Rare Variant Analysis and Molecular Dynamics Simulation in Alzheimer’s Disease Identifies Exonic Variants in FLG
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Statistical Analysis
2.2.1. Demographic Comparisons
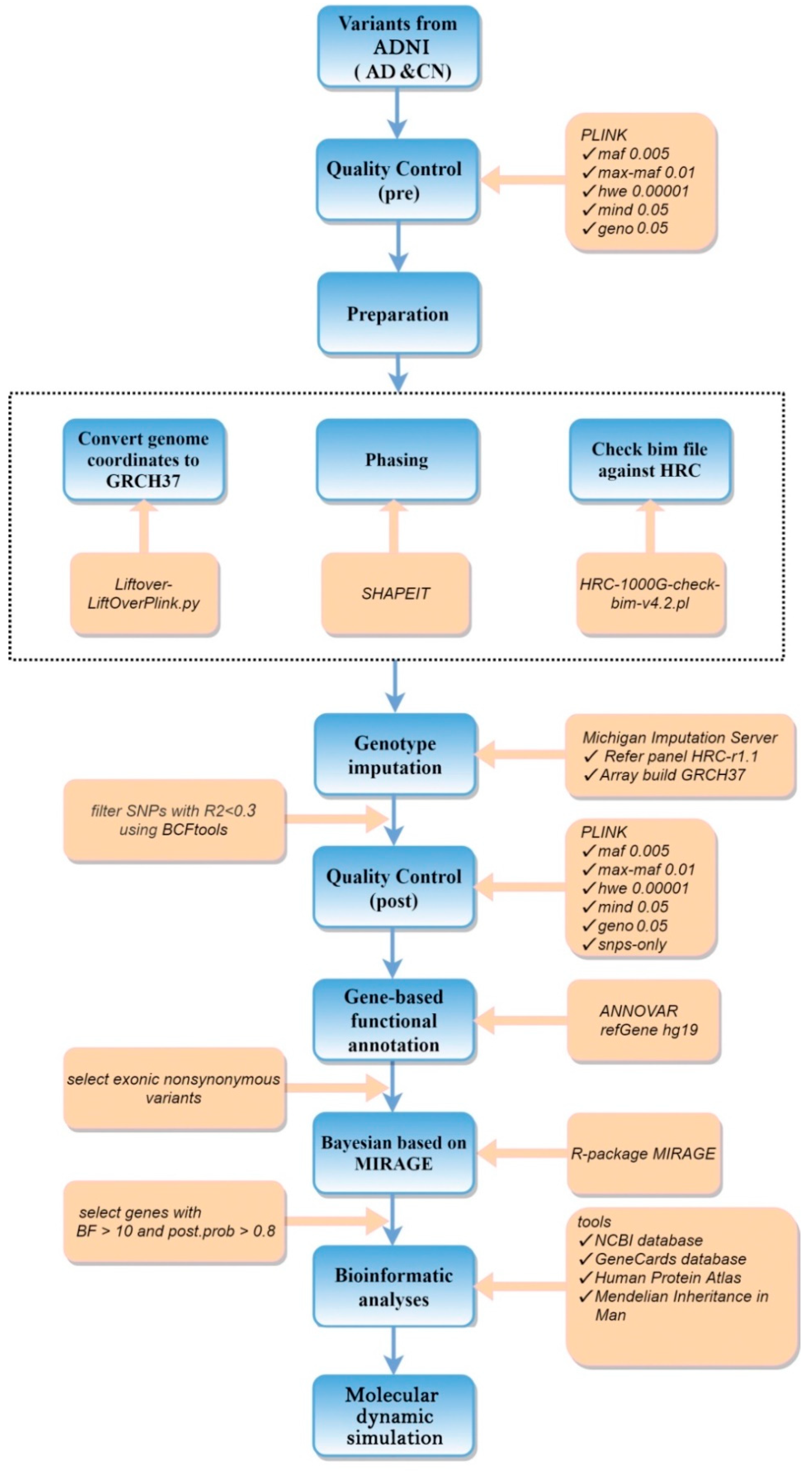
