Genetic Variants of MIR27A, MIR196A2 May Impact the Risk for the Onset of Coronary Artery Disease in the Pakistani Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Inclusion and Exclusion Criteria for CAD Patients and Healthy Controls
2.3. Blood Samples Collection and Genomic DNA Extraction
2.4. Primers for Allele-Specific PCR and Genotyping of Rs895819, Rs11614913, and Rs2168518
2.5. Statistical Analysis
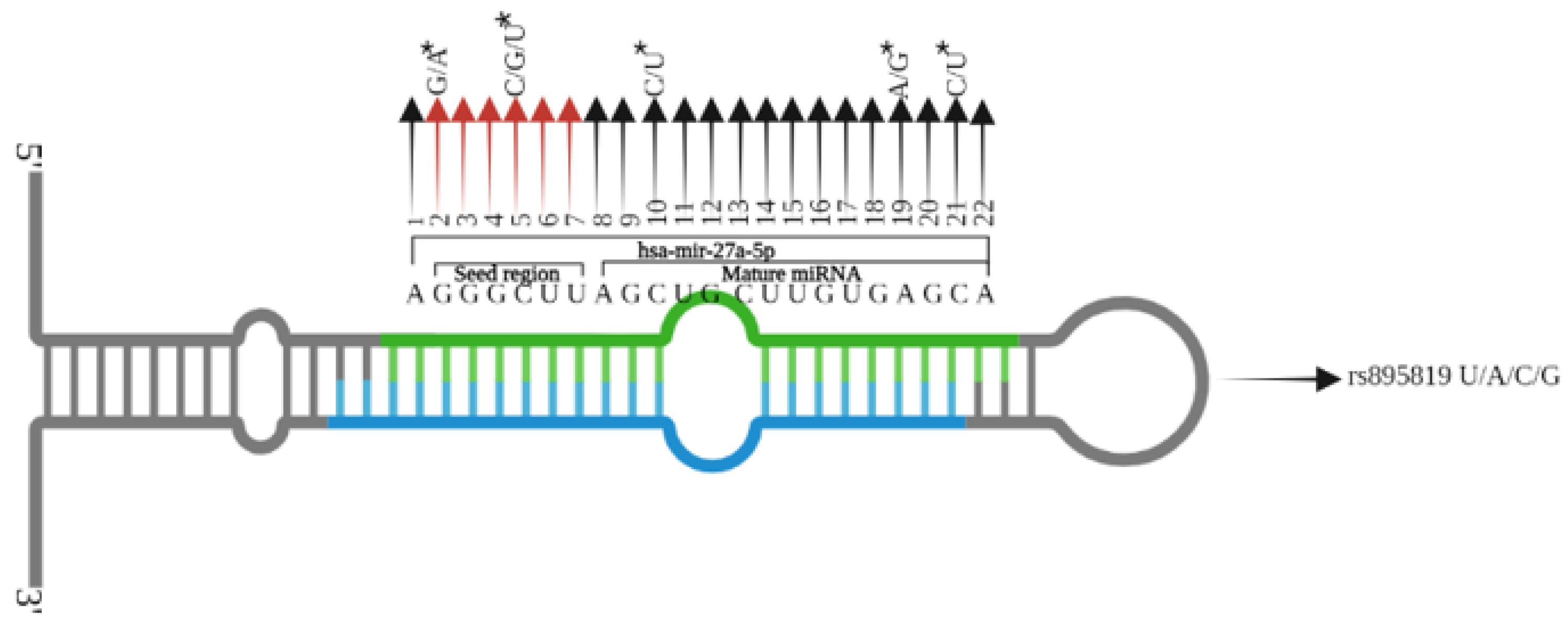
2.6. In-Silico Analyses of the Primary Structure of miRNAs
3. Results
3.1. Association of Rs2168518, Rs895819, and Rs11614913 with CAD
3.2. Consequences of Variant Rs2168518, Rs895819, Rs11614913 on miRNA Structure and Properties
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hill, C.G.; Jabbari, N.; Matyunina, L.V.; McDonald, J.F. Functional and evolutionary significance of human microRNA seed region mutations. PLoS ONE 2014, 9, e115241. [Google Scholar] [CrossRef]
- Klum, S.M.; Chandradoss, S.D.; Schirle, N.T.; Joo, C.; MacRae, I.J. Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition. EMBO J. 2018, 37, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Nigita, G.; Acunzo, M.; Romano, G.; Veneziano, D.; Lagana, A.; Vitiello, M.; Wernicke, D.; Ferro, A.; Croce, C.M. microRNA editing in seed region aligns with cellular changes in hypoxic conditions. Nucleic Acids Res. 2016, 44, 6298–6308. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.; Chen, L.; Luo, Z.; Zhang, T.; Chen, L.; Lou, P.; Sun, W.; Long, X.; Lan, J.; Wang, J.; et al. Spontaneous single nucleotide polymorphism in porcine microRNA-378 seed region leads to functional alteration. Biosci. Biotechnol. Biochem. 2018, 82, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Bronze-da-Rocha, E. MicroRNAs expression profiles in cardiovascular diseases. BioMed Res. Int. 2014, 2014, 985408. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Ullah, S.; John, P.; Bhatti, A. MicroRNAs with a role in gene regulation and in human diseases. Mol. Biol. Rep. 2014, 41, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, N.; Becker, M.; Schumann, T.; Speer, T.; Fehlmann, T.; Keller, A.; Meese, E. Bias in recent miRBase annotations potentially associated with RNA quality issues. Sci. Rep. 2017, 7, 5162. [Google Scholar] [CrossRef]
- van den Berg, A.; Mols, J.; Han, J. RISC-target interaction: Cleavage and translational suppression. Biochim. Biophys. Acta 2008, 1779, 668–677. [Google Scholar] [CrossRef]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar] [CrossRef]
- Tanzer, A.; Stadler, P.F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004, 339, 327–335. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef]
