Genome-Wide Identification and Expression Analysis of the Basic Leucine Zipper (bZIP) Transcription Factor Gene Family in Fusarium graminearum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Physicochemical Analysis of bZIP TFs in F. graminearum
2.2. Evolution, Diversity, and Species Tree Analysis of bZIP in Fungal Species
2.3. Gene Structure Analysis and Chromosomal Distribution of FgbZIP Genes
2.4. Examination of FgbZIP Protein Motifs and Subcellular Localization
2.5. Protein-Protein Interaction Network and Protein Homology Modeling
2.6. Biological Sample, Culture Conditions, Abiotic Treatments, and Total RNA Extractions
2.7. cDNA Synthesis and Quantitative Polymerase Chain Reaction (RT-qPCR)
3. Results
3.1. Gene Features and Physiochemistry of bZIPs
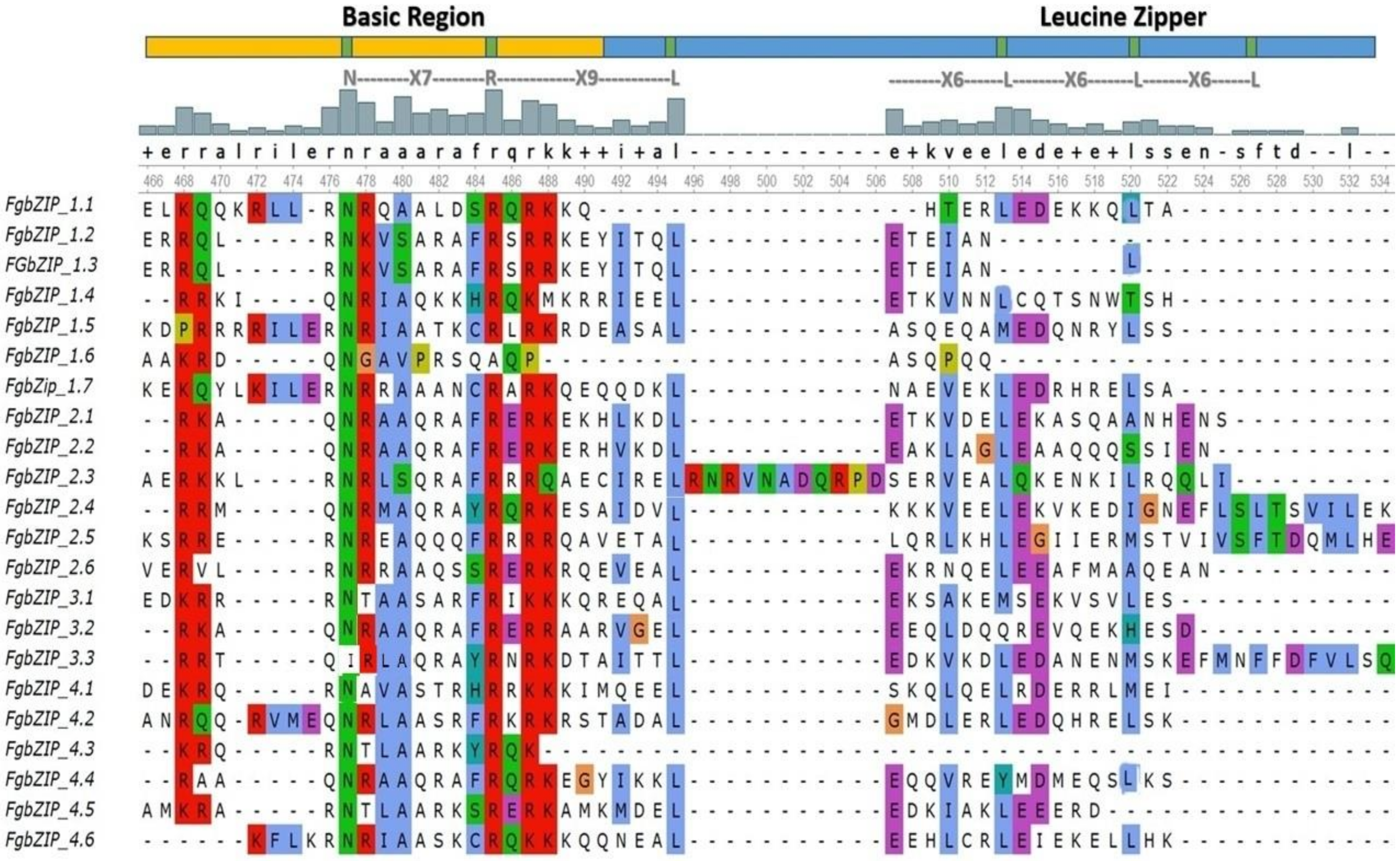
3.2. Multiple Sequence Alignment and Evolutionary Analysis
3.3. Gene Structure Analysis and Chromosomal Distribution of FgbZIP Genes
3.4. Conserved Domains and Motifs of bZIPs
3.5. Protein–Protein Interaction Network and Subcellular Localization of bZIPs
3.6. Homology Modeling of bZIPs
3.7. Expression Analysis of bZIP Genes in Various Developmental Stages and Conidiogenesis
3.8. Expression Patterns of bZIP Genes in Response to Abiotic Stress
3.9. Circos and Synteny Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rauwane, M.E.; Ogugua, U.V.; Kalu, C.M.; Ledwaba, L.K.; Woldesemayat, A.A.; Ntushelo, K. Pathogenicity and virulence factors of Fusarium graminearum including factors discovered using next generation sequencing technologies and proteomics. Microorganisms 2020, 8, 305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birr, T.; Hasler, M.; Verreet, J.-A.; Klink, H. Composition and predominance of Fusarium species causing Fusarium head blight in winter wheat grain depending on cultivar susceptibility and meteorological factors. Microorganisms 2020, 8, 617. [Google Scholar] [CrossRef] [PubMed]
- Alisaac, E.; Rathgeb, A.; Karlovsky, P.; Mahlein, A.-K. Fusarium head blight: Effect of infection timing on spread of Fusarium graminearum and spatial distribution of deoxynivalenol within wheat spikes. Microorganisms 2021, 9, 79. [Google Scholar] [CrossRef]
- Dweba, C.; Figlan, S.; Shimelis, H.; Motaung, T.; Sydenham, S.; Mwadzingeni, L.; Tsilo, T. Fusarium head blight of wheat: Pathogenesis and control strategies. Crop Prot. 2017, 91, 114–122. [Google Scholar] [CrossRef]
- Reiter, F.; Wienerroither, S.; Stark, A. Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 2017, 43, 73–81. [Google Scholar] [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The human transcription factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [Green Version]
- Wingender, E.; Schoeps, T.; Haubrock, M.; Krull, M.; Dönitz, J. TFClass: Expanding the classification of human transcription factors to their mammalian orthologs. Nucleic Acids Res. 2018, 46, D343–D347. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [Green Version]
