.-K.; Yan, Z.; Wei, S. Analysis of Terpene Synthase Family Genes in Camellia sinensis with an Emphasis on Abiotic Stress Conditions. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhao, C.; Zhang, G.; da Silva, J.A.T.; Duan, J. Genome-Wide Identification and Expression Profile of TPS Gene Family in Dendrobium officinale and the Role of DoTPS10 in Linalool Biosynthesis. Int. J. Mol. Sci. 2020, 21, 5419. [Google Scholar] [CrossRef]
- Li, R.-S.; Zhu, J.-H.; Guo, D.; Li, H.-L.; Wang, Y.; Ding, X.-P.; Mei, W.-L.; Chen, Z.-B.; Dai, H.-F.; Peng, S.-Q. Genome-wide identification and expression analysis of terpene synthase gene family in Aquilaria sinensis. Plant Physiol. Biochem. 2021, 164, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.-i.; Kawaide, H.; Notomi, M.; Sakigi, Y.; Matsuo, A.; Nozaki, H. Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. Febs Lett. 2006, 580, 6175–6181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guterman, I.; Shalit, M.; Menda, N.; Piestun, D.; Dafny-Yelin, M.; Shalev, G.; Bar, E.; Davydov, O.; Ovadis, M.; Emanuel, M.; et al. Rose scent: Genomics approach to discovering novel floral fragrance-related genes. Plant Cell 2002, 14, 2325–2338. [Google Scholar] [CrossRef] [Green Version]
- Raymond, O.; Gouzy, J.; Just, J.; Badouin, H.; Verdenaud, M.; Lemainque, A.; Vergne, P.; Moja, S.; Choisne, N.; Pont, C.; et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018, 50, 772. [Google Scholar] [CrossRef]
- Huang, P.; Lin, F.; Li, B.; Zheng, Y. Hybrid-Transcriptome Sequencing and Associated Metabolite Analysis Reveal Putative Genes Involved in Flower Color Difference in Rose Mutants. Plants 2019, 8, 267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendahmane, M.; Dubois, A.; Raymond, O.; Le Bris, M. Genetics and genomics of flower initiation and development in roses. J. Exp. Bot. 2013, 64, 847–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magnard, J.-L.; Roccia, A.; Caissard, J.-C.; Vergne, P.; Sun, P.; Hecquet, R.; Dubois, A.; Hibrand-Saint Oyant, L.; Jullien, F.; Nicole, F.; et al. Biosynthesis of monoterpene scent compounds in roses. Science 2015, 349, 81–83. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.-C.; Horvath, G.; Molnar, P.; Turcsi, E.; Deli, J.; Schrader, J.; Sandmann, G.; Schmidt, H.; Schwab, W. Substrate promiscuity of RdCCD1, a carotenoid cleavage oxygenase from Rosa damascena. Phytochemistry 2009, 70, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Fu, L.; Geng, Z.; Zhao, X.; Liu, Q.; Jiang, X. Physiological Characteristic Changes and Full-Length Transcriptome of Rose (Rosa chinensis) Roots and Leaves in Response to Drought Stress. Plant Cell Physiol. 2020, 61, 2153–2166. [Google Scholar] [CrossRef]
- Qi, W.; Zhang, C.; Wang, W.; Cao, Z.; Li, S.; Li, H.; Zhu, W.; Huang, Y.; Bao, M.; He, Y.; et al. Comparative transcriptome analysis of different heat stress responses between self-root grafting line and heterogeneous grafting line in rose. Hortic. Plant J. 2021, 7, 243–255. [Google Scholar] [CrossRef]
- Chimonidou-Pavlidou, D. Malformation of roses due to drought stress. Sci. Hortic. 2004, 99, 79–87. [Google Scholar] [CrossRef]
- Dela, G.; Or, E.; Ovadia, R.; Nissim-Levi, A.; Weiss, D.; Oren-Shamir, M. Changes in anthocyanin concentration and composition in 'Jaguar' rose flowers due to transient high-temperature conditions. Plant Sci. 2003, 164, 333–340. [Google Scholar] [CrossRef]
- Xie, L.; Zhang, H.; Li, D. Physiological responses of garden roses to hot and humid conditions. Hortic. Sci. 2019, 46, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Sharkey, T.D.; Singsaas, E.L. Why Plants Emit Isoprene. Nature 1995, 374, 769. [Google Scholar] [CrossRef]