- Wilk, G.; Braun, R. regQTLs: Single nucleotide polymorphisms that modulate microRNA regulation of gene expression in tumors. PLoS Genet. 2018, 14, e1007837. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell. 2016, 64, 320–333. [Google Scholar] [CrossRef]
- Vasudevan, S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip. Rev. RNA 2012, 3, 311–330. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Methods Mol. Biol. 2017, 1617, 57–67. [Google Scholar] [PubMed]
- Moazeni-Roodi, A.; Aftabi, S.; Sarabandi, S.; Karami, S.; Hashemi, M.; Ghavami, S.; Taheri, M. Association Between miR-146a rs2910164 Polymorphism and Breast Cancer Susceptibility: An Updated Meta-analysis of 9545 Cases and 10030 Controls. Microrna 2021, 10, 191–199. [Google Scholar]
- Massignam, E.T.; Dieter, C.; Pellenz, F.M.; Assmann, T.S.; Crispim, D. Involvement of miR-126 rs4636297 and miR-146a rs2910164 polymorphisms in the susceptibility for diabetic retinopathy: A case-control study in a type 1 diabetes population. Acta Ophthalmol. 2021, 99, e461–e469. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.; Elfaki, I.; Jha, C.; Javid, J.; Rehman, S.; Banu, S.; Mir, M.M.; Babakr, A.T.; Chahal, S.M.S. Molecular Evaluation of MicroRNA-146 Gene Variability (rs2910164 C > G) and its Association with Increased Susceptibility to Coronary Artery Disease. Microrna 2020, 9, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Knuuti, J.; Wijns, W.; Saraste, A.; Capodanno, D.; Barbato, E.; Funck-Brentano, C.; Prescott, E.; Storey, R.F.; Deaton, C.; Cuisset, T. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes: The Task Force for the diagnosis and management of chronic coronary syndromes of the European Society of Cardiology (ESC). Eur. Heart J. 2020, 41, 407–477. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Wang, Q.W.; Zhao, J.; Liu, X.; He, Z. miR-149 and miR-499 gene polymorphism and the incident of ischemic stroke in the Asian population: From a case-control study to meta-analysis. Clin. Neurol. Neurosurg. 2020, 193, 105789. [Google Scholar] [CrossRef]
- Ahmed, U.; Khaliq, S.; Ahmad, H.U.; Ahmad, I.; Ashfaq, U.A.; Qasim, M.; Masoud, M.S. Pathogenesis of Diabetic Cardiomyopathy and Role of miRNA. Crit. Rev. Eukaryot. Gene Expr. 2021, 31, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Fawzy, M.S.; Toraih, E.A.; Hamed, E.O.; Hussein, M.H.; Ismail, H.M. Association of MIR-499a expression and seed region variant (rs3746444) with cardiovascular disease in Egyptian patients. Acta Cardiol. 2018, 73, 131–140. [Google Scholar]
- Mullany, L.E.; Herrick, J.S.; Wolff, R.K.; Slattery, M.L. MicroRNA Seed Region Length Impact on Target Messenger RNA Expression and Survival in Colorectal Cancer. PLoS ONE 2016, 11, e0154177. [Google Scholar] [CrossRef] [PubMed]
