On the Forensic Use of Y-Chromosome Polymorphisms
Abstract
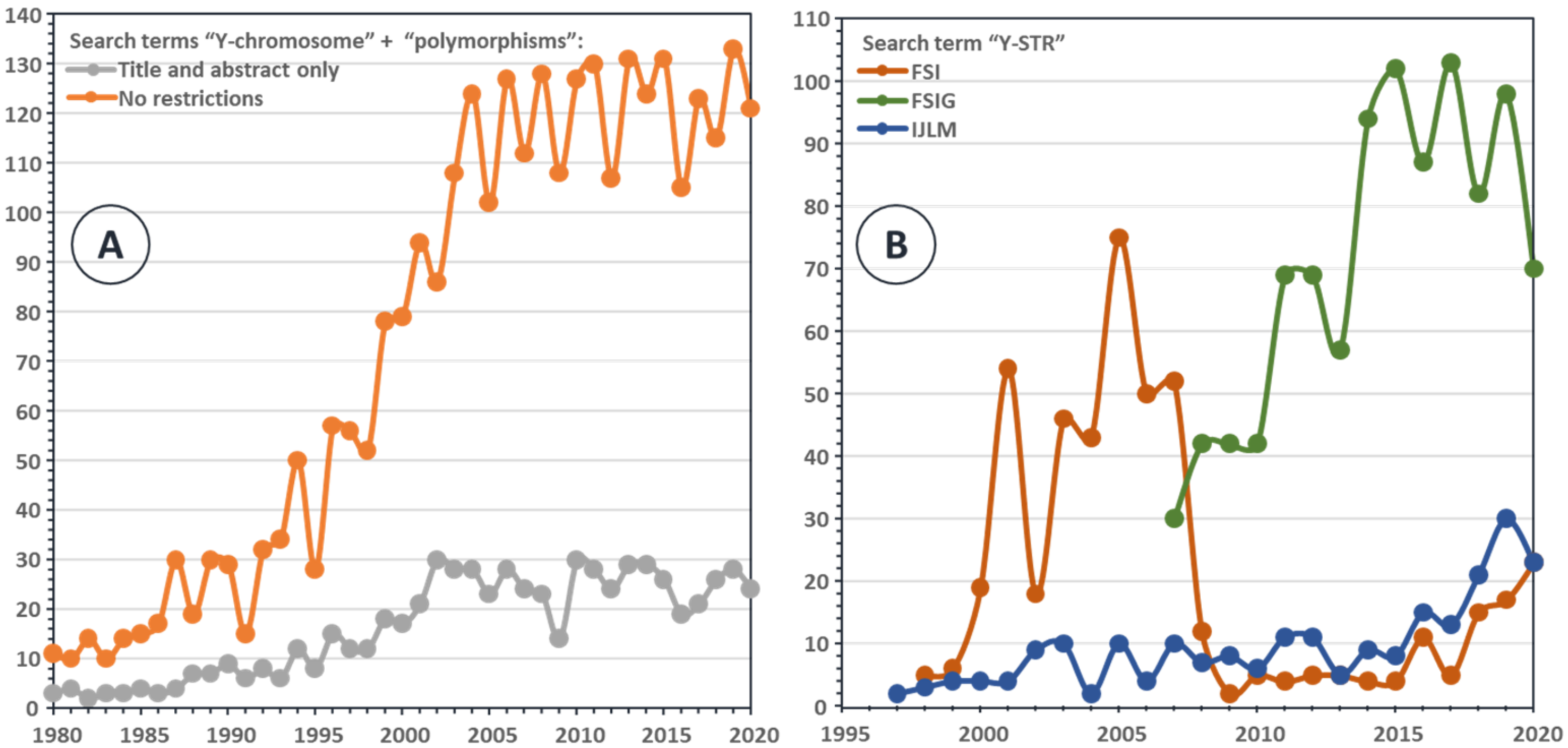
:1. A Brief History of Y-Chromosome Polymorphisms
2. Y-STRs and Y-SNPs
2.1. Haplogroups and Haplotypes
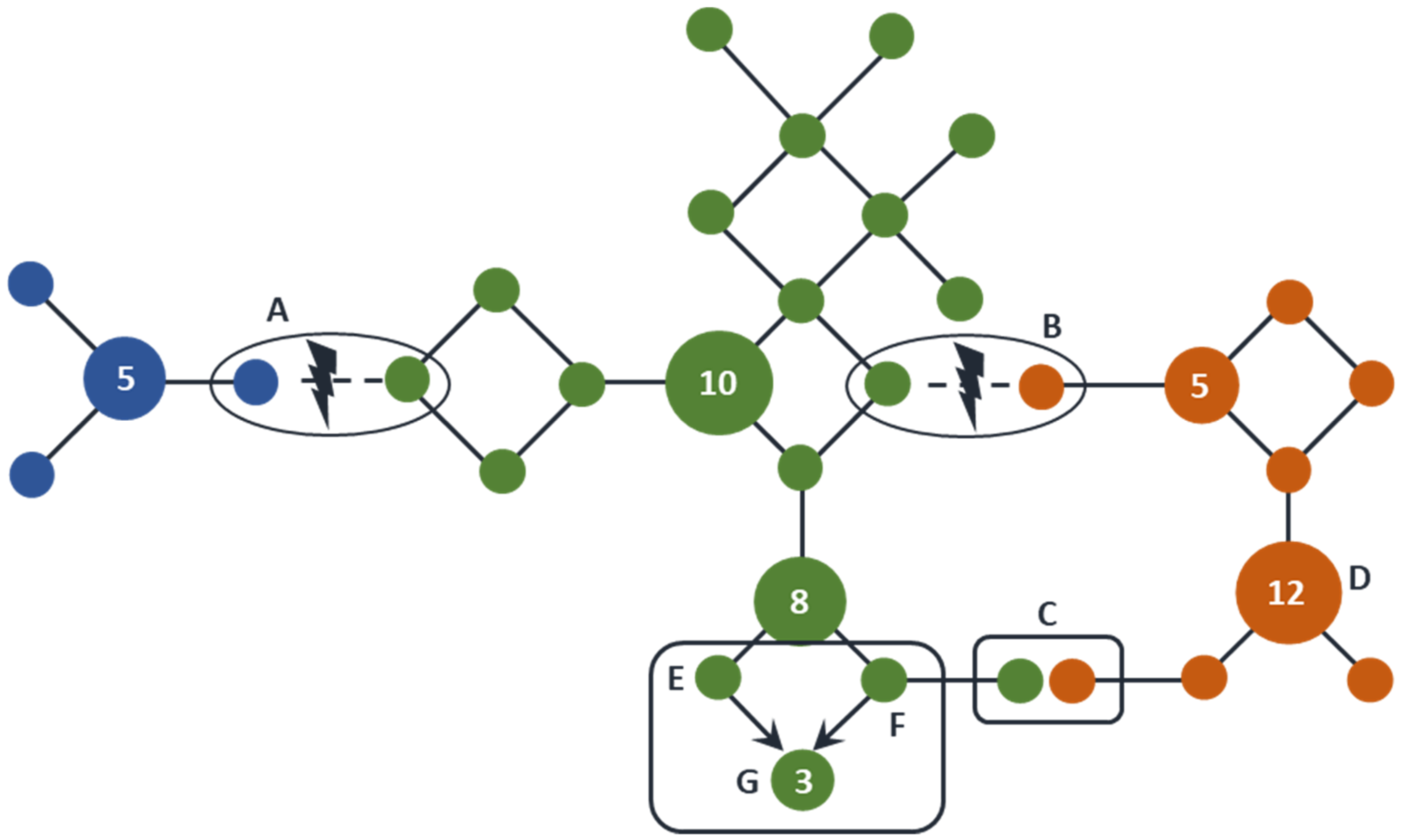
2.2. Identical by State (IBS) and Identical by Descent (IBD)
2.3. Database Frequencies
3. The Various Applications of Y-STRs and Y-SNPs
3.1. Biogeographic Ancestry Reconstruction Using Y-Chromosome Polymorphisms
3.2. Y-Chromosome-Based Dragnets
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Casanova, M.; Leroy, P.; Boucekkine, C.; Weissenbach, J.; Bishop, C.; Fellous, M.; Purrello, M.; Fiori, G.; Siniscalco, M. A Human Y-Linked DNA Polymorphism and Its Potential for Estimating Genetic and Evolutionary Distance. Science 1985, 230, 1403–1406. [Google Scholar] [CrossRef] [PubMed]
- Lucotte, G.; Ngo, N.Y. P49f, a Highly Polymorphic Probe, that Detects Taq1 RFLPs on the Human Y Chromosome. Nucl. Acids Res. 1985, 13, 8285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flam, F. Random Samples. Science 1993, 261, 679. [Google Scholar] [CrossRef]
- Page, D.C. Is ZFY the Sex-Determining Gene on the Human Y Chromosome? Philos. Trans. R. Soc. London. B 1988, 322, 155–157. [Google Scholar] [CrossRef]
- Dorit, R.L.; Akashi, H.; Gilbert, W. Absence of Polymorphism at the ZFY Locus on the Human Y Chromosome. Science 1995, 268, 1183–1185. [Google Scholar] [CrossRef] [Green Version]
- Pääbo, S. The Y Chromosome and the Origin of All of Us (Men). Science 1995, 268, 1141–1142. [Google Scholar] [CrossRef] [Green Version]
- Jobling, M.A.; Tyler-Smith, C. Fathers and Sons: The Y Chromosome and Human Evolution. Trends Genet. 1995, 11, 449–456. [Google Scholar] [CrossRef]
- Roewer, L.; Arnemann, J.; Spurr, N.K.; Grzeschik, K.-H.; Epplen, J.T. Simple Repeat Sequences on the Human Y Chromosome Are Equally Polymorphic as Their Autosomal Counterparts. Hum. Genet. 1992, 89, 389–394. [Google Scholar] [CrossRef]
- Mathias, N.; Bayes, M.; Tyler-Smith, C. Highly Informative Compound Haplotypes for the Human Y Chromosome. Hum. Mol. Genet. 1994, 3, 115–123. [Google Scholar] [CrossRef]
- Roewer, L.; Epplen, J.T. Rapid and Sensitive Typing of Forensic Stains by PCR Amplification of Polymorphic Simple Repeat Sequences in Case Work. Forensic Sci. Int. 1992, 53, 163–171. [Google Scholar] [CrossRef]