2.2.2. Data Preprocessing
2.2.3. RVAS Analysis
2.3. Bioinformatic Analyses
2.4. Molecular Dynamics Simulation
3. Results
3.1. Demographic Comparisons
3.2. RVAS Analysis
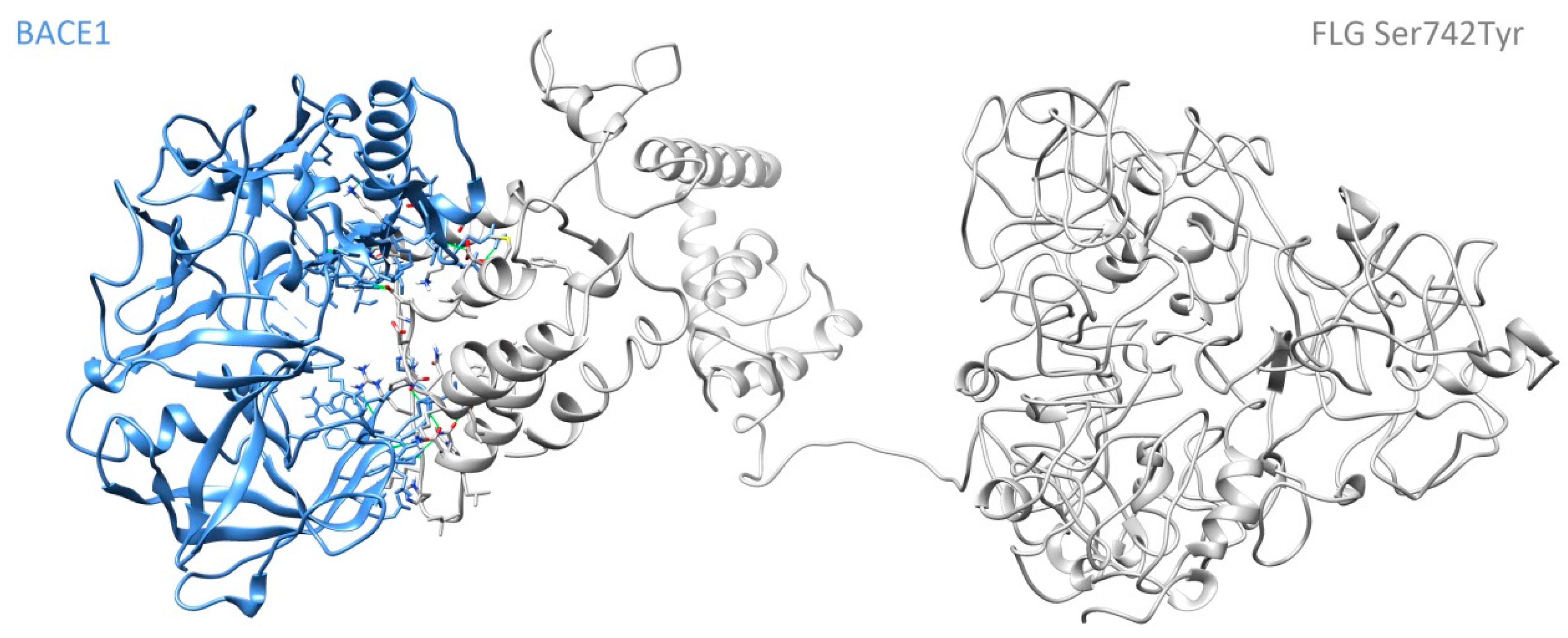
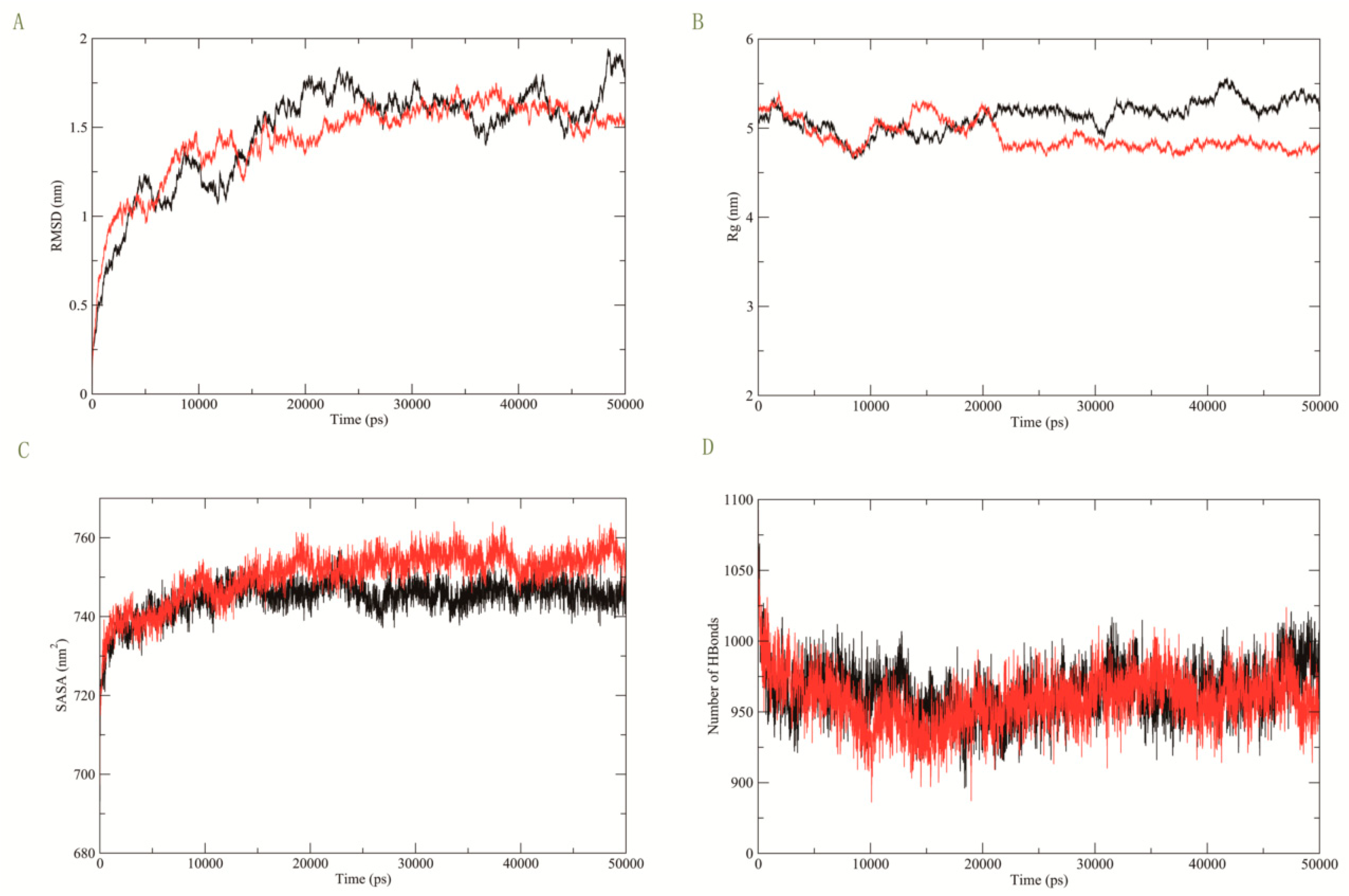