- Thom, T.; Haase, N.; Rosamond, W.; Howard, V.J.; Rumsfeld, J.; Manolio, T.; Zheng, Z.J.; Flegal, K.; O’Donnell, C.; Kittner, S.; et al. Heart disease and stroke statistics—2006 update: A report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 2006, 113, e85–e151. [Google Scholar]
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; de Ferranti, S.; Despres, J.P.; Fullerton, H.J.; Howard, V.J.; et al. Heart disease and stroke statistics—2015 update: A report from the American Heart Association. Circulation 2015, 131, e29–e322. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Kristensen, K.K.; Ploug, M.; Winther, A.L. The Importance of Lipoprotein Lipase Regulation in Atherosclerosis. Biomedicines 2021, 9, 782. [Google Scholar] [CrossRef]
- Pella, Z.; Pella, D.; Paralic, J.; Vanko, J.I.; Fedacko, J. Analysis of Risk Factors in Patients with Subclinical Atherosclerosis and Increased Cardiovascular Risk Using Factor Analysis. Diagnostics 2021, 11, 1284. [Google Scholar] [CrossRef]
- Cagle, S.D., Jr.; Cooperstein, N. Coronary Artery Disease: Diagnosis and Management. Prim. Care 2018, 45, 45–61. [Google Scholar] [CrossRef]
- Hughes, M.F.; Lenighan, Y.M.; Godson, C.; Roche, H.M. Exploring Coronary Artery Disease GWAs Targets With Functional Links to Immunometabolism. Front. Cardiovasc. Med. 2018, 5, 148. [Google Scholar] [CrossRef] [PubMed]
- Nikpay, M.; Goel, A.; Won, H.H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar]
- Cavarretta, E.; Frati, G. MicroRNAs in Coronary Heart Disease: Ready to Enter the Clinical Arena? BioMed Res. Int. 2016, 2016, 2150763. [Google Scholar] [CrossRef]
- Schulte, C.; Zeller, T. microRNA-based diagnostics and therapy in cardiovascular disease-Summing up the facts. Cardiovasc. Diagn. Ther. 2015, 5, 17–36. [Google Scholar]
- Thanikachalam, P.V.; Ramamurthy, S.; Wong, Z.W.; Koo, B.J.; Wong, J.Y.; Abdullah, M.F.; Chin, Y.H.; Chia, C.H.; Tan, J.Y.; Neo, W.T.; et al. Current attempts to implement microRNA-based diagnostics and therapy in cardiovascular and metabolic disease: A promising future. Drug Discov. Today 2018, 23, 460–480. [Google Scholar] [CrossRef]
- Churov, A.; Summerhill, V.; Grechko, A.; Orekhova, V.; Orekhov, A. MicroRNAs as potential biomarkers in atherosclerosis. Int. J. Mol. Sci. 2019, 20, 5547. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Gholipour, M.; Taheri, M. Role of microRNAs in the pathogenesis of coronary artery disease. Front. Cardiovasc. Med. 2021, 8, 632392. [Google Scholar] [CrossRef]
- Wang, M.; Ji, Y.; Cai, S.; Ding, W. MiR-206 Suppresses the Progression of Coronary Artery Disease by Modulating Vascular Endothelial Growth Factor (VEGF) Expression. Med. Sci. Monit. 2016, 22, 5011–5020. [Google Scholar] [CrossRef]