- Roewer, L.; Kayser, M.; Dieltjes, P.; Nagy, M.; Bakker, E.; Krawczak, M.; de Knijff, P. Analysis of Molecular Variance (AMOVA) of Y-Chromosome-Specific Microsatellites in Two Closely Related Human Populations. Hum. Mol. Genet. 1997, 6, 828, Erratum in Hum. Mol. Genet. 1996, 5, 1029–1033. [Google Scholar] [CrossRef] [Green Version]
- Kayser, M.; Caglià, A.; Corach, D.; Fretwell, N.; Gehrig, C.; Graziosi, G.; Heidorn, F.; Herrmann, S.; Herzog, B.; Hidding, M.; et al. Evaluation of Y-Chromosomal STRs: A Multicenter Study. Int. J. Leg. Med. 1997, 110, 125–133. [Google Scholar] [CrossRef]
- de Knijff, P.; Kayser, M.; Caglià, A.; Corach, D.; Fretwell, N.; Gehrig, C.; Graziosi, G.; Heidorn, F.; Herrmann, S.; Herzog, B.; et al. Chromosome Y Microsatellites: Population Genetic and Evolutionary Aspects. Int. J. Leg. Med. 1997, 110, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Jobling, M.A.; Pandya, A.; Tyler-Smith, C. The Y Chromosome in Forensic Analysis and Paternity Testing. Int. J. Leg. Med. 1997, 110, 118–124. [Google Scholar] [CrossRef]
- Heyer, E.; Puymirat, J.; Dieltjes, P.; Bakker, E.; de Knijff, P. Estimating Y Chromosome Specific Microsatellite Mutation Frequencies Using Deep Rooting Pedigrees. Hum. Mol. Genet. 1997, 6, 799–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, K.; Roewer, L.; de Knijff, P. Male DNA Typing from 25-Year-Old Vaginal Swabs Using Y Chromosomal STR Polymorphisms in a Retrial Request Case. J. Forensic Sci. 1999, 44, 868–872. [Google Scholar] [CrossRef]
- Kayser, M.; Krawczak, M.; Excoffier, L.; Dieltjes, P.; Corach, D.; Pascali, V.; Gehrig, C.; Bernini, L.F.; Jespersen, J.; Bakker, E.; et al. An Extensive Analysis of Y-Chromosomal Microsatellite Haplotypes in Globally Dispersed Human Populations. Am. J. Hum. Genet. 2001, 68, 990–1018. [Google Scholar] [CrossRef] [Green Version]
- Dettlaff-Kakol, A.; Pawlowski, R. First Polish DNA “Manhunt”—An Application of Y-Chromosome STRs. Int. J. Leg. Med. 2002, 116, 289–291. [Google Scholar] [CrossRef]
- Sinha, S.K. Forensic Casework Applications Using Y-PLEX™ 6 and Y-PLEX™ 5 Systems. Forensic Sci. Rev. 2003, 15, 199–203. [Google Scholar]
- Krenke, B.E.; Viculis, L.; Richard, M.L.; Prinz, M.; Milne, S.C.; Ladd, C.; Gross, A.M.; Gornall, T.; Frappier, J.R.H.; Eisenberg, A.J.; et al. Validation of a Male-Specific, 12-Locus Fluorescent Short Tandem Repeat (STR) Multiplex. Forensic Sci. Int. 2005, 148, 1–14, Erratum in Forensic Sci. Int. 2005, 151, 111–124. [Google Scholar] [CrossRef]
- PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 10 October 2021).
- YHRD. Available online: https://yhrd.org/ (accessed on 10 October 2021).
- Roewer, L.; Krawczak, M.; Willuweit, S.; Nagy, M.; Alves, C.; Amorim, A.; Anslinger, K.; Augustin, C.; Betz, A.; Bosch, E.; et al. Online Reference Database of European Y-Chromosomal Short Tandem Repeat (STR) Haplotypes. Forensic Sci. Int. 2001, 118, 106–113. [Google Scholar] [CrossRef] [Green Version]
- Jobling, M.A.; Tyler-Smith, C. The Human Y Chromosome: An Evolutionary Marker Comes of Age. Nat. Rev. Genet. 2003, 4, 598–612. [Google Scholar] [CrossRef]
- Jobling, M.A.; Tyler-Smith, C. Human Y-Chromosome Variation in the Genome-Sequencing Era. Nat. Rev. Genet. 2017, 18, 485–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kayser, M. Forensic Use of Y-Chromosome DNA: A General Overview. Hum. Genet. 2017, 136, 621–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kittler, R.; Erler, A.; Brauer, S.; Stoneking, M.; Kayser, M. Apparent Intrachromosomal Exchange on the Human Y Chromosome Explained by Population History. Eur. J. Hum. Genet. 2003, 11, 304–314. [Google Scholar] [CrossRef] [Green Version]
- Rolf, B.; Meyer, E.; Brinkmann, B.; de Knijff, P. Polymorphism at the Tetranucleotide Repeat Locus DYS389 in 10 Populations Reveals Strong Geographic Clustering. Eur. J. Hum. Genet. 1998, 6, 583–588. [Google Scholar] [CrossRef]
- Ballantyne, K.N.; Keerl, V.; Wollstein, A.; Choi, Y.; Zuniga, S.B.; Ralf, A.; Vermeulen, M.; de Knijff, P.; Kayser, M. A New Future of Forensic Y-Chromosome Analysis: Rapidly Mutating Y-STRs for Differentiating Male Relatives and Paternal Lineages. For. Sci. Int. Genet. 2012, 6, 208–218. [Google Scholar] [CrossRef]
- Ralf, A.; Lubach, D.; Kousouri, N.; Winkler, C.; Schulz, I.; Roewer, L.; Purps, J.; Lessig, R.; Krajewski, P.; Ploski, R.; et al. Identification and Characterization of Novel Rapidly Mutating Y-chromosomal Short Tandem Repeat Markers. Hum. Mutat. 2020, 41, 1680–1696. [Google Scholar] [CrossRef]
- Westen, A.A.; Kraaijenbrink, T.; Clarisse, L.; Grol, L.J.W.; Willemse, P.; Zuniga, S.B.; Robles de Medina, E.A.; Schouten, R.; van der Gaag, K.J.; Weiler, N.E.C.; et al. Analysis of 36 Y-STR Marker Units Including a Concordance Study among 2085 Dutch Males. For. Sci. Int. Genet. 2015, 14, 174–181. [Google Scholar] [CrossRef] [Green Version]
