Gnas Promoter Hypermethylation in the Basolateral Amygdala Regulates Reconsolidation of Morphine Reward Memory in Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Drugs
2.3. Experimental Procedures
Conditioned Place Preference
2.4. Experimental Design and Statistical Analysis
2.5. Statistical Analysis
3. Results
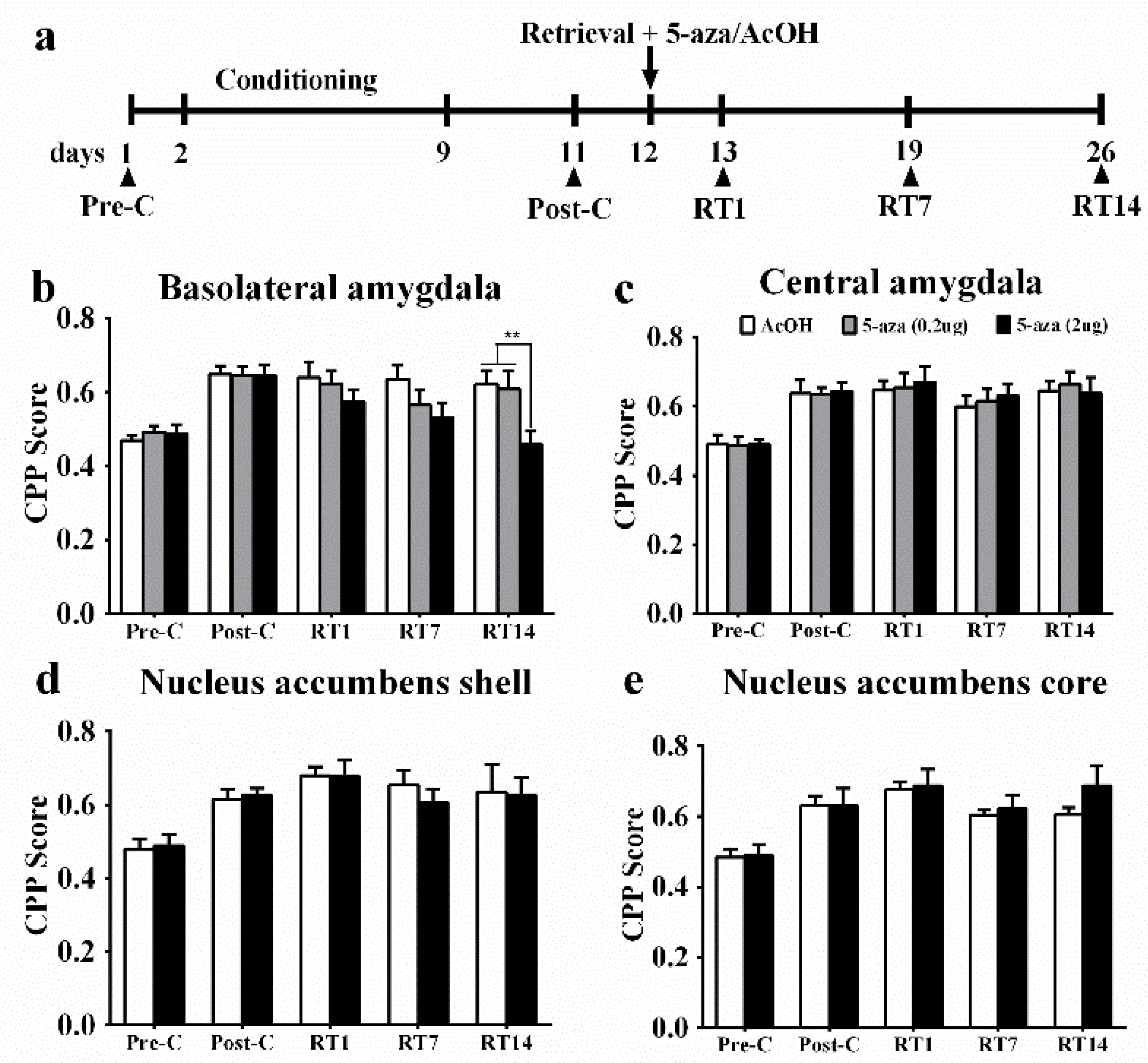
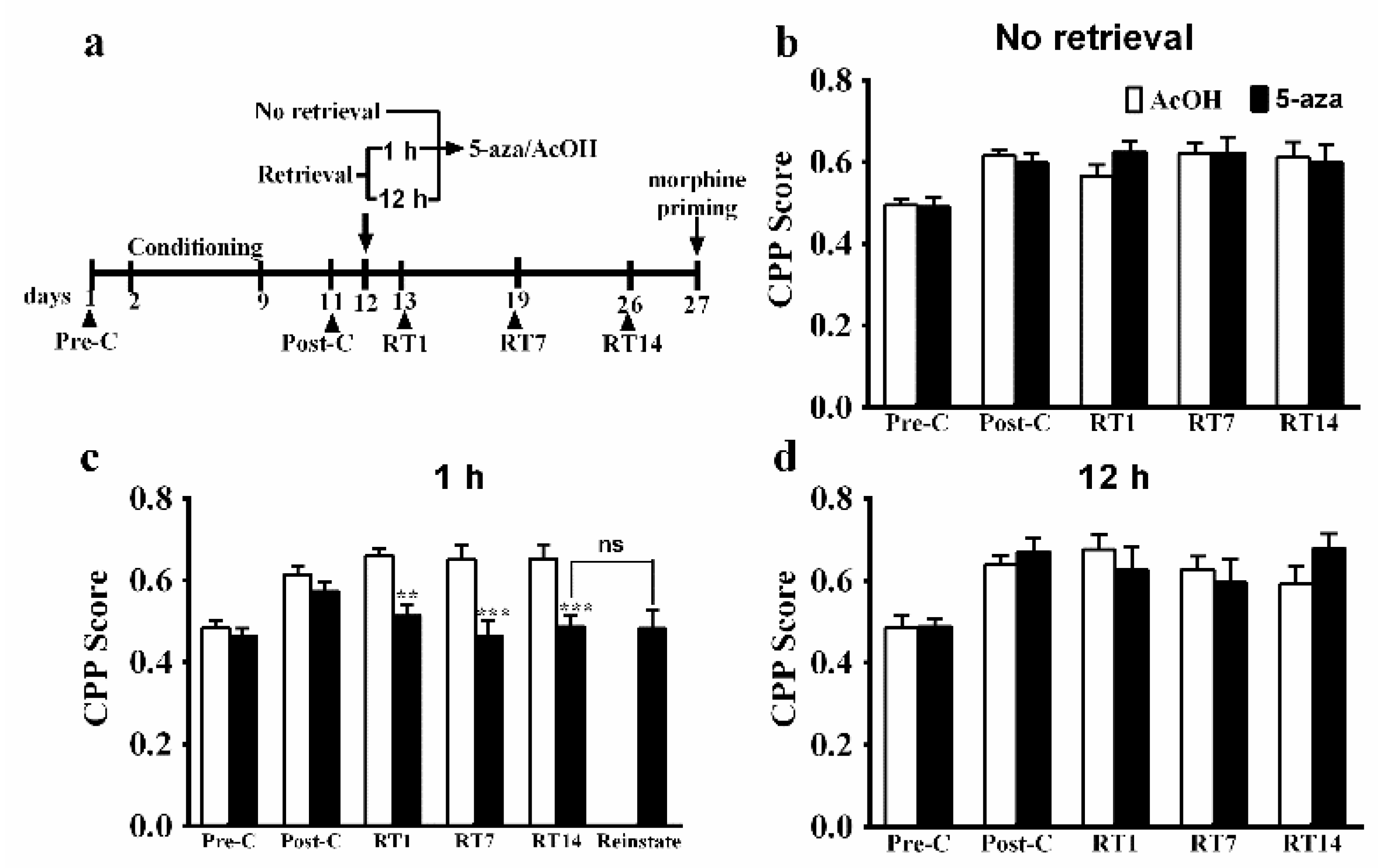
3.1. Region-Specific Effects of Dnmts Inhibition on the Reconsolidation of Morphine Reward Memory
3.2. Upregulation of Dnmt3a and Dnmt3b mRNA Expression in the BLA after Memory Retrieval
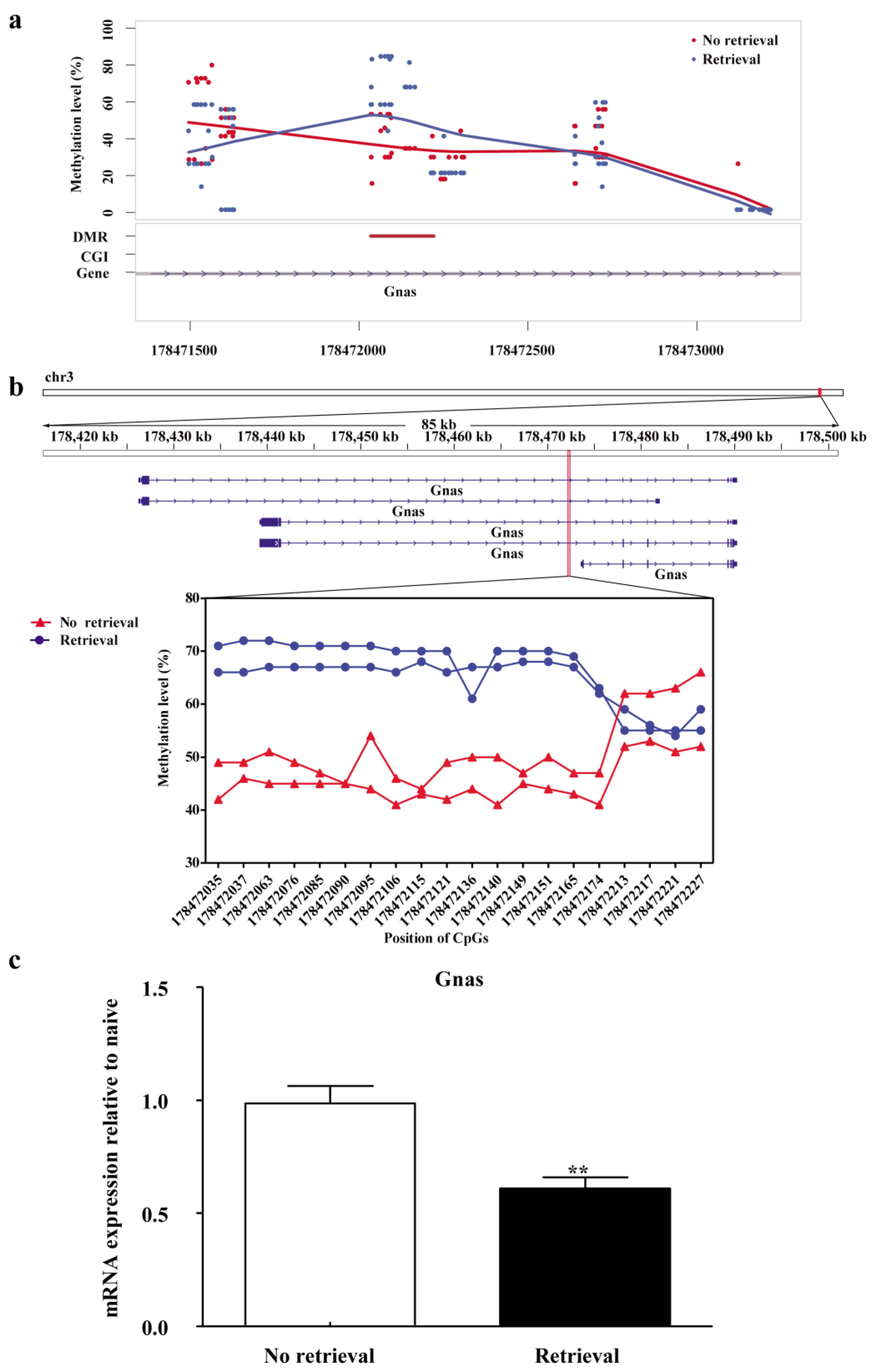
3.3. Gnas Is the Key Gene in the BLA That Is Methylated during Reconsolidation of Morphine-related Memories
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, J.L.; Di Ciano, P.; Thomas, K.L.; Everitt, B.J. Disrupting reconsolidation of drug memories reduces cocaine-seeking behavior. Neuron 2005, 47, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, J.J.; Yu, L.C. Post-retrieval extinction training enhances or hinders the extinction of morphine-induced conditioned place preference in rats dependent on the retrieval-extinction interval. Psychopharmacology 2012, 221, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.X.; Luo, Y.X.; Wu, P.; Shi, H.S.; Xue, L.F.; Chen, C.; Zhu, W.L.; Ding, Z.B.; Bao, Y.P.; Shi, J.; et al. A memory retrieval-extinction procedure to prevent drug craving and relapse. Science 2012, 336, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Das, R.K.; Hindocha, C.; Freeman, T.P.; Lazzarino, A.I.; Curran, H.V.; Kamboj, S.K. Assessing the translational feasibility of pharmacological drug memory reconsolidation blockade with memantine in quitting smokers. Psychopharmacology 2015, 232, 3363–3374. [Google Scholar] [CrossRef]
- Nader, K.; Schafe, G.E.; Le Doux, J.E. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature 2000, 406, 722–726. [Google Scholar] [CrossRef]
- Sorg, B.A. Reconsolidation of drug memories. Neurosci. Biobehav. Rev. 2012, 36, 1400–1417. [Google Scholar] [CrossRef]
- Milekic, M.H.; Brown, S.D.; Castellini, C.; Alberini, C.M. Persistent disruption of an established morphine conditioned place preference. J. Neurosci. 2006, 26, 3010–3020. [Google Scholar] [CrossRef][Green Version]
- Valjent, E.; Corbille, A.G.; Bertran-Gonzalez, J.; Herve, D.; Girault, J.A. Inhibition of ERK pathway or protein synthesis during reexposure to drugs of abuse erases previously learned place preference. Proc. Natl. Acad. Sci. USA 2006, 103, 2932–2937. [Google Scholar] [CrossRef]