3.3. Molecular Dynamics Simulation
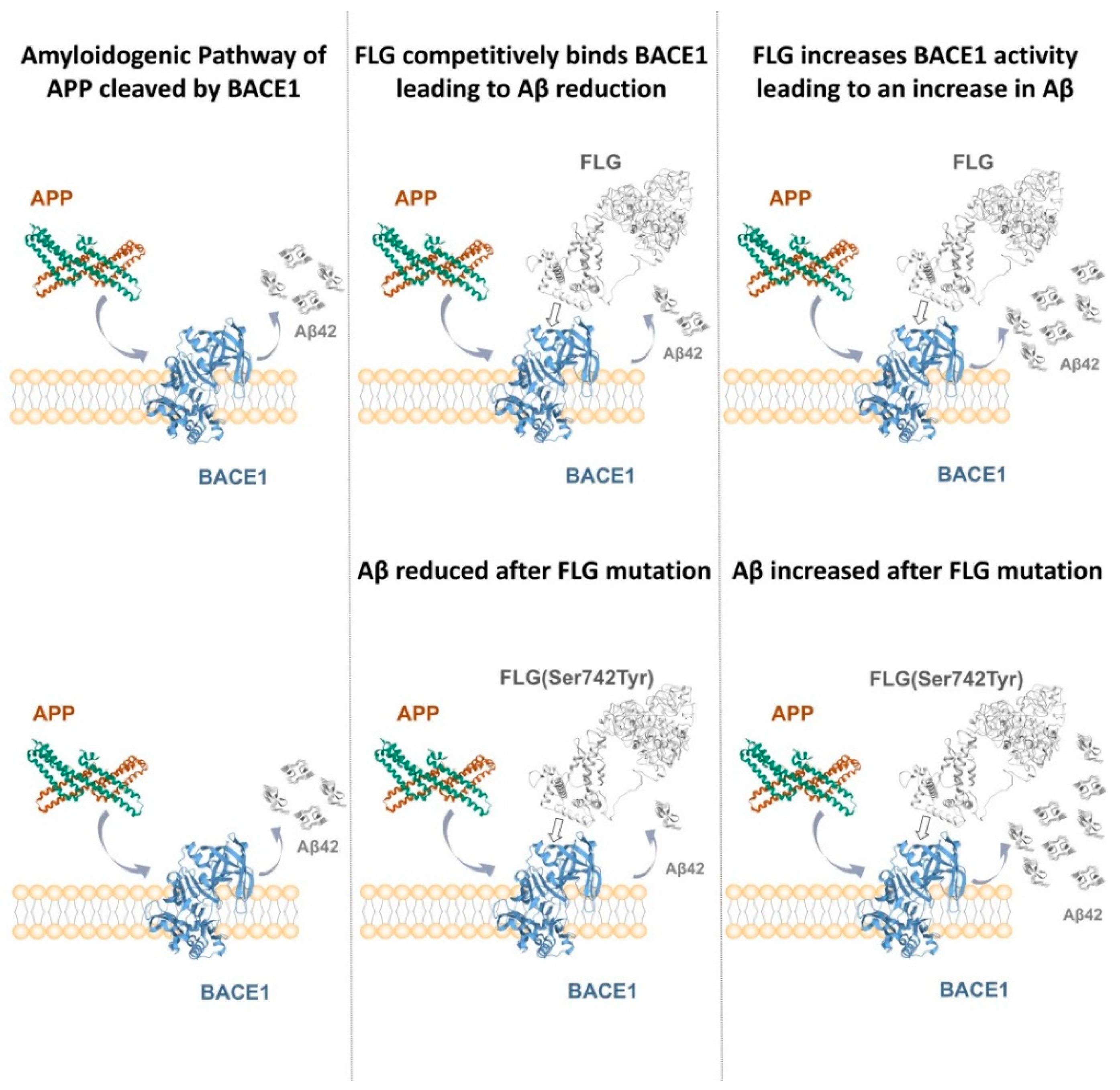
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aggleton, J.P.; Pralus, A.; Nelson, A.J.; Hornberger, M. Thalamic pathology and memory loss in early Alzheimer’s disease: Moving the focus from the medial temporal lobe to Papez circuit. Brain 2016, 139, 1877–1890. [Google Scholar] [CrossRef] [PubMed]
- Key Facts of Dementia. Available online: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed on 2 September 2021).