- Purps, J.; Siegert, S.; Willuweit, S.; Nagy, M.; Alves, C.; Salazar, R.; Angustia, S.M.T.; Santos, L.H.; Anslinger, K.; Bayer, B.; et al. A Global Analysis of Y-Chromosomal Haplotype Diversity for 23 STR Loci. For. Sci. Int. Genet. 2014, 12, 12–23. [Google Scholar] [CrossRef]
- Gopinath, S.; Zhong, C.; Nguyen, V.; Ge, J.; Lagacé, R.E.; Short, M.L.; Mulero, J.J. Developmental Validation of the Yfiler® Plus PCR Amplification Kit: An Enhanced Y-STR Multiplex for Casework and Database Applications. For. Sci. Int. Genet. 2016, 24, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Steffen, C.R.; Huszar, T.I.; Borsuk, L.A.; Vallone, P.M.; Gettings, K.B. A Multi-Dimensional Evaluation of the ‘NIST 1032’ Sample Set across Four Forensic Y-STR Multiplexes. For. Sci. Int. Genet. 2022, 57, 102655. [Google Scholar] [CrossRef]
- Claerhout, S.; Verstraete, P.; Warnez, L.; Vanpaemel, S.; Larmuseau, M.; Decorte, R. CSYseq: The First Y-Chromosome Sequencing Tool Typing a Large Number of Y-SNPs and Y-STRs to Unravel Worldwide Human Population Genetics. PLoS Genet. 2021, 17, e1009758. [Google Scholar] [CrossRef] [PubMed]
- Jobling, M.A.; Tyler-Smith, C. New Uses for New Haplotypes. Trends Genet. 2000, 16, 356–362. [Google Scholar] [CrossRef]
- Kalaydjieva, L.; Calafell, F.; Jobling, M.A.; Angelicheva, D.; de Knijff, P.; Rosser, Z.; Hurles, M.E.; Underhill, P.; Tournev, I.; Marushiakova, E.; et al. Patterns of Inter- and Intra-Group Genetic Diversity in the Vlax Roma as Revealed by Y Chromosome and Mitochondrial DNA Lineages. Eur. J. Hum. Genet. 2001, 9, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Underhill, P.A.; Shen, P.; Lin, A.A.; Jin, L.; Passarino, G.; Yang, W.H.; Kauffman, E.; Bonné-Tamir, B.; Bertranpetit, J.; Francalacci, P.; et al. Y Chromosome Sequence Variation and the History of Human Populations. Nat. Genet. 2000, 26, 358–361. [Google Scholar] [CrossRef]
- Hammer, M.F.; Karafet, T.M.; Redd, A.J.; Jarjanazi, H.; Santachiara-Benerecetti, S.; Soodyall, H.; Zegura, S.L. Hierarchical Patterns of Global Human Y-Chromosome Diversity. Mol. Biol. Evol. 2001, 18, 1189–1203. [Google Scholar] [CrossRef] [Green Version]