- Siwko, M.E.; Marrink, S.J.; de Vries, A.H.; Kozubek, A.; Uiterkamp, A.J.M.S.; Mark, A.E. Does isoprene protect plant membranes from thermal shock? A molecular dynamics study. Biochim. Et Biophys. Acta-Biomembr. 2007, 1768, 198–206. [Google Scholar] [CrossRef] [Green Version]
- Loreto, F.; Schnitzler, J.-P. Abiotic stresses and induced BVOCs. Trends Plant Sci. 2010, 15, 154–166. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The arabidopsis information resource: Making and mining the ‘gold standard’ annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wan, H.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. Comparative RNA-seq analysis of transcriptome dynamics during petal development in Rosa chinensis. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Yu, J.; Zhao, T.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. Dissecting the Genome-Wide Evolution and Function of R2R3-MYB Transcription Factor Family in Rosa chinensis. Genes 2019, 10, 823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Sun, J.; Jiang, A.; Bai, M.; Fan, C.; Liu, J.; Ning, G.; Wang, C. Alternate expression of CONSTANS-LIKE 4 in short days and CONSTANS in long days facilitates day-neutral response in Rosa chinensis. J. Exp. Bot. 2020, 71, 4057–4068. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Q.; Yang, S.; Lin, S.; Bao, M.; Bendahmane, M.; Wu, Q.; Wang, C.; Fu, X. Identification and Characterization of the MADS-Box Genes and Their Contribution to Flower Organ in Carnation (Dianthus caryophyllus L.). Genes 2018, 9, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, X.; Xiao, X.; Chou, K.-C. pLoc-mPlant: Predict subcellular localization of multi-location plant proteins by incorporating the optimal GO information into general PseAAC. Mol. Biosyst. 2017, 13, 1722–1727. [Google Scholar] [CrossRef]
- Rynkiewicz, M.J.; Cane, D.E.; Christianson, D.W. Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade. Proc. Natl. Acad. Sci. USA 2001, 98, 13543–13548. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.-P.; Lin, J.-J.; Li, W.-H. Positional distribution of transcription factor binding sites in Arabidopsis thaliana. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Brandt, S.; Knight, M.R. A history of stress alters drought calcium signalling pathways in Arabidopsis. Plant J. 1998, 16, 681–687. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yu, D.; Qanmber, G.; Lu, L.; Wang, L.; Zheng, L.; Liu, Z.; Wu, H.; Liu, X.; Chen, Q.; et al. GhKLCR1, a kinesin light chain-related gene, induces drought-stress sensitivity in Arabidopsis. Sci. China-Life Sci. 2019, 62, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Jin, Z.; Xu, B.; Li, J.; Li, Y.; Wang, X.; Wang, A.; Hu, W.; Huang, D.; Wei, Q.; et al. Identification of transcription factors interacting with a 1274 bp promoter of MaPIP1;1 which confers high-level gene expression and drought stress Inducibility in transgenic Arabidopsis thaliana. Bmc Plant Biol. 2020, 20. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Chen, X.; Fang, P.; Shi, S.; Li, J.; Liu, X.; Cao, X.; Zhao, N.; Hao, H.; Li, Y.; et al. Genomic and transcriptomic sequencing of Rosa hybrida provides microsatellite markers for breeding, flower trait improvement and taxonomy studies. Bmc Plant Biol. 2018, 18. [Google Scholar] [CrossRef]