- Gai, Y.; Li, L.; Liu, B.; Ma, H.; Chen, Y.; Zheng, F.; Sun, X.; Wang, M.; Jiao, C.; Li, H. Distinct and essential roles of bZIP transcription factors in the stress response and pathogenesis in Alternaria alternata. Microbiol. Res. 2022, 256, 126915. [Google Scholar] [CrossRef]
- Lei, Y.; Liu, G.; Yao, G.; Li, Z.; Qin, Y.; Qu, Y. A novel bZIP transcription factor ClrC positively regulates multiple stress responses, conidiation and cellulase expression in Penicillium oxalicum. Res. Microbiol. 2016, 167, 424–435. [Google Scholar] [CrossRef]
- Fujii, Y.; Shimizu, T.; Toda, T.; Yanagida, M.; Hakoshima, T. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat. Struct. Biol. 2000, 7, 889–893. [Google Scholar] [CrossRef]
- Ellenberger, T.E.; Brandl, C.J.; Struhl, K.; Harrison, S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: Crystal structure of the protein-DNA complex. Cell 1992, 71, 1223–1237. [Google Scholar] [CrossRef]
- Vinson, C.R.; Sigler, P.B.; McKnight, S.L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science 1989, 246, 911–916. [Google Scholar] [CrossRef]
- Liao, Y.; Zou, H.-F.; Wei, W.; Hao, Y.-J.; Tian, A.-G.; Huang, J.; Liu, Y.-F.; Zhang, J.-S.; Chen, S.-Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.-M.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef] [Green Version]
- Jindrich, K.; Degnan, B.M. The diversification of the basic leucine zipper family in eukaryotes correlates with the evolution of multicellularity. BMC Evol. Biol. 2016, 16, 28. [Google Scholar] [CrossRef] [Green Version]
- Alves, M.S.; Dadalto, S.P.; Gonçalves, A.B.; De Souza, G.B.; Barros, V.A.; Fietto, L.G. Plant bZIP Transcription Factors Responsive to Pathogens: A Review. Int. J. Mol. Sci. 2013, 14, 7815–7828. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.-H.; Yang, S.L.; Chung, K.-R. The YAP1 homolog–mediated oxidative stress tolerance is crucial for pathogenicity of the necrotrophic fungus Alternaria alternata in citrus. Mol. Plant-Microbe Interact. 2009, 22, 942–952. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Chen, Y.; Du, Y.; Dong, Y.; Guo, W.; Zhai, S.; Zhang, H.; Dong, S.; Zhang, Z.; Wang, Y. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2011, 7, e1001302. [Google Scholar] [CrossRef] [Green Version]
- Etxebeste, O.; Ni, M.; Garzia, A.; Kwon, N.-J.; Fischer, R.; Yu, J.-H.; Espeso, E.A.; Ugalde, U. Basic-zipper-type transcription factor FlbB controls asexual development in Aspergillus nidulans. Eukaryot. Cell 2008, 7, 38–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, P.; Shin, K.-S.; Wang, T.; Yu, J.-H. Aspergillus fumigatus flbB encodes two basic leucine zipper domain (bZIP) proteins required for proper asexual development and gliotoxin production. Eukaryot. Cell 2010, 9, 1711–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, C.; Kasuga, T.; Sachs, M.S.; Glass, N.L. Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot. Cell 2007, 6, 1018–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, W.-B.; Reinke, A.W.; Szilagyi, M.; Emri, T.; Chiang, Y.-M.; Keating, A.E.; Pocsi, I.; Wang, C.C.; Keller, N.P. bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans. Microbiology 2013, 159, 77. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wu, F.; Liu, L.; Liu, X.; Che, Y.; Keller, N.P.; Guo, L.; Yin, W.-B. The bZIP transcription factor PfZipA regulates secondary metabolism and oxidative stress response in the plant endophytic fungus Pestalotiopsis fici. Fungal Genet. Biol. 2015, 81, 221–228. [Google Scholar] [CrossRef]
- Son, H.; Seo, Y.-S.; Min, K.; Park, A.R.; Lee, J.; Jin, J.-M.; Lin, Y.; Cao, P.; Hong, S.-Y.; Kim, E.-K. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog. 2011, 7, e1002310. [Google Scholar] [CrossRef]
- Hortschansky, P.; Ando, E.; Tuppatsch, K.; Arikawa, H.; Kobayashi, T.; Kato, M.; Haas, H.; Brakhage, A.A. Deciphering the combinatorial DNA-binding code of the CCAAT-binding complex and the iron-regulatory basic region leucine zipper (bZIP) transcription factor HapX. J. Biol. Chem. 2015, 290, 6058–6070. [Google Scholar] [CrossRef] [Green Version]
- Tudzynski, B. Nitrogen regulation of fungal secondary metabolism in fungi. Front. Microbiol. 2014, 5, 656. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Ma, Y.; Zhao, J.; Geng, X.; Chen, W.; Ding, S.; Li, H.; Li, H. The bZIP transcription factor FpAda1 is essential for fungal growth and conidiation in Fusarium pseudograminearum. Curr. Genet. 2020, 66, 507–515. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Wang, Y.; Tian, C. bZIP transcription factor CgAP1 is essential for oxidative stress tolerance and full virulence of the poplar anthracnose fungus Colletotrichum gloeosporioides. Fungal Genet. Biol. 2016, 95, 58–66. [Google Scholar] [CrossRef]
- Cheon, S.A.; Jung, K.-W.; Chen, Y.-L.; Heitman, J.; Bahn, Y.-S.; Kang, H.A. Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog. 2011, 7, e1002177. [Google Scholar] [CrossRef] [Green Version]
- Montenegro-Montero, A.; Goity, A.; Larrondo, L.F. The bZIP transcription factor HAC-1 is involved in the unfolded protein response and is necessary for growth on cellulose in Neurospora crassa. PLoS ONE 2015, 10, e0131415. [Google Scholar]
- Natorff, R.; Sieńko, M.; Brzywczy, J.; Paszewski, A. The Aspergillus nidulans metR gene encodes a bZIP protein which activates transcription of sulphur metabolism genes. Mol. Microbiol. 2003, 49, 1081–1094. [Google Scholar] [CrossRef]
- Jain, S.; Sekonyela, R.; Knox, B.P.; Palmer, J.M.; Huttenlocher, A.; Kabbage, M.; Keller, N.P. Selenate sensitivity of a laeA mutant is restored by overexpression of the bZIP protein MetR in Aspergillus fumigatus. Fungal Genet. Biol. 2018, 117, 1–10. [Google Scholar] [CrossRef]