- Karafet, T.; Xu, L.; Du, R.; Wang, W.; Feng, S.; Wells, R.S.; Redd, A.J.; Zegura, S.L.; Hammer, M.F. Paternal Population History of East Asia: Sources, Patterns, and Microevolutionary Processes. Am. J. Hum. Genet. 2001, 69, 615–628. [Google Scholar] [CrossRef] [Green Version]
- Semino, O.; Passarino, G.; Oefner, P.J.; Lin, A.A.; Arbuzova, S.; Beckman, L.E.; Benedictis, G.D.; Francalacci, P.; Kouvatsi, A.; Limborska, S.; et al. The Genetic Legacy of Paleolithic Homo Sapiens Sapiens in Extant Europeans: A Y Chromosome Perspective. Science 2000, 290, 1155–1159. [Google Scholar] [CrossRef]
- Su, B.; Xiao, J.; Underhill, P.; Deka, R.; Zhang, W.; Akey, J.; Huang, W.; Shen, D.; Lu, D.; Luo, J.; et al. Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age. Am. J. Hum. Genet. 1999, 65, 1718–1724. [Google Scholar] [CrossRef] [Green Version]
- Capelli, C.; Wilson, J.F.; Richards, M.; Stumpf, M.P.H.; Gratrix, F.; Oppenheimer, S.; Underhill, P.; Pascali, V.L.; Ko, T.-M.; Goldstein, D.B. A Predominantly Indigenous Paternal Heritage for the Austronesian-Speaking Peoples of Insular Southeast Asia and Oceania. Am. J. Hum. Genet. 2001, 68, 432–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Y Chromosome Consortium. A Nomenclature System for the Tree of Human Y-Chromosomal Binary Haplogroups. Genome Res. 2002, 12, 339–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karafet, T.M.; Mendez, F.L.; Meilerman, M.B.; Underhill, P.A.; Zegura, S.L.; Hammer, M.F. New Binary Polymorphisms Reshape and Increase Resolution of the Human Y Chromosomal Haplogroup Tree. Genome Res. 2008, 18, 830–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ralf, A.; van Oven, M.; Montiel González, D.; de Knijff, P.; van der Beek, K.; Wootton, S.; Lagacé, R.; Kayser, M. Forensic Y-SNP Analysis beyond SNaPshot: High-Resolution Y-Chromosomal Haplogrouping from Low Quality and Quantity DNA Using Ion AmpliSeq and Targeted Massively Parallel Sequencing. For. Sci. Int. Genet. 2019, 41, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Rohrlach, A.B.; Papac, L.; Childebayeva, A.; Rivollat, M.; Villalba-Mouco, V.; Neumann, G.U.; Penske, S.; Skourtanioti, E.; van de Loosdrecht, M.; Akar, M.; et al. Using Y-Chromosome Capture Enrichment to Resolve Haplogroup H2 Shows New Evidence for a Two-Path Neolithic Expansion to Western Europe. Sci. Rep. 2021, 11, 15005. [Google Scholar] [CrossRef]