- Wong, C.C.; Mill, J.; Fernandes, C. Drugs and addiction: An introduction to epigenetics. Addiction 2011, 106, 480–489. [Google Scholar] [CrossRef]
- Graff, J.; Joseph, N.F.; Horn, M.E.; Samiei, A.; Meng, J.; Seo, J.; Rei, D.; Bero, A.W.; Phan, T.X.; Wagner, F.; et al. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell 2014, 156, 261–276. [Google Scholar] [CrossRef]
- Shi, L.; Wu, J. Epigenetic regulation in mammalian preimplantation embryo development. Reprod. Biol. Endocrinol. 2009, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Beckers, T.; Kindt, M. Memory Reconsolidation Interference as an Emerging Treatment for Emotional Disorders: Strengths, Limitations, Challenges, and Opportunities. Annu. Rev. Clin. Psychol. 2017, 13, 99–121. [Google Scholar] [CrossRef] [PubMed]
- Villain, H.; Benkahoul, A.; Drougard, A.; Lafragette, M.; Muzotte, E.; Pech, S.; Bui, E.; Brunet, A.; Birmes, P.; Roullet, P. Effects of Propranolol, a β-noradrenergic Antagonist, on Memory Consolidation and Reconsolidation in Mice. Front. Behav. Neurosci. 2016, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.; Whalen, P.J. The amygdala: Vigilance and emotion. Mol. Psychiatry 2001, 6, 13–34. [Google Scholar] [CrossRef]
- Wells, A.M.; Arguello, A.A.; Xie, X.; Blanton, M.A.; Lasseter, H.C.; Reittinger, A.M.; Fuchs, R.A. Extracellular signal-regulated kinase in the basolateral amygdala, but not the nucleus accumbens core, is critical for context-response-cocaine memory reconsolidation in rats. Neuropsychopharmacology 2013, 38, 753–762. [Google Scholar] [CrossRef]
- LaPlant, Q.; Vialou, V.; Covington, H.E., 3rd; Dumitriu, D.; Feng, J.; Warren, B.L.; Maze, I.; Dietz, D.M.; Watts, E.L.; Iniguez, S.D.; et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 2010, 13, 1137–1143. [Google Scholar] [CrossRef]
- Feng, J.; Zhou, Y.; Campbell, S.L.; Le, T.; Li, E.; Sweatt, J.D.; Silva, A.J.; Fan, G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010, 13, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.A.; Sweatt, J.D. Covalent modification of DNA regulates memory formation. Neuron 2007, 53, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Massart, R.; Barnea, R.; Dikshtein, Y.; Suderman, M.; Meir, O.; Hallett, M.; Kennedy, P.; Nestler, E.J.; Szyf, M.; Yadid, G. Role of DNA methylation in the nucleus accumbens in incubation of cocaine craving. J. Neurosci. 2015, 35, 8042–8058. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, H.; Sun, Z. Lamarck rises from his grave: Parental environment-induced epigenetic inheritance in model organisms and humans. Biol. Rev. Camb. Philos. Soc. 2017, 92, 2084–2111. [Google Scholar] [CrossRef]