- Dubois, A.; Raymond, O.; Maene, M.; Baudino, S.; Langlade, N.B.; Boltz, V.; Vergne, P.; Bendahmane, M. Tinkering with the C-Function: A Molecular Frame for the Selection of Double Flowers in Cultivated Roses. PLoS ONE 2010, 5, e9288. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cao, X.; Shi, S.; Zhao, N.; Li, D.; Fang, P.; Chen, X.; Qi, W.; Zhang, Z. Comparative RNA-Seq analysis reveals a critical role for brassinosteroids in rose (Rosa hybrida) petal defense against Botrytis cinerea infection. Bmc Genet. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.M.; Zerbe, P. Specialized diterpenoid metabolism in monocot crops: Biosynthesis and chemical diversity. Phytochemistry 2020, 172. [Google Scholar] [CrossRef]
- Campbell, D.R.; Sosenski, P.; Raguso, R.A. Phenotypic plasticity of floral volatiles in response to increasing drought stress. Ann. Bot. 2019, 123, 601–610. [Google Scholar] [CrossRef]
- Vaughan, M.M.; Christensen, S.; Schmelz, E.A.; Huffaker, A.; McAuslane, H.J.; Alborn, H.T.; Romero, M.; Allen, L.H.; Teal, P.E.A. Accumulation of terpenoid phytoalexins in maize roots is associated with drought tolerance. Plant Cell Environ. 2015, 38, 2195–2207. [Google Scholar] [CrossRef] [PubMed]
- Kodama, O.; Suzuki, T.; Miyakawa, J.; Akatsuka, T. Ultraviolet-Induced Accumulation of Phytoalexins in Rice Leaves. Agric. Biol. Chem. 1988, 52, 2469–2473. [Google Scholar] [CrossRef]
- Vaughan, M.M.; Huffaker, A.; Schmelz, E.A.; Dafoe, N.J.; Christensen, S.; Sims, J.; Martins, V.F.; Swerbilow, J.; Romero, M.; Alborn, H.T.; et al. Effects of elevated [CO2] on maize defence against mycotoxigenic Fusarium verticillioides. Plant Cell Environ. 2014, 37, 2691–2706. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tholl, D.; D'Auria, J.C.; Farooq, A.; Pichersky, E.; Gershenzon, J. Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 2003, 15, 481–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, D.M.; Toub, O.; Chiang, A.; Lo, B.C.; Ohse, S.; Lund, S.T.; Bohlmann, J. The bouquet of grapevine (Vitis vinifera L. cv. Cabernet Sauvignon) flowers arises from the biosynthesis of sesquiterpene volatiles in pollen grains. Proc. Natl. Acad. Sci. USA 2009, 106, 7245–7250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sofia Lima, A.; Schimmel, J.; Lukas, B.; Novak, J.; Barroso, J.G.; Cristina Figueiredo, A.; Pedro, L.G.; Degenhardt, J.; Trindade, H. Genomic characterization, molecular cloning and expression analysis of two terpene synthases from Thymus caespititius (Lamiaceae). Planta 2013, 238, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Abbas, F.; Ke, Y.; Yu, R.; Fan, Y. Functional characterization and expression analysis of two terpene synthases involved in floral scent formation in Lilium “Siberia”. Planta 2019, 249, 71–93. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Guo, Y.; Shi, X.; Yang, Y.; Chen, J.; Zhang, Q.; Sun, M. Overexpression of LiTPS2 from a cultivar of lily (Lilium 'Siberia') enhances the monoterpenoids content in tobacco flowers. Plant Physiol. Biochem. 2020, 151, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Priya, P.; Yadav, A.; Chand, J.; Yadav, G. Terzyme: A tool for identification and analysis of the plant terpenome. Plant Methods 2018, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapp, S.C.; Croteau, R.B. Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 2001, 158, 811–832. [Google Scholar] [CrossRef]
- Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Starks, C.M.; Back, K.W.; Chappell, J.; Noel, J.P. Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science 1997, 277, 1815–1820. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Li, W.; Lin, Y.-J.; Pickett, J.A.; Birkett, M.A.; Wu, K.; Wang, G.; Zhou, J.-J. Expression of lima bean terpene synthases in rice enhances recruitment of a beneficial enemy of a major rice pest. Plant Cell Environ. 