- Sekonyela, R.; Palmer, J.M.; Bok, J.-W.; Jain, S.; Berthier, E.; Forseth, R.; Schroeder, F.; Keller, N.P. RsmA regulates Aspergillus fumigatus gliotoxin cluster metabolites including cyclo (L-Phe-L-Ser), a potential new diagnostic marker for invasive aspergillosis. PLoS ONE 2013, 8, e62591. [Google Scholar] [CrossRef]
- Voigt, O.; Herzog, B.; Jakobshagen, A.; Pöggeler, S. bZIP transcription factor SmJLB1 regulates autophagy-related genes Smatg8 and Smatg4 and is required for fruiting-body development and vegetative growth in Sordaria macrospora. Fungal Genet. Biol. 2013, 61, 50–60. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Y.; Zhao, H.; Wu, M.; Zhang, J.; Chen, W.; Li, G.; Yang, L. Genome-wide identification and expression analysis of the bZIP transcription factors in the mycoparasite Coniothyrium minitans. Microorganisms 2020, 8, 1045. [Google Scholar] [CrossRef]
- Yin, W.; Cui, P.; Wei, W.; Lin, Y.; Luo, C. Genome-wide identification and analysis of the basic leucine zipper (bZIP) transcription factor gene family in Ustilaginoidea virens. Genome 2017, 60, 1051–1059. [Google Scholar] [CrossRef] [Green Version]
- Kong, S.; Park, S.Y.; Lee, Y.H. Systematic characterization of the bZIP transcription factor gene family in the rice blast fungus, M agnaporthe oryzae. Environ. Microbiol. 2015, 17, 1425–1443. [Google Scholar] [CrossRef]
- Lysøe, E.; Bone, K.R.; Klemsdal, S.S. Real-time quantitative expression studies of the zearalenone biosynthetic gene cluster in Fusarium graminearum. Phytopathology 2009, 99, 176–184. [Google Scholar] [CrossRef] [Green Version]
- Montibus, M.; Ducos, C.; Bonnin-Verdal, M.-N.; Bormann, J.; Ponts, N.; Richard-Forget, F.; Barreau, C. The bZIP transcription factor Fgap1 mediates oxidative stress response and trichothecene biosynthesis but not virulence in Fusarium graminearum. PLoS ONE 2013, 8, e83377. [Google Scholar]
- Seong, K.; Hou, Z.; Tracy, M.; Kistler, H.C.; Xu, J.-R. Random insertional mutagenesis identifies genes associated with virulence in the wheat scab fungus Fusarium graminearum. Phytopathology 2005, 95, 744–750. [Google Scholar] [CrossRef] [Green Version]
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999, 294, 1351–1362. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Team, U. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0. PLoS ONE 2010, 5, e9931. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Rabinal, C.; Bhat, S. Identification of differentially expressed genes in Trichoderma koningii IABT1252 during its interaction with Sclerotium rolfsii. Curr. Microbiol. 2020, 77, 396–404. [Google Scholar] [CrossRef]
- Kim, H.-K.; Yun, S.-H. Evaluation of potential reference genes for quantitative RT-PCR analysis in Fusarium graminearum under different culture conditions. Plant Pathol. J. 2011, 27, 301–309. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-Y.; Choi, J.; Lim, S.-E.; Lee, G.-W.; Park, J.; Kim, Y.; Kong, S.; Kim, S.R.; Rho, H.-S.; Jeon, J. Global expression profiling of transcription factor genes provides new insights into pathogenicity and stress responses in the rice blast fungus. PLoS Pathog. 