- Tillmar, A.; Sturk-Andreaggi, K.; Daniels-Higginbotham, J.; Thomas, J.T.; Marshall, C. The FORCE Panel: An All-in-One SNP Marker Set for Confirming Investigative Genetic Genealogy Leads and for General Forensic Applications. Genes 2021, 12, 1968. [Google Scholar] [CrossRef]
- Ralf, A.; Montiel González, D.; Zhong, K.; Kayser, M. Yleaf: Software for Human Y-Chromosomal Haplogroup Inference from Next-Generation Sequencing Data. Mol. Biol. Evol. 2018, 35, 1291–1294. [Google Scholar] [CrossRef]
- de Knijff, P. Messages through Bottlenecks: On the Combined Use of Slow and Fast Evolving Polymorphic Markers on the Human Y Chromosome. Am. J. Hum. Genet. 2000, 67, 1055–1061. [Google Scholar] [CrossRef]
- de Knijff, P. Y chromosomes shared by descent or by state. In Archaeogenetics: DNA and the Population Prehistory of Europe; McDonald Institute Monographs; Renfrew, C., Boyle, K.V., Eds.; McDonald Institute for Archaeological Research: Cambridge, UK, 2000; pp. 301–305. ISBN 978-1-902937-08-3. [Google Scholar]
- Larmuseau, M.H.D.; Vanderheyden, N.; Van Geystelen, A.; Oven, M.; Knijff, P.; Decorte, R. Recent Radiation within Y-chromosomal Haplogroup R-M269 Resulted in High Y-STR Haplotype Resemblance. Ann. Hum. Genet. 2014, 78, 92–103. [Google Scholar] [CrossRef]
- Solé-Morata, N.; Bertranpetit, J.; Comas, D.; Calafell, F. Recent Radiation of R-M269 and High Y-STR Haplotype Resemblance Confirmed. Ann. Hum. Genet. 2014, 78, 253–254. [Google Scholar] [CrossRef] [Green Version]
- Yoshimoto, T.; Tamaki, K.; Katsumata, S.; Huang, X.-L.; Uchihi, R.; Tanaka, M.; Uchida, H.; Yamamoto, T.; Chen, S.; Armour, J.A.L.; et al. Sequence Analysis of Alleles at a Microsatellite Locus D14S299 (Wg1c5) and Population Genetic Comparisons. Int. J. Leg. Med. 1999, 113, 15–18. [Google Scholar] [CrossRef]
- Claerhout, S.; Van der Haegen, M.; Vangeel, L.; Larmuseau, M.H.D.; Decorte, R. A Game of Hide and Seq: Identification of Parallel Y-STR Evolution in Deep-Rooting Pedigrees. Eur. J. Hum. Genet. 2019, 27, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Huszar, T.I.; Jobling, M.A.; Wetton, J.H. A Phylogenetic Framework Facilitates Y-STR Variant Discovery and Classification via Massively Parallel Sequencing. For. Sci. Int. Genet. 2018, 35, 97–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, C.; Gelabert-Besada, M.; Fernandez-Formoso, L.; García-Magariños, M.; Santos, C.; Fondevila, M.; Ballard, D.; Syndercombe Court, D.; Carracedo, Á.; Victoria Lareu, M. “New Turns from Old STaRs”: Enhancing the Capabilities of Forensic Short Tandem Repeat Analysis: Nucleic Acids. Electrophoresis 2014, 35, 3173–3187. [Google Scholar] [CrossRef]