- Han, J.; Li, Y.; Wang, D.; Wei, C.; Yang, X.; Sui, N. Effect of 5-aza-2-deoxycytidine microinjecting into hippocampus and prelimbic cortex on acquisition and retrieval of cocaine-induced place preference in C57BL/6 mice. Eur. J. Pharmacol. 2010, 642, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Han, J.; Sui, N. Okadaic acid blocks the effects of 5-aza-2-deoxycytidine on consolidation, acquisition and retrieval of morphine-induced place preference in rats. Neuropharmacology 2014, 86, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Zhao, M.; Li, M.; Song, T.; Zhang, M.; Quan, L.; Li, S.; Sun, Z.S. Reversal of cocaine-conditioned place preference through methyl supplementation in mice: Altering global DNA methylation in the prefrontal cortex. PLoS ONE 2012, 7, e33435. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhang, J.; Li, M.; Sui, N. Distinctive Roles of 5-aza-2’-deoxycytidine in Anterior Agranular Insular and Basolateral Amygdala in Reconsolidation of Aversive Memory Associated with Morphine in Rats. Front. Behav. Neurosci. 2016, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates, 5th ed.; Elsevier Academic Press: Amsterdam, The Netherlands, 2005. [Google Scholar]
- Wu, Y.; Li, Y.; Gao, J.; Sui, N. Differential effect of NMDA receptor antagonist in the nucleus accumbens on reconsolidation of morphine -related positive and aversive memory in rats. Eur. J. Pharmacol. 2012, 674, 321–326. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- Wang, Z.; Li, X.; Jiang, Y.; Shao, Q.; Liu, Q.; Chen, B.; Huang, D. swDMR: A Sliding Window Approach to Identify Differentially Methylated Regions Based on Whole Genome Bisulfite Sequencing. PLoS ONE 2015, 10, e0132866. [Google Scholar] [CrossRef]
- Anier, K.; Malinovskaja, K.; Aonurm-Helm, A.; Zharkovsky, A.; Kalda, A. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacology 2010, 35, 2450–2461. [Google Scholar] [CrossRef]
- Le, Q.; Yan, B.; Yu, X.; Li, Y.; Song, H.; Zhu, H.; Hou, W.; Ma, D.; Wu, F.; Zhou, Y.; et al. Drug-seeking motivation level in male rats determines offspring susceptibility or resistance to cocaine-seeking behaviour. Nat. Commun. 2017, 8, 15527. [Google Scholar] [CrossRef]
- Miller, C.A.; Gavin, C.F.; White, J.A.; Parrish, R.R.; Honasoge, A.; Yancey, C.R.; Rivera, I.M.; Rubio, M.D.; Rumbaugh, G.; Sweatt, J.D. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 2010, 13, 664–666. [Google Scholar] [CrossRef]