2013, 9, e1003350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chen, W.; Zhou, J.; He, H.; Chen, L.; Chen, H.; Deng, X.W. Basic leucine zipper transcription factor OsbZIP16 positively regulates drought resistance in rice. Plant Sci. 2012, 193, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Park, A.R.; Son, H.; Min, K.; Park, J.; Goo, J.H.; Rhee, S.; Chae, S.K.; Lee, Y.W. Autoregulation of ZEB 2 expression for zearalenone production in F usarium graminearum. Mol. Microbiol. 2015, 97, 942–956. [Google Scholar] [CrossRef] [Green Version]
- Nagygyörgy, E.D.; Hornok, L.; Ádám, A.L. Role of MAP kinase signaling in secondary metabolism and adaptation to abiotic/fungicide stress in Fusarium. In Fungicides-Beneficial and Harmful Aspects; IntechOpen: London, UK, 2011. [Google Scholar]
- Pareek, M.; Rajam, M.V. RNAi-mediated silencing of MAP kinase signalling genes (Fmk1, Hog1, and Pbs2) in Fusarium oxysporum reduces pathogenesis on tomato plants. Fungal Biol. 2017, 121, 775–784. [Google Scholar] [CrossRef]
- Sabaratnam, K.; Renner, M.; Paesen, G.; Harlos, K.; Nair, V.; Owens, R.J.; Grimes, J.M. Insights from the crystal structure of the chicken CREB3 bZIP suggest that members of the CREB3 subfamily transcription factors may be activated in response to oxidative stress. Protein Sci. 2019, 28, 779–787. [Google Scholar] [CrossRef]
- Toda, T.; Shimanuki, M.; Yanagida, M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991, 5, 60–73. [Google Scholar] [CrossRef] [Green Version]
- Toone, W.M.; Kuge, S.; Samuels, M.; Morgan, B.A.; Toda, T.; Jones, N. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: Requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 1998, 12, 1453–1463. [Google Scholar] [CrossRef] [Green Version]
- Calvo, I.A.; Ayté, J.; Hidalgo, E. Reversible thiol oxidation in the H2O2-dependent activation of the transcription factor Pap1. J. Cell Sci. 2013, 126, 2279–2284. [Google Scholar] [CrossRef] [Green Version]
- Martínez, M.; Ramírez Albuquerque, L.; Arata, A.F.; Biganzoli, F.; Fernández Pinto, V.; Stenglein, S.A. Effects of Fusarium graminearum and Fusarium poae on disease parameters, grain quality and mycotoxins contamination in bread wheat (Part I). J. Sci. Food Agric. 2020, 100, 863–873. [Google Scholar] [CrossRef]
- Guo, C.; Guo, R.; Xu, X.; Gao, M.; Li, X.; Song, J.; Zheng, Y.; Wang, X. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 2014, 65, 1513–1528. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Y.; Yao, J.; Zhang, S.; Wang, L.; Guo, C.; Van Nocker, S.; Wang, X. Genome-wide identification and expression analyses of the homeobox transcription factor family during ovule development in seedless and seeded grapes. Sci. Rep. 2017, 7, 12638. [Google Scholar] [CrossRef] [Green Version]
- Bensassi, F.; Zaied, C.; Abid, S.; Hajlaoui, M.R.; Bacha, H. Occurrence of deoxynivalenol in durum wheat in Tunisia. Food Control 2010, 21, 281–285. [Google Scholar] [CrossRef]