- Gatz, M.; Reynolds, C.A.; Fratiglioni, L.; Johansson, B.; Mortimer, J.A.; Berg, S.; Fiske, A.; Pedersen, N.L. Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry 2006, 63, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Ridge, P.G.; Hoyt, K.B.; Boehme, K.; Mukherjee, S.; Crane, P.K.; Haines, J.L.; Mayeux, R.; Farrer, L.A.; Pericak-Vance, M.A.; Schellenberg, G.D. Assessment of the genetic variance of late-onset Alzheimer’s disease. Neurobiol. Aging 2016, 41, 200.e213–200.e220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghavan, N.S.; Dumitrescu, L.; Mormino, E.; Mahoney, E.R.; Lee, A.J.; Gao, Y.; Bilgel, M.; Goldstein, D.; Harrison, T.; Engelman, C.D. Association between common variants in RBFOX1, an RNA-binding protein, and brain amyloidosis in early and preclinical Alzheimer disease. JAMA Neurol. 2020, 77, 1288–1298. [Google Scholar] [CrossRef]
- Michaelson, D.M. APOE ε4: The most prevalent yet understudied risk factor for Alzheimer’s disease. Alzheimer’s Dement. 2014, 10, 861–868. [Google Scholar] [CrossRef]
- Zuk, O.; Schaffner, S.F.; Samocha, K.; Do, R.; Hechter, E.; Kathiresan, S.; Daly, M.J.; Neale, B.M.; Sunyaev, S.R.; Lander, E.S. Searching for missing heritability: Designing rare variant association studies. Proc. Natl. Acad. Sci. USA 2014, 111, E455–E464. [Google Scholar] [CrossRef] [Green Version]
- Cruchaga, C.; Chakraverty, S.; Mayo, K.; Vallania, F.L.; Mitra, R.D.; Faber, K.; Williamson, J.; Bird, T.; Diaz-Arrastia, R.; Foroud, T.M. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s disease families. PLoS ONE 2012, 7, e31039. [Google Scholar] [CrossRef]
- So, H.C.; Gui, A.H.; Cherny, S.S.; Sham, P.C. Evaluating the heritability explained by known susceptibility variants: A survey of ten complex diseases. Genet. Epidemiol. 2011, 35, 310–317. [Google Scholar] [CrossRef]
- Bagyinszky, E.; Giau, V.V.; An, S.A. Transcriptomics in Alzheimer’s Disease: Aspects and Challenges. Int. J. Mol. Sci. 2020, 21, 3517. [Google Scholar] [CrossRef]
- Cirulli, E.T.; White, S.; Read, R.W.; Elhanan, G.; Metcalf, W.J.; Tanudjaja, F.; Fath, D.M.; Sandoval, E.; Isaksson, M.; Schlauch, K.A. Genome-wide rare variant analysis for thousands of phenotypes in over 70,000 exomes from two cohorts. Nat. Commun. 2020, 11, 542. [Google Scholar] [CrossRef]
- Huffman, J.E.; De Vries, P.S.; Morrison, A.C.; Sabater-Lleal, M.; Kacprowski, T.; Auer, P.L.; Brody, J.A.; Chasman, D.I.; Chen, M.-H.; Guo, X. Rare and low-frequency variants and their association with plasma levels of fibrinogen, FVII, FVIII, and vWF. Blood 2015, 126, e19–e29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerreiro, R.; Wojtas, A.; Bras, J.; Carrasquillo, M.; Rogaeva, E.; Majounie, E.; Cruchaga, C.; Sassi, C.; Kauwe, J.S.; Younkin, S. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2013, 368, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Van Cauwenberghe, C.; Van Broeckhoven, C.; Sleegers, K. The genetic landscape of Alzheimer disease: Clinical implications and perspectives. Genet. Med. 2016, 18, 421–430. [Google Scholar] [CrossRef] [Green Version]
- Hollingworth, P.; Harold, D.; Sims, R.; Gerrish, A.; Lambert, J.-C.; Carrasquillo, M.M.; Abraham, R.; Hamshere, M.L.; Pahwa, J.S.; Moskvina, V. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 2011, 43, 429–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Wang, P.; Meng, J.; Chen, L.; Zhu, W.; Ma, W. A permutation method for detecting trend correlations in rare variant association studies. Genet. Res. 2019, 101, E13. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Abecasis, G.R.; Boehnke, M.; Lin, X. Rare-variant association analysis: Study designs and statistical tests. Am. J. Hum. Genet. 2014, 95, 5–23. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Knoblauch, N.; Wang, G.; Zhao, S.; Liu, Y.; Xie, Y.; Sheng, W.; Nguyen, H.T.; He, X. A Bayesian method for rare variant analysis using functional annotations and its application to Autism. bioRxiv 2019, 828061. [Google Scholar]