- Willuweit, S.; Roewer, L. The New Y Chromosome Haplotype Reference Database. For. Sci. Int. Genet. 2015, 15, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Willuweit, S.; Roewer, L. Y Chromosome Haplotype Reference Database (YHRD): Update. For. Sci. Int. Genet. 2007, 1, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Willuweit, S.; Caliebe, A.; Andersen, M.M.; Roewer, L. Y-STR Frequency Surveying Method: A Critical Reappraisal. For. Sci. Int. Genet. 2011, 5, 84–90. [Google Scholar] [CrossRef]
- Andersen, M.M.; Balding, D.J. Assessing the Forensic Value of DNA Evidence from Y Chromosomes and Mitogenomes. Genes 2021, 12, 1209. [Google Scholar] [CrossRef]
- Andersen, M.M.; Balding, D.J. How Convincing Is a Matching Y-Chromosome Profile? PLoS Genet. 2017, 13, e1007028. [Google Scholar] [CrossRef] [Green Version]
- Roewer, L.; Andersen, M.M.; Ballantyne, J.; Butler, J.M.; Caliebe, A.; Corach, D.; D’Amato, M.E.; Gusmão, L.; Hou, Y.; de Knijff, P.; et al. DNA Commission of the International Society of Forensic Genetics (ISFG): Recommendations on the Interpretation of Y-STR Results in Forensic Analysis. For. Sci. Int. Genet. 2020, 48, 102308. [Google Scholar] [CrossRef]
- Freeman, L.; Brimacombe, C.S.; Elhaik, E. AYChr-DB: A Database of Ancient Human Y Haplogroups. NAR Genom. Bioinform. 2020, 2, lqaa081. [Google Scholar] [CrossRef] [PubMed]
- de la Chapelle, A. The Use and Misuse of Sex Chromatin Screening for ‘Gender Identification’ of Female Athletes. JAMA 1986, 256, 1920–1923. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.R.; Pandya, A.; Tyler-Smith, C. Reliability of DNA-Based Sex Tests. Nat. Genet. 1998, 18, 103. [Google Scholar] [CrossRef] [PubMed]
- Thangaraj, K.; Reddy, A.G.; Singh, L. Is the Amelogenin Gene Reliable for Gender Identification in Forensic Casework and Prenatal Diagnosis? Int. J. Legal. Med. 2002, 116, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, C. Sex Redefined. Nature 2015, 518, 288–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reardon, S. The Spectrum of Sex Development: Eric Vilain and the Intersex Controversy. Nature 2016, 533, 160–163. [Google Scholar] [CrossRef] [Green Version]