- Becher, R.; Miedaner, T.; Wirsel, S.G. 8 Biology, diversity, and management of FHB-causing Fusarium species in small-grain cereals. In Agricultural Applications; Springer: Berlin/Heidelberg, Germany, 2013; pp. 199–241. [Google Scholar]
- Zuriegat, Q.; Zheng, Y.; Liu, H.; Wang, Z.; Yun, Y. Current progress on pathogenicity-related transcription factors in Fusarium oxysporum. Mol. Plant Pathol. 2021, 22, 882–895. [Google Scholar] [CrossRef]
- Leiter, É.; Emri, T.; Pákozdi, K.; Hornok, L.; Pócsi, I. The impact of bZIP Atf1ortholog global regulators in fungi. Appl. Microbiol. Biotechnol. 2021, 105, 5769–5783. [Google Scholar] [CrossRef]
- Liu, Y.; Chai, M.; Zhang, M.; He, Q.; Su, Z.; Priyadarshani, S.V.G.N.; Liu, L.; Dong, G.; Qin, Y. Genome-Wide Analysis, Characterization, and Expression Profile of the Basic Leucine Zipper Transcription Factor Family in Pineapple. Int. J. Genom. 2020, 2020, 3165958. [Google Scholar] [CrossRef]
- Liu, M.; Wen, Y.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, phylogeny, evolutionary expansion and expression analyses of bZIP transcription factor family in tartary buckwheat. BMC Genom. 2019, 20, 483. [Google Scholar] [CrossRef] [Green Version]
- Sornaraj, P.; Luang, S.; Lopato, S.; Hrmova, M. Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: A molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta—Gen. Subj. 2016, 1860, 46–56. [Google Scholar] [CrossRef]
- Liu, C.; Schläppi, M.R.; Mao, B.; Wang, W.; Wang, A.; Chu, C. The bZIP 73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol. J. 2019, 17, 1834–1849. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Pan, R.; Tan, L.; Zhang, Z.; Guo, M. Pleiotropic roles of O-mannosyltransferase MoPmt4 in development and pathogenicity of Magnaporthe oryzae. Curr. Genet. 2019, 65, 223–239. [Google Scholar] [CrossRef]
- Guo, M.; Guo, W.; Chen, Y.; Dong, S.; Zhang, X.; Zhang, H.; Song, W.; Wang, W.; Wang, Q.; Lv, R.; et al. The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 2010, 23, 1053–1068. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Xu, D.; Jia, L.; Huang, X.; Ma, G.; Wang, S.; Zhu, M.; Zhang, A.; Guan, M.; Lu, K. Genome-wide identification and structural analysis of bZIP transcription factor genes in Brassica napus. Genes 2017, 8, 288. [Google Scholar] [CrossRef] [Green Version]
- Hwang, I.; Manoharan, R.K.; Kang, J.-G.; Chung, M.-Y.; Kim, Y.-W.; Nou, I.-S. Genome-Wide Identification and Characterization of bZIP Transcription Factors in Brassica oleracea under Cold Stress. BioMed Res. Int. 2016, 2016, 4376598. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Cheng, K.; Wan, L.; Yan, L.; Jiang, H.; Liu, S.; Lei, Y.; Liao, B. Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genom. 2015, 16, 1053. [Google Scholar] [CrossRef] [Green Version]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-Wide Analysis of bZIP-Encoding Genes in Maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [Green Version]