- Maddox, S.A.; Schafe, G.E. Epigenetic alterations in the lateral amygdala are required for reconsolidation of a Pavlovian fear memory. Learn. Mem. 2011, 18, 579–593. [Google Scholar] [CrossRef]
- Zhang, J.J.; Jiang, F.Z.; Zheng, W.; Duan, Y.; Jin, S.B.; Shen, F.; Liang, J.; Li, M.; Sui, N. DNMT3a in the hippocampal CA1 is crucial in the acquisition of morphine self-administration in rats. Addict. Biol. 2019, 25, e12730. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.S.; Luo, Y.X.; Yin, X.; Wu, H.H.; Xue, G.; Geng, X.H.; Hou, Y.N. Reconsolidation of a cocaine associated memory requires DNA methyltransferase activity in the basolateral amygdala. Sci. Rep. 2015, 5, 13327. [Google Scholar] [CrossRef] [PubMed]
- Milton, A.L.; Lee, J.L.; Butler, V.J.; Gardner, R.; Everitt, B.J. Intra-amygdala and systemic antagonism of NMDA receptors prevents the reconsolidation of drug-associated memory and impairs subsequently both novel and previously acquired drug-seeking behaviors. J. Neurosci. 2008, 28, 8230–8237. [Google Scholar] [CrossRef]
- Kinney, S.R.; Pradhan, S. Regulation of expression and activity of DNA (cytosine-5) methyltransferases in mammalian cells. Prog. Mol. Biol. Transl. Sci. 2011, 101, 311–333. [Google Scholar] [CrossRef] [PubMed]
- Kemenes, G.; Kemenes, I.; Michel, M.; Papp, A.; Muller, U. Phase-dependent molecular requirements for memory reconsolidation: Differential roles for protein synthesis and protein kinase A activity. J. Neurosci. Off. J. Soc. Neurosci. 2006, 26, 6298–6302. [Google Scholar] [CrossRef]
- Pol Bodetto, S.; Carouge, D.; Fonteneau, M.; Dietrich, J.-B.; Zwiller, J.; Anglard, P. Cocaine represses protein phosphatase-1Cbeta through DNA methylation and Methyl-CpG Binding Protein-2 recruitment in adult rat brain. Neuropharmacology 2013, 73, 31–40. [Google Scholar] [CrossRef]
- Brown, A.N.; Feng, J. Drug Addiction and DNA Modifications. Adv. Exp. Med. Biol. 2017, 978, 105–125. [Google Scholar] [CrossRef]
- Barros, M.; Dempster, E.L.; Illott, N.; Chabrawi, S.; Maior, R.S.; Tomaz, C.; Silva, M.A.; Huston, J.P.; Mill, J.; Muller, C.P. Decreased methylation of the NK3 receptor coding gene (TACR3) after cocaine-induced place preference in marmoset monkeys. Addict. Biol. 2013, 18, 452–454. [Google Scholar] [CrossRef]
- Nieratschker, V.; Grosshans, M.; Frank, J.; Strohmaier, J.; von der Goltz, C.; El-Maarri, O.; Witt, S.H.; Cichon, S.; Nothen, M.M.; Kiefer, F.; et al. Epigenetic alteration of the dopamine transporter gene in alcohol-dependent patients is associated with age. Addict. Biol. 2014, 19, 305–311. [Google Scholar] [CrossRef]