- Hu, G.; Di Paola, L.; Liang, Z.; Giuliani, A. Comparative study of elastic network model and protein contact network for protein complexes: The hemoglobin case. BioMed Res. Int. 2017, 2017, 2483264. [Google Scholar] [CrossRef]
- Von Mering, C.; Krause, R.; Snel, B.; Cornell, M.; Oliver, S.G.; Fields, S.; Bork, P. Comparative assessment of large-scale data sets of protein–protein interactions. Nature 2002, 417, 399–403. [Google Scholar] [CrossRef]
- Koelsch, G. BACE1 function and inhibition: Implications of intervention in the amyloid pathway of Alzheimer’s disease pathology. Molecules 2017, 22, 1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, R.; Vassar, R. Targeting the β secretase BACE1 for Alzheimer’s disease therapy. Lancet Neurol. 2014, 13, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Franzmeier, N.; Dewenter, A.; Frontzkowski, L.; Dichgans, M.; Rubinski, A.; Neitzel, J.; Smith, R.; Strandberg, O.; Ossenkoppele, R.; Buerger, K. Patient-centered connectivity-based prediction of tau pathology spread in Alzheimer’s disease. Sci. Adv. 2020, 6, eabd1327. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, G.; Zhu, W.; Plowey, E.D. BECN1/Beclin 1 sorts cell-surface APP/amyloid β precursor protein for lysosomal degradation. Autophagy 2016, 12, 2404–2419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakravarthy, M.; Veedu, R.N. BACE1 inhibition using 2′-OMePS steric blocking antisense oligonucleotides. Genes 2019, 10, 705. [Google Scholar] [CrossRef] [Green Version]
- Spooner, A.; Chen, E.; Sowmya, A.; Sachdev, P.; Kochan, N.A.; Trollor, J.; Brodaty, H. A comparison of machine learning methods for survival analysis of high-dimensional clinical data for dementia prediction. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Osswald, H.L. BACE1 (β-secretase) inhibitors for the treatment of Alzheimer’s disease. Chem. Soc. Rev. 2014, 43, 6765–6813. [Google Scholar] [CrossRef] [Green Version]
- Agashe, D.; Sane, M.; Phalnikar, K.; Diwan, G.D.; Habibullah, A.; Martinez-Gomez, N.C.; Sahasrabuddhe, V.; Polachek, W.; Wang, J.; Chubiz, L.M. Large-effect beneficial synonymous mutations mediate rapid and parallel adaptation in a bacterium. Mol. Biol. Evol. 2016, 33, 1542–1553. [Google Scholar] [CrossRef] [Green Version]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [Green Version]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Bassot, C.; Elofsson, A. Accurate contact-based modelling of repeat proteins predicts the structure of new repeats protein families. PLoS Comput. Biol. 2021, 17, e1008798. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Dominguez, C.; Boelens, R.; Bonvin, A.M. HADDOCK: A protein—Protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Zhao, S.; Liu, B.; Zhang, Q.; Li, Y.; Liu, J.; Shen, Y.; Ding, X.; Lin, J.; Wu, Y. Perturbations of BMP/TGF-β and VEGF/VEGFR signalling pathways in non-syndromic sporadic brain arteriovenous malformations (BAVM). J. Med. Genet. 2018, 55, 675–684. [Google Scholar] [CrossRef]
- Hart, M.; Merz, P.; Bennett-Gray, J.; Menezes, A.; Goeken, J.; Schelper, R.; Wisniewski, H. β-amyloid protein of Alzheimer’s disease is found in cerebral and spinal cord vascular malformations. Am. J. Pathol. 1988, 132, 167. [Google Scholar]