- Deppmann, C.D.; Acharya, A.; Rishi, V.; Wobbes, B.; Smeekens, S.; Taparowsky, E.J.; Vinson, C. Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: A comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 2004, 32, 3435–3445. [Google Scholar] [CrossRef]
- Fassler, J.; Landsman, D.; Acharya, A.; Moll, J.R.; Bonovich, M.; Vinson, C. B-ZIP proteins encoded by the Drosophila genome: Evaluation of potential dimerization partners. Genome Res. 2002, 12, 1190–1200. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, P.; Baranwal, V.K.; Khurana, P. Genome-wide analysis of bZIP transcription factors in wheat and functional characterization of a TabZIP under abiotic stress. Sci. Rep. 2019, 9, 4608. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.P.; Wang, M.; Xiang, Y.; Qiu, L.; Hu, S.; Zhang, Z.; Mattjus, P.; Zhu, X.; Zhang, Y. Nach Is a Novel Subgroup at an Early Evolutionary Stage of the CNC-bZIP Subfamily Transcription Factors from the Marine Bacteria to Humans. Int. J. Mol. Sci. 2018, 19, 2927. [Google Scholar] [CrossRef] [Green Version]
- Albataineh, M.T.; Kadosh, D. Regulatory roles of phosphorylation in model and pathogenic fungi. Med. Mycol. 2015, 54, 333–352. [Google Scholar] [CrossRef] [Green Version]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Sun, J.; Chen, Y.; Zhu, P.; Zhang, L.; Wu, S.; Ma, D.; Cao, Q.; Li, Z.; Xu, T. Genome-wide identification, structural and gene expression analysis of the bZIP transcription factor family in sweet potato wild relative Ipomoea trifida. BMC Genet. 2019, 20, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurst, H.C. Transcription factors. 1: bZIP proteins. Protein Profile 1994, 1, 123–168. [Google Scholar] [PubMed]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021, 21, 122. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhao, S.; Hu, C.; Mao, C.; Guo, L.; Yu, H.; Yu, H. Whole Genome Sequence of an Edible Mushroom Stropharia rugosoannulata (Daqiugaigu). J. Fungi 2022, 8, 99. [Google Scholar] [CrossRef]
- Deppmann, C.D.; Alvania, R.S.; Taparowsky, E.J. Cross-species annotation of basic leucine zipper factor interactions: Insight into the evolution of closed interaction networks. Mol. Biol. Evol. 2006, 23, 1480–1492. [Google Scholar] [CrossRef] [Green Version]
- Nimmanee, P.; Woo, P.C.; Vanittanakom, P.; Youngchim, S.; Vanittanakom, N. Functional analysis of atfA gene to stress response in pathogenic thermal dimorphic fungus Penicillium marneffei. PLoS ONE 2014, 9, e111200. [Google Scholar] [CrossRef] [Green Version]
- Orosz, E.; Antal, K.; Gazdag, Z.; Szabó, Z.; Han, K.-H.; Yu, J.-H.; Pócsi, I.; Emri, T. Transcriptome-based modeling reveals that oxidative stress induces modulation of the AtfA-dependent signaling networks in Aspergillus nidulans. Int. J. Genom. 2017, 2017, 6923849. [Google Scholar] [CrossRef]