- Kaur, A.; Mackin, S.T.; Schlosser, K.; Wong, F.L.; Elharram, M.; Delles, C.; Stewart, D.J.; Dayan, N.; Landry, T.; Pilote, L. Systematic review of microRNA biomarkers in acute coronary syndrome and stable coronary artery disease. Cardiovasc. Res. 2020, 116, 1113–1124. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Q.; Hosen, M.R.; Zietzer, A.; Flender, A.; Levermann, P.; Schmitz, T.; Frühwald, D.; Goody, P.; Nickenig, G. Atherosclerotic conditions promote the packaging of functional microRNA-92a-3p into endothelial microvesicles. Circ. Res. 2019, 124, 575–587. [Google Scholar] [CrossRef]
- Ren, J.; Ma, R.; Zhang, Z.B.; Li, Y.; Lei, P.; Men, J.L. Effects of microRNA-330 on vulnerable atherosclerotic plaques formation and vascular endothelial cell proliferation through the WNT signaling pathway in acute coronary syndrome. J. Cell. Biochem. 2018, 119, 4514–4527. [Google Scholar] [CrossRef]
- Yang, B.; Lin, H.; Xiao, J.; Lu, Y.; Luo, X.; Li, B.; Zhang, Y.; Xu, C.; Bai, Y.; Wang, H. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 2007, 13, 486–491. [Google Scholar] [CrossRef]
- Sayed, D.; Hong, C.; Chen, I.-Y.; Lypowy, J.; Abdellatif, M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ. Res. 2007, 100, 416–424. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Bui, A.V.; Diesch, J.; Manasseh, R.; Hausding, C.; Rivera, J.; Haviv, I.; Agrotis, A.; Htun, N.M.; Jowett, J. A novel mouse model of atherosclerotic plaque instability for drug testing and mechanistic/therapeutic discoveries using gene and microRNA expression profiling. Circ. Res. 2013, 113, 252–265. [Google Scholar] [CrossRef]
- Ghanbari, M.; de Vries, P.S.; de Looper, H.; Peters, M.J.; Schurmann, C.; Yaghootkar, H.; Dorr, M.; Frayling, T.M.; Uitterlinden, A.G.; Hofman, A.; et al. A genetic variant in the seed region of miR-4513 shows pleiotropic effects on lipid and glucose homeostasis, blood pressure, and coronary artery disease. Hum. Mutat. 2014, 35, 1524–1531. [Google Scholar] [CrossRef]
- Mir, R.; Jha, C.K.; Elfaki, I.; Javid, J.; Rehman, S.; Khullar, N.; Banu, S.; Chahal, S.M.S. Incidence of MicroR-4513C/T Gene Variability in Coronary Artery Disease—A Case-Control Study. Endocr. Metab. Immune Disord.-Drug Targets 2019, 19, 1216–1223. [Google Scholar] [CrossRef]
- Mashayekhi, S.; Saeidi Saedi, H.; Salehi, Z.; Soltanipour, S.; Mirzajani, E. Effects of miR-27a, miR-196a2 and miR-146a polymorphisms on the risk of breast cancer. Br. J. Biomed. Sci. 2018, 75, 76–81. [Google Scholar] [CrossRef]
- Ding, Y.; Chan, C.Y.; Lawrence, C.E. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA 2005, 11, 1157–1166. [Google Scholar] [CrossRef]