2018, 41, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.; Schaff, C.; Zhang, Z.; Lipka, A.E.; Tian, F.; Kollner, T.G.; Schnee, C.; Preiss, S.; Irmisch, S.; Jander, G.; et al. Characterization of Biosynthetic Pathways for the Production of the Volatile Homoterpenes DMNT and TMTT in Zea mays. Plant Cell 2016, 28, 2651–2665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arimura, G.-I.; Garms, S.; Maffei, M.; Bossi, S.; Schulze, B.; Leitner, M.; Mithofer, A.; Boland, W. Herbivore-induced terpenoid emission in Medicago truncatula: Concerted action of jasmonate, ethylene and calcium signaling. Planta 2008, 227, 453–464. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Yu, C.; Cheng, B.; Han, Y.; Luo, L.; Pan, H.; Zhang, Q. Studies on the volatile compounds in flower extracts of Rosa odorata and R. chinensis. Ind. Crop. Prod. 2020, 146. [Google Scholar] [CrossRef]
- Magnard, J.-L.; Bony, A.R.; Bettini, F.; Campanaro, A.; Blerot, B.; Baudino, S.; Jullien, F. Linalool and linalool nerolidol synthases in roses, several genes for little scent. Plant Physiol. Biochem. 2018, 127, 74–87. [Google Scholar] [CrossRef]
- McCormick, A.C.; Unsicker, S.B.; Gershenzon, J. The specificity of herbivore-induced plant volatiles in attracting herbivore enemies. Trends Plant Sci. 2012, 17, 303–310. [Google Scholar] [CrossRef] [PubMed]





| Gene | Accession Number | Chr | AA (aa) | MW (kDa) | pI | Subcellular Localization | Subfamily |
|---|---|---|---|---|---|---|---|
| RcTPS01 | RchiOBHm_Chr6g0265741 | 6 | 567 | 65.92 | 5.16 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS02 | RchiOBHm_Chr5g0023641 | 5 | 724 | 81.88 | 5.72 | Chloroplast a, b | TPS-e/f |
| RcTPS03 | RchiOBHm_Chr5g0044191 | 5 | 565 | 65.40 | 5.25 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS04 | RchiOBHm_Chr7g0210371 | 7 | 565 | 65.34 | 5.07 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS05 | RchiOBHm_Chr3g0475221 | 3 | 560 | 64.87 | 5.17 | Chloroplast a, b | TPS-a |
| RcTPS06 | RchiOBHm_Chr5g0059511 | 5 | 557 | 64.72 | 5.95 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS07 | RchiOBHm_Chr6g0246011 | 6 | 455 | 52.89 | 6.07 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS08 | RchiOBHm_Chr5g0059541 | 5 | 544 | 62.75 | 5.19 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS09 | RchiOBHm_Chr5g0004631 | 5 | 583 | 67.00 | 5.14 | Chloroplast a, b | TPS-b |
| RcTPS10 | RchiOBHm_Chr1g0326061 | 1 | 580 | 66.90 | 5.22 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS11 | RchiOBHm_Chr3g0474501 | 3 | 560 | 64.58 | 5.17 | Chloroplast a, b | TPS-a |
| RcTPS12 | RchiOBHm_Chr7g0228501 | 7 | 539 | 62.16 | 5.43 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS13 | RchiOBHm_Chr6g0252721 | 6 | 569 | 65.74 | 5.59 | Chloroplast a/Cytoplasm b | TPS-b |
| RcTPS14 | RchiOBHm_Chr5g0065101 | 5 | 557 | 64.23 | 5.18 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS15 | RchiOBHm_Chr6g0245751 | 6 | 557 | 64.36 | 5.35 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS16 | RchiOBHm_Chr5g0038101 | 5 | 565 | 65.25 | 5.66 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS17 | RchiOBHm_Chr1g0326251 | 1 | 556 | 64.54 | 5.21 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS18 | RchiOBHm_Chr6g0246001 | 6 | 559 | 64.70 | 5.57 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS19 | RchiOBHm_Chr3g0484891 | 3 | 566 | 64.99 | 5.49 | Chloroplast a/Cytoplasm b | TPS-b |
| RcTPS20 | RchiOBHm_Chr2g0160561 | 2 | 570 | 65.43 | 5.62 | Chloroplast a/Cytoplasm b | TPS-b |
| RcTPS21 | RchiOBHm_Chr1g0313881 | 1 | 562 | 65.05 | 5.11 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS22 | RchiOBHm_Chr3g0474411 | 3 | 560 | 64.74 | 5.15 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS23 | RchiOBHm_Chr1g0326391 | 1 | 556 | 64.54 | 5.37 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS24 | RchiOBHm_Chr6g0305391 | 6 | 570 | 65.88 | 5.07 | Chloroplast a, b | TPS-a |