- Tsonou, E. Exploring the Importance of PREX Proteins in Glucose Homeostasis and Insulin Signalling. Ph.D. Thesis, University of Cambridge, Cambridge, UK, 2020. [Google Scholar]
- Galea, E.; Weinstock, L.D.; Larramona-Arcas, R.; Pybus, A.F.; Giménez-Llort, L.; Escartin, C.; Wood, L.B. Multi-transcriptomic analysis points to early organelle dysfunction in human astrocytes in Alzheimer’s disease. Neurobiol. Dis. 2022, 166, 105655. [Google Scholar] [CrossRef]
- Duke, C.G.; Kennedy, A.J.; Gavin, C.F.; Day, J.J.; Sweatt, J.D. Experience-dependent epigenomic reorganization in the hippocampus. Learn. Mem. 2017, 24, 278–288. [Google Scholar] [CrossRef]
- Fernandez, V.; Huey, D.; Budde, J.P.; Farias, F.H.; Harari, O.; Norton, J.; Gentsch, J.; Morris, J.C.; Goate, A.; Rademakers, R. O2-10-02: Whole Exome Sequencing Analysis in Early Onset Alzheimer Reveals Novel Candidate Genes. Alzheimer’s Dement. 2019, 15, P564. [Google Scholar] [CrossRef]
- Li, C. Understanding Rare Neurological Disorders Using Drosophila Models: Mechanistic Characterization of Neurotoxicity in Snyder-Robinson Syndrome. Ph.D. Thesis, University of Miami, Coral Gables, FL, USA, 2018. [Google Scholar]
- Hawkes, C.A.; Gatherer, M.; Sharp, M.M.; Dorr, A.; Yuen, H.M.; Kalaria, R.; Weller, R.O.; Carare, R.O. Regional differences in the morphological and functional effects of aging on cerebral basement membranes and perivascular drainage of amyloid-β from the mouse brain. Aging Cell 2013, 12, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Chen, X.; Ning, C.; Zhu, Q.; Yao, Y.; Zhao, Y.; Luan, F. Methylation of the genes ROD1, NLRC5, and HKR1 is associated with aging in Hainan centenarians. BMC Med. Genom. 2018, 11, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altuna, M.; Urdánoz-Casado, A.; Sánchez-Ruiz de Gordoa, J.; Zelaya, M.V.; Labarga, A.; Lepesant, J.M.; Roldán, M.; Blanco-Luquin, I.; Perdones, Á.; Larumbe, R. DNA methylation signature of human hippocampus in Alzheimer’s disease is linked to neurogenesis. Clin. Epigenetics 2019, 11, 91. [Google Scholar] [CrossRef] [Green Version]
- van Doorn, J.; van den Bergh, D.; Böhm, U.; Dablander, F.; Derks, K.; Draws, T.; Etz, A.; Evans, N.J.; Gronau, Q.F.; Haaf, J.M. The JASP guidelines for conducting and reporting a Bayesian analysis. Psychon. Bull. Rev. 2021, 28, 813–826. [Google Scholar] [CrossRef]
- Hewett, K.; Sanders, D.B.; Grove, R.A.; Broderick, C.L.; Rudo, T.J.; Bassiri, A.; Zvartau-Hind, M.; Bril, V.; Group, B.S. Randomized study of adjunctive belimumab in participants with generalized myasthenia gravis. Neurology 2018, 90, e1425–e1434. [Google Scholar] [CrossRef] [Green Version]
- Hooper, C.; Meimaridou, E.; Tavassoli, M.; Melino, G.; Lovestone, S.; Killick, R. p53 is upregulated in Alzheimer’s disease and induces tau phosphorylation in HEK293a cells. Neurosci. Lett. 2007, 418, 34–37. [Google Scholar] [CrossRef] [Green Version]
- Black, S.K. An Investigation into the Roles of p53, Nodal/Activin and Fibroblast Growth Factor Signalling in Early Heart Development. Ph.D. Thesis, Cardiff University, Cardiff, UK, 2016. [Google Scholar]
- Marei, H.E.; Althani, A.; Afifi, N.; Abd-Elmaksoud, A.; Bernardini, C.; Michetti, F.; Barba, M.; Pescatori, M.; Maira, G.; Paldino, E. Over-expression of hNGF in adult human olfactory bulb neural stem cells promotes cell growth and oligodendrocytic differentiation. PLoS ONE 2013, 8, e82206. [Google Scholar] [CrossRef] [Green Version]
- Soltani, S.; Saghazadeh, A.; Movahedi, M.; Tavakol, M.; Sadr, M.; Farhadi, E.; Rezaei, N. FLG single nucleotide polymorphisms in chronic idiopathic urticaria. Allergol. Immunopathol. 2016, 44, 341–345. [Google Scholar] [CrossRef]