- Previtali, E.; Bucciarelli, P.; Passamonti, S.M.; Martinelli, I. Risk factors for venous and arterial thrombosis. Blood Transfus. 2011, 9, 120–138. [Google Scholar] [PubMed]
- Libby, P.; Buring, J.E.; Badimon, L.; Hansson, G.K.; Deanfield, J.; Bittencourt, M.S.; Tokgözoğlu, L.; Lewis, E.F. Atherosclerosis. Nat. Rev. Dis. Primers 2019, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Haverich, A.; Boyle, E.C. Atherosclerosis Pathogenesis and Microvascular Dysfunction; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Choi, G.; Lee, J.M.; Kim, H.-J.; Park, J.-B.; Sankaran, S.; Otake, H.; Doh, J.-H.; Nam, C.-W.; Shin, E.-S.; Taylor, C.A. Coronary artery axial plaque stress and its relationship with lesion geometry: Application of computational fluid dynamics to coronary CT angiography. Cardiovasc. Imaging 2015, 8, 1156–1166. [Google Scholar]
- Ratiu, M.; Chitu, M.; Benedek, I.; Benedek, T.; Kovacs, I.; Rat, N.; Rezus, C. Impact of coronary plaque geometry on plaque vulnerability and its association with the risk of future cardiovascular events in patients with chest pain undergoing coronary computed tomographic angiography—The GEOMETRY study: Protocol for a prospective clinical trial. Medicine 2018, 97, e13498. [Google Scholar]
- Sachidanandam, R.; Weissman, D.; Schmidt, S.C.; Kakol, J.M.; Stein, L.D.; Marth, G.; Sherry, S.; Mullikin, J.C.; Mortimore, B.J.; Willey, D.L.; et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 2001, 409, 928–933. [Google Scholar]
- Hu, G.; Modrek, B.; Riise Stensland, H.M.; Saarela, J.; Pajukanta, P.; Kustanovich, V.; Peltonen, L.; Nelson, S.F.; Lee, C. Efficient discovery of single-nucleotide polymorphisms in coding regions of human genes. Pharm. J. 2002, 2, 236–242. [Google Scholar] [CrossRef][Green Version]
- Robert, F.; Pelletier, J. Exploring the Impact of Single-Nucleotide Polymorphisms on Translation. Front. Genet. 2018, 9, 507. [Google Scholar] [CrossRef]
- Condorelli, G.; Latronico, M.V.; Cavarretta, E. microRNAs in cardiovascular diseases: Current knowledge and the road ahead. J. Am. Coll. Cardiol. 2014, 63, 2177–2187. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Krol, J.; Sobczak, K.; Wilczynska, U.; Drath, M.; Jasinska, A.; Kaczynska, D.; Krzyzosiak, W.J. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J. Biol. Chem. 2004, 279, 42230–42239. [Google Scholar] [CrossRef]
- Kawamata, T.; Seitz, H.; Tomari, Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009, 16, 953–960. [Google Scholar] [CrossRef]
- Li, Q.; Chen, L.; Chen, D.; Wu, X.; Chen, M. Influence of microRNA-related polymorphisms on clinical outcomes in coronary artery disease. Am. J. Transl. Res. 2015, 7, 393–400. [Google Scholar]