- Rao, V.V.; Schnittger, S.; Hansmann, I. G protein Gs α (GNAS 1), the probable candidate gene for Albright hereditary osteodystrophy, is assigned to human chromosome 20q12-q13.2. Genomics 1991, 10, 257–261. [Google Scholar] [PubMed]
- Chao, C.C.; Ma, Y.L.; Lee, E.H. Protein kinase CK2 impairs spatial memory formation through differential cross talk with PI-3 kinase signaling: Activation of Akt and inactivation of SGK1. J. Neurosci. Off. J. Soc. Neurosci. 2007, 27, 6243–6248. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Zhang, C.; Qu, L.; Huang, J.; Jiang, L.; Liu, J.; Pinello, L.; Yuan, G.C.; Shou, C. Gene regulatory pattern analysis reveals essential role of core transcriptional factors’ activation in triple-negative breast cancer. Oncotarget 2017, 8, 21938–21953. [Google Scholar] [CrossRef]
- Giese, K.P.; Mizuno, K. The roles of protein kinases in learning and memory. Learn. Mem. 2013, 20, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.P.; Cheung, Y.F.; Favilla, C.; Siegel, S.J.; Kanes, S.J.; Houslay, M.D.; Abel, T. Constitutive activation of the G-protein subunit Galphas within forebrain neurons causes PKA-dependent alterations in fear conditioning and cortical Arc mRNA expression. Learn. Mem. 2008, 15, 75–83. [Google Scholar] [CrossRef]
- Kelsey, G. Epigenetics and imprinted genes: Insights from the imprinted Gnas locus. Horm. Res. 2009, 71 (Suppl. 2), 22–29. [Google Scholar] [CrossRef] [PubMed]
- Kaut, O.; Sharma, A.; Schmitt, I.; Wullner, U. DNA methylation of imprinted loci of autosomal chromosomes and IGF2 is not affected in Parkinson’s disease patients’ peripheral blood mononuclear cells. Neurol. Res. 2017, 39, 281–284. [Google Scholar] [CrossRef]
- Schanen, N.C. Epigenetics of autism spectrum disorders. Hum. Mol. Genet. 2006, 15 (Suppl. 2), R138–R150. [Google Scholar] [CrossRef]
- Bourtchouladze, R.; Patterson, S.L.; Kelly, M.P.; Kreibich, A.; Kandel, E.R.; Abel, T. Chronically increased Gsalpha signaling disrupts associative and spatial learning. Learn. Mem. 2006, 13, 745–752. [Google Scholar] [CrossRef][Green Version]
- MacDonald, J.F.; Jackson, M.F.; Beazely, M.A. G protein-coupled receptors control NMDARs and metaplasticity in the hippocampus. Biochim. Biophys. Acta 2007, 1768, 941–951. [Google Scholar] [CrossRef]
- Li, Y.; Ge, S.; Li, N.; Chen, L.; Zhang, S.; Wang, J.; Wu, H.; Wang, X.; Wang, X. NMDA and dopamine D1 receptors within NAc-shell regulate IEG proteins expression in reward circuit during cocaine memory reconsolidation. Neuroscience 2016, 315, 45–69. [Google Scholar] [CrossRef] [PubMed]