| Gene Name | Gene ID | Length (nt) | MW(Da) | PI | Subcellular Localization | Chr No. | No. of Phosphorylation Sites | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Ser | Thr | Tyr | Total | |||||||
| FgbZIP_1.1 | 23548003 | 1901 | 66620.22 | 4.970 | Nucleus | 1 | 24 | 5 | 3 | 32 |
| FgbZIP_1.2 | 23548989 | 2295 | 62349.02 | 4.490 | Nucleus | 1 | 26 | 9 | 2 | 37 |
| FgbZIP_1.3 | 23549271 | 786 | 53346.27 | 4.610 | Nucleus | 1 | 22 | 7 | 2 | 31 |
| FgbZIP_1.4 | 23549776 | 1048 | 37664.64 | 5.750 | Nucleus | 1 | 20 | 2 | 4 | 26 |
| FgbZIP_1.5 | 23550109 | 1113 | 40664.44 | 7.150 | Nucleus | 1 | 22 | 6 | 2 | 30 |
| FgbZIP_1.6 | 23557062 | 2519 | 55444.4 | 9.010 | ER, Nucleus | 1 | 28 | 12 | 3 | 43 |
| FgbZip_1.7 | 23560565 | 788 | 28795.86 | 5.730 | Nucleus | 1 | 14 | 5 | 3 | 22 |
| FgbZIP_2.1 | 23555786 | 2456 | 63564.24 | 4.950 | Nucleus | 2 | 44 | 9 | 4 | 57 |
| FgbZIP_2.2 | 23560120 | 1260 | 33990.35 | 6.670 | Nucleus | 2 | 14 | 2 | 2 | 18 |
| FgbZIP_2.3 | 23559160 | 1655 | 55819.27 | 5.780 | Nucleus | 2 | 20 | 3 | 6 | 29 |
| FgbZIP_2.4 | 23550377 | 1737 | 61730.63 | 5.010 | Nucleus | 2 | 30 | 8 | 8 | 46 |
| FgbZIP_2.5 | 23550286 | 1279 | 38592.17 | 5.710 | Nucleus | 2 | 15 | 7 | 2 | 24 |
| FgbZIP_2.6 | 23558445 | 1318 | 46629.52 | 4.520 | Nucleus | 2 | 33 | 10 | 1 | 44 |
| FgbZIP_3.1 | 23552363 | 2372 | 31208.33 | 5.330 | Nucleus | 3 | 17 | 3 | 2 | 22 |
| FgbZIP_3.2 | 23553081 | 2218 | 65908.15 | 8.380 | Nucleus | 3 | 40 | 17 | 3 | 60 |
| FgbZIP_3.3 | 23553554 | 1910 | 66021.86 | 5.230 | Cyt, Nucleus | 3 | 30 | 20 | 3 | 53 |
| FgbZIP_4.1 | 23553774 | 1674 | 60396.63 | 9.960 | Nucleus | 4 | 43 | 12 | 5 | 60 |
| FgbZIP_4.2 | 23554841 | 1017 | 33766.39 | 5.750 | Nucleus | 4 | 16 | 10 | 3 | 29 |
| FgbZIP_4.3 | 23554911 | 1280 | 18012.13 | 9.040 | Nucleus | 4 | 22 | 8 | 0 | 30 |
| FgbZIP_4.4 | 23556762 | 1299 | 30497.77 | 6.670 | Nucleus | 4 | 10 | 2 | 1 | 13 |
| FgbZIP_4.5 | 23556246 | 1582 | 24980.68 | 4.730 | ER, Nucleus | 4 | 18 | 1 | 2 | 21 |
| FgbZIP_4.6 | 23555970 | 1008 | 35630.37 | 6.020 | Nucleus | 4 | 25 | 2 | 1 | 28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, S.; Tai, B.; Hussain, A.; Jahan, I.; Yang, B.; Xing, F. Genome-Wide Identification and Expression Analysis of the Basic Leucine Zipper (bZIP) Transcription Factor Gene Family in Fusarium graminearum. Genes 2022, 13, 607. https://doi.org/10.3390/genes13040607
Hussain S, Tai B, Hussain A, Jahan I, Yang B, Xing F. Genome-Wide Identification and Expression Analysis of the Basic Leucine Zipper (bZIP) Transcription Factor Gene Family in Fusarium graminearum. Genes. 2022; 13(4):607. https://doi.org/10.3390/genes13040607
Chicago/Turabian StyleHussain, Sarfaraz, Bowen Tai, Athar Hussain, Israt Jahan, Bolei Yang, and Fuguo Xing. 2022. "Genome-Wide Identification and Expression Analysis of the Basic Leucine Zipper (bZIP) Transcription Factor Gene Family in Fusarium graminearum" Genes 13, no. 4: 607. https://doi.org/10.3390/genes13040607