| RcTPS25 | RchiOBHm_Chr5g0037011 | 5 | 555 | 64.33 | 5.25 | Chloroplast a, b | TPS-a |
| RcTPS26 | RchiOBHm_Chr5g0036921 | 5 | 561 | 65.14 | 5.36 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS27 | RchiOBHm_Chr5g0060571 | 5 | 473 | 54.87 | 5.86 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS28 | RchiOBHm_Chr4g0418071 | 4 | 564 | 64.74 | 5.16 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS29 | RchiOBHm_Chr2g0162311 | 2 | 852 | 96.91 | 6.12 | Chloroplast a, b | TPS-e/f |
| RcTPS30 | RchiOBHm_Chr6g0274871 | 6 | 553 | 63.26 | 5.49 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS31 | RchiOBHm_Chr5g0023471 | 5 | 799 | 90.14 | 5.58 | Chloroplast a, b | TPS-e/f |
| RcTPS32 | RchiOBHm_Chr1g0326071 | 1 | 546 | 62.85 | 5.37 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS33 | RchiOBHm_Chr1g0331211 | 1 | 583 | 67.49 | 5.28 | Chloroplast a, b | TPS-b |
| RcTPS34 | RchiOBHm_Chr5g0038021 | 5 | 565 | 65.52 | 5.63 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS35 | RchiOBHm_Chr5g0004761 | 5 | 580 | 66.13 | 6.17 | Chloroplast a/Cytoplasm b | TPS-g |
| RcTPS36 | RchiOBHm_Chr5g0059501 | 5 | 557 | 64.72 | 5.95 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS37 | RchiOBHm_Chr5g0004711 | 5 | 544 | 62.64 | 5.09 | Chloroplast a, b | TPS-g |
| RcTPS38 | RchiOBHm_Chr1g0326051 | 1 | 581 | 67.11 | 5.34 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS39 | RchiOBHm_Chr7g0227831 | 7 | 571 | 66.02 | 5.26 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS40 | RchiOBHm_Chr5g0004731 | 5 | 509 | 58.29 | 5.26 | Chloroplast a/Cytoplasm b | TPS-g |
| RcTPS41 | RchiOBHm_Chr5g0004801 | 5 | 580 | 66.55 | 6.05 | Chloroplast a/Cytoplasm b | TPS-g |
| RcTPS42 | RchiOBHm_Chr6g0270581 | 6 | 564 | 65.34 | 5.5 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS43 | RchiOBHm_Chr3g0474541 | 3 | 557 | 64.31 | 5.06 | Chloroplast a, b | TPS-a |
| RcTPS44 | RchiOBHm_Chr7g0212441 | 7 | 558 | 64.77 | 5.46 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS45 | RchiOBHm_Chr5g0037601 | 5 | 549 | 63.56 | 5.23 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS46 | RchiOBHm_Chr5g0004591 | 5 | 580 | 66.74 | 5.16 | Chloroplast a, b | TPS-b |
| RcTPS47 | RchiOBHm_Chr5g0023441 | 5 | 522 | 59.69 | 8.91 | Chloroplast a, b | TPS-e/f |
| RcTPS48 | RchiOBHm_Chr3g0474441 | 3 | 560 | 64.75 | 5.08 | Chloroplast a/Cytoplasm b | TPS-a |
| RcTPS49 | RchiOBHm_Chr6g0290871 | 6 | 857 | 98.52 | 5.76 | Chloroplast a, b | TPS-c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Li, M.; Zhang, X.; Kong, W.; Bendahmane, M.; Bao, M.; Fu, X. Tissue-Specific Expression of the Terpene Synthase Family Genes in Rosa chinensis and Effect of Abiotic Stress Conditions. Genes 2022, 13, 547. https://doi.org/10.3390/genes13030547
Yan Y, Li M, Zhang X, Kong W, Bendahmane M, Bao M, Fu X. Tissue-Specific Expression of the Terpene Synthase Family Genes in Rosa chinensis and Effect of Abiotic Stress Conditions. Genes. 2022; 13(3):547. https://doi.org/10.3390/genes13030547
Chicago/Turabian StyleYan, Yuhang, Mouliang Li, Xiaoni Zhang, Weilong Kong, Mohammed Bendahmane, Manzhu Bao, and Xiaopeng Fu. 2022. "Tissue-Specific Expression of the Terpene Synthase Family Genes in Rosa chinensis and Effect of Abiotic Stress Conditions" Genes 13, no. 3: 547. https://doi.org/10.3390/genes13030547
APA StyleYan, Y., Li, M., Zhang, X., Kong, W., Bendahmane, M., Bao, M., & Fu, X. (2022). Tissue-Specific Expression of the Terpene Synthase Family Genes in Rosa chinensis and Effect of Abiotic Stress Conditions. Genes, 13(3), 547. https://doi.org/10.3390/genes13030547