- Auber, A.; Tedesco, V.; Jones, C.E.; Monfils, M.H.; Chiamulera, C. Post-retrieval extinction as reconsolidation interference: Methodological issues or boundary conditions? Psychopharmacology 2013, 226, 631–647. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.J.; Chang, C.H.; Gean, P.W. AMPA receptor endocytosis in the amygdala is involved in the disrupted reconsolidation of Methamphetamine-associated contextual memory. Neurobiol. Learn. Mem. 2013, 103, 72–81. [Google Scholar] [CrossRef] [PubMed]

| Gene | Primer Sequence | |
|---|---|---|
| Forward Sequence | Reverse Sequence | |
| Dnmt1 | TATTGCAGTCGCGGTCACTT | CTGATTGATTGGCCCCAGGT |
| Dnmt3a | TCTTGCTCACAAAGACCACGA | TACCACGGTTCTCCTCCTGT |
| Dnmt3b | ACCAGGCCTTGAAAACCTCAG | CATGGTTTCCTGCAAGTCCCT |
| GAPDH | ACCTTTGATGCTGGGGCTGGC | GGGCTGAGTTGGGATGGGGACT |
| Gnas | GGAGGACAACCAGACCAACC | GACCTTCTCAGCAAGCAGATC |
| Genes | Differentially Methylated Regions | Hypermethylated or Hypomethylated | p Value |
|---|---|---|---|
| Zrsr1 | Chr14:107824490-107824759 | Hypermethylated | 7.57 × 10−6 |
| Gnas | chr3:178472035-1784722218 | Hypermethylated | 2.43 × 10−5 |
| Pced1b | Chr7:138329413-138329534 | Hypermethylated | 0.003298 |
| Amigo2 | Chr7:138329413-138329534 | Hypermethylated | 0.003298 |
| Sox10 | chr7:120397931-120398118 | Hypermethylated | 0.0102 |
| Pik3r1 | chr2:50965442-50965530 | Hypermethylated | 0.0107 |
| Tnnc2 | Chr3:167428575-167428739 | Hypermethylated | 0.01381041 |
| Cxcr5 | Chr8:47457610-47457753 | Hypermethylated | 0.024671 |
| Fkbpl | chr20:6490315-6490406 | Hypermethylated | 0.0248 |
| RGD1563015 | chr5:142077035-142077175 | Hypermethylated | 0.0258 |
| Rps10 | chr20:9422414-9422470 | Hypomethylated | 1.41 × 10−9 |
| RGD1560608 | chr6:144559138-144559194 | Hypomethylated | 0.0019 |
| Crb2 | chr3:27274071-27274146 | Hypomethylated | 0.0224 |
| Serpinh1 | chr1:170510550-170510601 | Hypomethylated | 0.0252 |
| Gene Name | Position of DMR | CpG Number of the DMR | p Value | Promoter | Gene Body | Upstream | Downstream |
|---|---|---|---|---|---|---|---|
| Aff1 | chr14:7218380-7218515 | 9 | 0.00207539 | Y | |||
| Amigo2 | chr7:138329413-138329534 | 10 | 0.003297764 | Y | Y | ||
| Ano4 | chr7:29919053-29919161 | 10 | 0.01020776 | Y | |||
| Arhgap31 | chr11:67463804-67463994 | 14 | 0.000473217 | Y | |||
| Arhgef2 | chr2:207404742-207404827 | 6 | 0.009214125 | Y | |||
| Bcl11b | chr6:141020775-141021154 | 15 | 8.95 × 10−10 | Y | |||
| Bcl11b | chr6:141052868-141053020 | 6 | 0.02759777 | Y | |||