- Yang, S.; Zheng, Y.; Zhou, L.; Jin, J.; Deng, Y.; Yao, J.; Yang, P.; Yao, L.; Wu, Y.; Zhai, Z.; et al. miR-499 rs3746444 and miR-196a-2 rs11614913 Are Associated with the Risk of Glioma, but Not the Prognosis. Mol. Ther. Nucleic Acids 2020, 22, 340–351. [Google Scholar] [CrossRef]
- Ahmad, M.; Shah, A.A. Predictive role of single nucleotide polymorphism (rs11614913) in the development of breast cancer in Pakistani population. Pers. Med. 2020, 17, 213–227. [Google Scholar] [CrossRef]
- Liu, Y.; He, A.; Liu, B.; Zhong, Y.; Liao, X.; Yang, J.; Chen, J.; Wu, J.; Mei, H. rs11614913 polymorphism in miRNA-196a2 and cancer risk: An updated meta-analysis. OncoTargets Ther. 2018, 11, 1121–1139. [Google Scholar] [CrossRef]
- Soltanian, A.R.; Hosseini, B.; Mahjub, H.; Bahreini, F.; Nazemalhosseini Mojarad, E.; Ghaffari, M.E. Association between rs11614913 Polymorphism of The MiR-196-a2 Gene and Colorectal Cancer in The Presence of Departure from Hardy-Weinberg Equilibrium. Cell J. 2021, 23, 313–318. [Google Scholar]
- Zhu, Z.; Zhang, Y.; Bai, R.; Yang, R.; Shan, Z.; Ma, C.; Yang, J.; Sun, D. Association of Genetic Polymorphisms in MicroRNAs With Type 2 Diabetes Mellitus in a Chinese Population. Front. Endocrinol. 2020, 11, 587561. [Google Scholar] [CrossRef]
- Zhang, S.; Han, Q.; Zhu, K.; Wang, Q. The association of miR-27a rs895819 polymorphism with colorectal cancer risk in Chinese population. J. Clin. Lab. Anal. 2020, 34, e23497. [Google Scholar] [CrossRef]
| Categories | Age (Year) | Gender | BMI (kg/m2) | RBS (mg/dL) | TC (mg/dL) | TG (mg/dL) | HDL (mg/dL) | LDL (mg/dL) |
|---|---|---|---|---|---|---|---|---|
| CAD | 55.2 (27–91) | Male = 183 | 23.3 (12.2–38.2) | 245.8 (114–415) | 222.7 (156–262) | 199.3 (110–395) | 38.2 (21–58) | 126.8 (24–271) |
| Female = 40 | ||||||||
| Controls | ±45 | Male = 138 | ±22.3 | ±112.4 | ±200 | ±162.5 | ±35 | ±100 |
| Female = 12 |
| SNP ID | miRNA Gene Name | Name of Mature miRNA Sequences | Chromosome No. | miRNA Location (Coordinates) | Coded Allele | Other Alleles | MAF |
|---|---|---|---|---|---|---|---|
| rs895819 | MIR27A | hsa-miR-27a-5p | 19 | 13836440-13836517 [−] | T | A/C/G | 0.50 |
| hsa-miR-27a-3p | |||||||
| rs11614913 | MIR196A2 | hsa-miR-196a-5p | 12 | 53991738-53991847 [+] | C | T | 0.49 |
| hsa-miR-196a-3p | |||||||
| rs2168518 | MIR4513 | hsa-miR-4513 | 15 | 74788672-74788757 [−] | G | A | 0.47 |
| Gene (Accession Number) | Statistical Models | Genotypes | Cases | Control | Odds Ratiο (95% CI) | χ2-Value, df | p-Value |
|---|---|---|---|---|---|---|---|
| MIR196A2 (rs11614913) | Co-dominant | CC CT TT | 24 40 16 | 50 19 11 | — | 54.4, 2 | <0.0001 |
| Dominant | CC CT + TT | 24 56 | 50 30 | 0.257 (0.133–0.496) | — | <0.0001 | |
| Recessive | TT CT + CC | 16 64 | 11 69 | 1.56 (0.677–0.632) | — | 0.398 | |