- Shafiq, M.; Zafar, S.; Younas, N.; Noor, A.; Puig, B.; Altmeppen, H.C.; Schmitz, M.; Matschke, J.; Ferrer, I.; Glatzel, M. Prion protein oligomers cause neuronal cytoskeletal damage in rapidly progressive Alzheimer’s disease. Mol. Neurodegener. 2021, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, I.E.; Tanabe, K.; Strittmatter, S.M. Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J. Neurosci. 2002, 22, 1303–1315. [Google Scholar] [CrossRef] [PubMed]
- Richens, J.L.; Spencer, H.L.; Butler, M.; Cantlay, F.; Vere, K.-A.; Bajaj, N.; Morgan, K.; O’Shea, P. Rationalising the role of Keratin 9 as a biomarker for Alzheimer’s disease. Sci. Rep. 2016, 6, 22962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| AD | CN | p-Value | ||
|---|---|---|---|---|
| No. of participants, n (%) | 175 (45%) | 214 (55%) | ||
| Sex, n (%) | M | 93 (53%) | 115 (54%) | 0.907 |
| F | 82 (47%) | 99 (46%) | ||
| Age, Median (IQR) | 75.8 | 75.5 | 0.815 | |
| (70.85, 81.10) | (72.12, 78.38) | |||
| Race, n (%) | Asian | 2 (1) | 2 (1) | 0.357 |
| Black | 8 (5) | 15 (7) | ||
| More than one | 2 (1) | 0 (0) | ||
| White | 163 (93) | 197 (92) | ||
| Education, Median (IQR) | 15 (12, 16) | 16 (14, 18) | <0.001 | |
| Participant residence, n (%) | House | 121 (69) | 162 (76) | 0.005 |
| Condo/Co-op (owned) | 17 (10) | 32 (15) | ||
| Apartment (rented) | 21 (12) | 13 (6) | ||
| Mobile Home | 3 (2) | 2 (1) | ||
| Retirement Community | 5 (3) | 3 (1) | ||
| Assisted Living | 7 (4) | 0 (0) | ||
| Other | 1 (1) | 2 (1) | ||
| APOE4, n (%) | 0 | 58 (33) | 156 (73) | <0.001 |
| 1 | 85 (49) | 53 (25) | ||
| 2 | 32 (18) | 5 (2) |
| Gene | BF | Post.Prob |
|---|---|---|
| FLG | 4040.782 | 0.996 |
| ZNF585B | 87.062 | 0.842 |
| ZNF875 | 9.331 | 0.364 |
| PREX2 | 6.746 | 0.293 |
| NID2 | 5.819 | 0.263 |
| DHX16 | 4.515 | 0.217 |
| BACE1-FLG | BACE1-FLG Ser742Tyr | |
|---|---|---|
| HADDOCK score | −77.500 ± 13.900 | −89.600 ± 16.000 |
| Root Mean Square Deviation from the overall lowest-energy structure | 0.500 ± 0.300 | 0.800 ± 0.500 |
| Van der Waals energy | −79.400 ± 5.400 | −83.800 ± 14.200 |
| Electrostatic energy | −516.500 ± 81.300 | −566.100 ± 71.800 |
| Desolvation energy | 2.400 ± 7.700 | 2.700 ± 3.400 |
| Buried Surface Area | 3250.100 ± 135.700 | 3318.500 ± 346.700 |
| Z-Score | −1.400 | −1.800 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, W.; Cai, J.; Li, R.; Wen, C.; Tan, H.; on behalf of the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Database. Rare Variant Analysis and Molecular Dynamics Simulation in Alzheimer’s Disease Identifies Exonic Variants in FLG. Genes 2022, 13, 838. https://doi.org/10.3390/genes13050838
Xiong W, Cai J, Li R, Wen C, Tan H, on behalf of the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Database. Rare Variant Analysis and Molecular Dynamics Simulation in Alzheimer’s Disease Identifies Exonic Variants in FLG. Genes. 2022; 13(5):838. https://doi.org/10.3390/genes13050838
Chicago/Turabian StyleXiong, Weixue, Jiahui Cai, Ruijia Li, Canhong Wen, Haizhu Tan, and on behalf of the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Database. 2022. "Rare Variant Analysis and Molecular Dynamics Simulation in Alzheimer’s Disease Identifies Exonic Variants in FLG" Genes 13, no. 5: 838. https://doi.org/10.3390/genes13050838
APA StyleXiong, W., Cai, J., Li, R., Wen, C., Tan, H., & on behalf of the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Database. (2022). Rare Variant Analysis and Molecular Dynamics Simulation in Alzheimer’s Disease Identifies Exonic Variants in FLG. Genes, 13(5), 838. https://doi.org/10.3390/genes13050838