| Bmp7 | chr3:176986680-176986773 | 5 | 0.003185848 | Y | |||
| Btbd17 | chr10:102994047-102994225 | 8 | 0.02664855 | Y | |||
| C4a | chr20:6490315-6490406 | 6 | 0.02475037 | Y | |||
| C4a | chr20:6530195-6530367 | 11 | 0.006960988 | Y | |||
| Cacna2d2 | chr8:115596160-115596246 | 6 | 0.003831569 | Y | |||
| Capn2 | chr13:105860689-105860886 | 7 | 0.000954839 | Y | |||
| Clstn1 | chr5:170207211-170207370 | 8 | 0.009061262 | Y | |||
| Col16a1 | chr5:152027022-152027215 | 6 | 0.001871846 | Y | |||
| Commd1 | chr14:107824490-107824759 | 27 | 7.57 × 10−6 | Y | |||
| Crb2 | chr3:27274071-27274146 | 5 | 0.02240409 | Y | Y | ||
| Cxcr5 | chr8:47457610-47457753 | 8 | 0.02467058 | Y | Y | ||
| Disc1 | chr19:68763258-68763457 | 18 | 0.000567238 | Y | |||
| Dpysl2 | chr15:48089025-48089178 | 14 | 3.45 × 10−13 | Y | |||
| Dscaml1 | chr8:48478120-48478263 | 7 | 0.000375843 | Y | |||
| Eif4b | chr7:141499843-141500003 | 8 | 0.0045236 | Y | |||
| Fkbpl | chr20:6490315-6490406 | 6 | 0.02475037 | Y | Y | ||
| Gipc2 | chr2:275973554-275973698 | 7 | 0.000207575 | Y | |||
| Gnas | chr3:178441267-178441419 | 19 | 1.28 × 10−12 | Y | |||
| Gnas | chr3:178472035-178472221 | 22 | 2.43 × 10−5 | Y | Y | Y | |
| Hm13 | chr3:154572338-154572523 | 17 | 1.25 × 10−6 | Y | |||
| Hs3st3b1 | chr10:50167234-50167300 | 8 | 0.001685876 | Y | |||
| Jak3 | chr16:19972561-19972636 | 5 | 0.01845945 | Y | |||
| Jup | chr10:88094186-88094336 | 7 | 0.006709436 | Y | |||
| Kazn | chr5:164412676-164412854 | 12 | 0.004983534 | Y | |||
| Kcnh3 | chrX:115381900-115382098 | 7 | 0.000139526 | Y | |||
| Kif19 | chr10:102994047-102994225 | 0.02664855 | Y | ||||
| Kif26b | chr13:101990776-101990963 | 5 | 0.02067161 | Y | |||
| Lrrk1 | chr1:128250485-128250670 | 10 | 0.001559071 | Y | |||
| Lrrn2 | chr13:54749407-54749591 | 5 | 0.01956404 | Y | |||
| Mcoln2 | chr2:270577752-270577951 | 12 | 0.000800436 | Y | |||
| Mecom | chr2:137270802-137270931 | 5 | 0.0126658 | Y | |||
| Mtss1 | chr7:99426687-99426843 | 6 | 0.01163478 | Y | |||
| Mvd | chr19:65970858-65970974 | 6 | 0.00726908 | Y | |||
| Narfl | chr10:14960107-14960271 | 16 | 7.19 × 10−6 | Y | |||
| Nfix | chr19:36874870-36874950 | 11 | 7.19 × 10−5 | Y | |||
| Oxct1 | chr2:72940605-72940773 | 5 | 0.0135552 | Y | |||
| Pced1b | chr7:138329413-138329534 | 10 | 0.003297764 | Y | Y | ||
| Per3 | chr5:171694018-171694214 | 12 | 0.003234661 | Y | |||
| Pik3r1 | chr2:50965442-50965530 | 6 | 0.01074433 | Y | Y | ||
| Prr5 | chr7:125324729-125324849 | 6 | 5.39 × 10−6 | Y | |||
| Ptpn11 | chr12:42770644-42770806 | 10 | 0.000300016 | Y | |||