| Additive | C T | 88 72 | 119 41 | 0.421 (0.262–0.675) | — | 0.0004 | |
| MIR27A (rs895819) | Co-dominant | AA AG GG | 10 46 4 | 28 35 5 | — | 9.669, 2 | <0.008 |
| Dominant | AA AG + GG | 10 50 | 28 40 | 0.285 (0.1242–0.6575) | — | <0.0034 | |
| Recessive | GG AG + AA | 4 56 | 5 63 | 0.900 (0.3202–3.519) | — | 1.000 | |
| Additive | A G | 66 54 | 91 45 | 0.604 (0.3640–1.002) | — | 0.05 | |
| MIR4513 (rs2168518) | Co-dominant | GG GA AA | 14 105 24 | 4 47 19 | — | 3.682, 2 | 0.1586 |
| Dominant | GG GA + AA | 14 129 | 4 66 | 1.791 (0.5668–5.658) | — | 0.4340 | |
| Recessive | AA GA + GG | 24 119 | 19 51 | 0.5414 (0.2727–1.075) | — | 0.1012 | |
| Additive | G A | 133 153 | 55 85 | 1.343 (0.8905–2.027) | — | 0.1773 |
| Parameters | Reference | Mutated | Reference | Mutated | Reference | Mutated |
|---|---|---|---|---|---|---|
| MIR4513 rs2168518 | MIR4513 rs2168518 | MIR27A rs895819 | MIR27A rs895819 | MIR196A2 rs11614913 | MIR196A2 rs11614913 | |
| Free energy of the thermodynamic ensemble | −41.83 kcal/mol | −42.34 kcal/mol | −38.24 kcal/mol | −38.28 kcal/mol. | −52.02 kcal/mol | −46.52 kcal/mol |
| The frequency of the MFE structure in the ensemble | 26.16% | 18.55% | 15.62%. | 14.84%. | 6.14% | 5.18% |
| The ensemble diversity | 3.58 | 3.50 | 4.41 | 4.55 | 7.18 | 7.49 |
| The optimal secondary structure with a minimum free energy | −41.00 kcal/mol | −41.30 kcal/mol | −34.40 kcal/mol | −34.40 kcal/mol | −50.30 kcal/mol | −44.70 kcal/mol |
| The centroid secondary structure | −41.00 kcal/mol | −41.30 kcal/mol | −37.10 kcal/mol | −37.10 kcal/mol | −49.90 kcal/mol | −44.30 kcal/mol |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haq, T.U.; Zahoor, A.; Ali, Y.; Chen, Y.; Jalil, F.; Shah, A.A. Genetic Variants of MIR27A, MIR196A2 May Impact the Risk for the Onset of Coronary Artery Disease in the Pakistani Population. Genes 2022, 13, 747. https://doi.org/10.3390/genes13050747
Haq TU, Zahoor A, Ali Y, Chen Y, Jalil F, Shah AA. Genetic Variants of MIR27A, MIR196A2 May Impact the Risk for the Onset of Coronary Artery Disease in the Pakistani Population. Genes. 2022; 13(5):747. https://doi.org/10.3390/genes13050747
Chicago/Turabian StyleHaq, Taqweem Ul, Abdul Zahoor, Yasir Ali, Yangchao Chen, Fazal Jalil, and Aftab Ali Shah. 2022. "Genetic Variants of MIR27A, MIR196A2 May Impact the Risk for the Onset of Coronary Artery Disease in the Pakistani Population" Genes 13, no. 5: 747. https://doi.org/10.3390/genes13050747
APA StyleHaq, T. U., Zahoor, A., Ali, Y., Chen, Y., Jalil, F., & Shah, A. A. (2022). Genetic Variants of MIR27A, MIR196A2 May Impact the Risk for the Onset of Coronary Artery Disease in the Pakistani Population. Genes, 13(5), 747. https://doi.org/10.3390/genes13050747