| Ptprm | chr9:114967504-114967678 | 8 | 0.009920264 | Y | |||
| Pygm | chr1:228746598-228746766 | 8 | 0.005512662 | Y | |||
| RGD1560608 | chr6:144559138-144559194 | 7 | 0.001928227 | Y | Y | ||
| RGD1563015 | chr5:142077035-142077175 | 6 | 0.02576382 | Y | Y | ||
| Rhbdd1 | chr9:87975914-87975949 | 8 | 0.02561427 | Y | |||
| Rps10 | chr20:9422414-9422470 | 8 | 1.41 × 10−9 | Y | Y | ||
| Sall1 | chr19:34396734-34396837 | 7 | 0.01097885 | Y | |||
| Sema5b | chr11:71436403-71436588 | 7 | 0.01134854 | Y | |||
| Serpinh1 | chr1:170510550-170510601 | 6 | 0.02524889 | Y | Y | ||
| Sfxn5 | chr4:181583937-181584095 | 10 | 0.000178655 | Y | |||
| Sox10 | chr7:120397931-120398118 | 8 | 0.01023295 | Y | Y | ||
| Spats2 | chrX:115381900-115382098 | 7 | 0.000139526 | Y | |||
| Stk19 | chr20:6490315-6490406 | 6 | 0.02475037 | Y | |||
| Stk19 | chr20:6530195-6530367 | 11 | 0.006960988 | Y | |||
| Tmem63a | chr13:104248401-104248590 | 10 | 0.01664342 | Y | |||
| Tnnc2 | chr3:167428575-167428739 | 15 | 0.01381041 | Y | Y | ||
| Tspan5 | chr2:262646069-262646263 | 8 | 0.002377802 | Y | |||
| Txndc15 | chr17:11251263-11251421 | 18 | 1.58 × 10−6 | Y | |||
| Wnt7b | chr7:126158090-126158176 | 8 | 0.002492002 | Y | |||
| Zfp503 | chr15:2361950-2362100 | 15 | 8.09 × 10−5 | Y | |||
| Zrsr1 | chr14:107824490-107824759 | 27 | 7.57 × 10−6 | Y | Y |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Liang, J.; Jiang, F.; Cai, W.; Shen, F.; Liang, J.; Zhang, J.; Sun, Z.; Sui, N. Gnas Promoter Hypermethylation in the Basolateral Amygdala Regulates Reconsolidation of Morphine Reward Memory in Rats. Genes 2022, 13, 553. https://doi.org/10.3390/genes13030553
Liu P, Liang J, Jiang F, Cai W, Shen F, Liang J, Zhang J, Sun Z, Sui N. Gnas Promoter Hypermethylation in the Basolateral Amygdala Regulates Reconsolidation of Morphine Reward Memory in Rats. Genes. 2022; 13(3):553. https://doi.org/10.3390/genes13030553
Chicago/Turabian StyleLiu, Peng, Jialong Liang, Fengze Jiang, Wanshi Cai, Fang Shen, Jing Liang, Jianjun Zhang, Zhongsheng Sun, and Nan Sui. 2022. "Gnas Promoter Hypermethylation in the Basolateral Amygdala Regulates Reconsolidation of Morphine Reward Memory in Rats" Genes 13, no. 3: 553. https://doi.org/10.3390/genes13030553
APA StyleLiu, P., Liang, J., Jiang, F., Cai, W., Shen, F., Liang, J., Zhang, J., Sun, Z., & Sui, N. (2022). Gnas Promoter Hypermethylation in the Basolateral Amygdala Regulates Reconsolidation of Morphine Reward Memory in Rats. Genes, 13(3), 553. https://doi.org/10.3390/genes13030553