- Syndercombe Court, D. The Y Chromosome and Its Use in Forensic DNA Analysis. Emerg. Top. Life Sci. 2021, 5, 427–441. [Google Scholar] [CrossRef]
- Jobling, M.A.; Gill, P. Encoded Evidence: DNA in Forensic Analysis. Nat. Rev. Genet. 2004, 5, 739–751. [Google Scholar] [CrossRef]
- Ruano, G.; Brash, D.E.; Kidd, K.K. PCR: The first few cycles. Amplifications 1991, 7, 1–4. [Google Scholar]
- Wallin, J.M.; Buoncristiani, M.R.; Lazaruk, K.D.; Fildes, N.; Holt, C.L.; Walsh, P.S. TWGDAM Validation of the AmpFISTRTM Blue PCR Amplification Kit for Forensic Casework Analysis. J. Forensic Sci. 1998, 43, 854–870. [Google Scholar] [CrossRef]
- Prinz, M.; Boll, K.; Baum, H.; Shaler, B. Multiplexing of Y Chromosome Specific STRs and Performance for Mixed Samples. Forensic Sci. Int. 1997, 85, 209–218. [Google Scholar] [CrossRef]
- Walsh, P.S.; Erlich, H.A.; Higuchi, R. Preferential PCR Amplification of Alleles: Mechanisms and Solutions. Genome Res. 1992, 1, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Hidding, M.; Staak, M.; Schmitt, C. Y-chromosomal STR systems; application of a triplex PCR in forensic stain analysis. In Progress in Forensic Genetics, 7; Olaisen, B., Brinkmann, B., Lincoln, P.J., Eds.; Elsevier: Amsterdam, The Netherlands, 1997; pp. 515–517. [Google Scholar]
- Shewale, J.G. Y-Short Tandem Repeat Multiplex Systems—Y-PLEXTM 6 and Y-PLEXTM 5. Forensic Sci. Rev. 2003, 15, 115–135. [Google Scholar] [PubMed]
- Shewale, J.G.; Sikka, S.C.; Schneida, E.; Sinha, S.D. DNA Profiling of Azoospermic Semen Samples from Vasectomized Males by Using Y-PLEXTM6 Amplification Kit. J. Forensic Sci. 2003, 48, 127–129. [Google Scholar] [CrossRef] [PubMed]
- Roewer, L. Y Chromosome STR Typing in Crime Casework. Forensic Sci. Med. Pathol. 2009, 5, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Roewer, L. Y-chromosome Short Tandem Repeats in Forensics—Sexing, Profiling, and Matching Male DNA. WIREs Forensic Sci. 2019, 1, e1336. [Google Scholar] [CrossRef] [Green Version]
- Tvedebrink, T. Review of the Forensic Applicability of Biostatistical Methods for Inferring Ancestry from Autosomal Genetic Markers. Genes 2022, 13, 141. [Google Scholar] [CrossRef]
- Underhill, P.A.; Kivisild, T. Use of Y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations. Annu. Rev. Genet. 2007, 41, 539–564. [Google Scholar] [CrossRef] [Green Version]
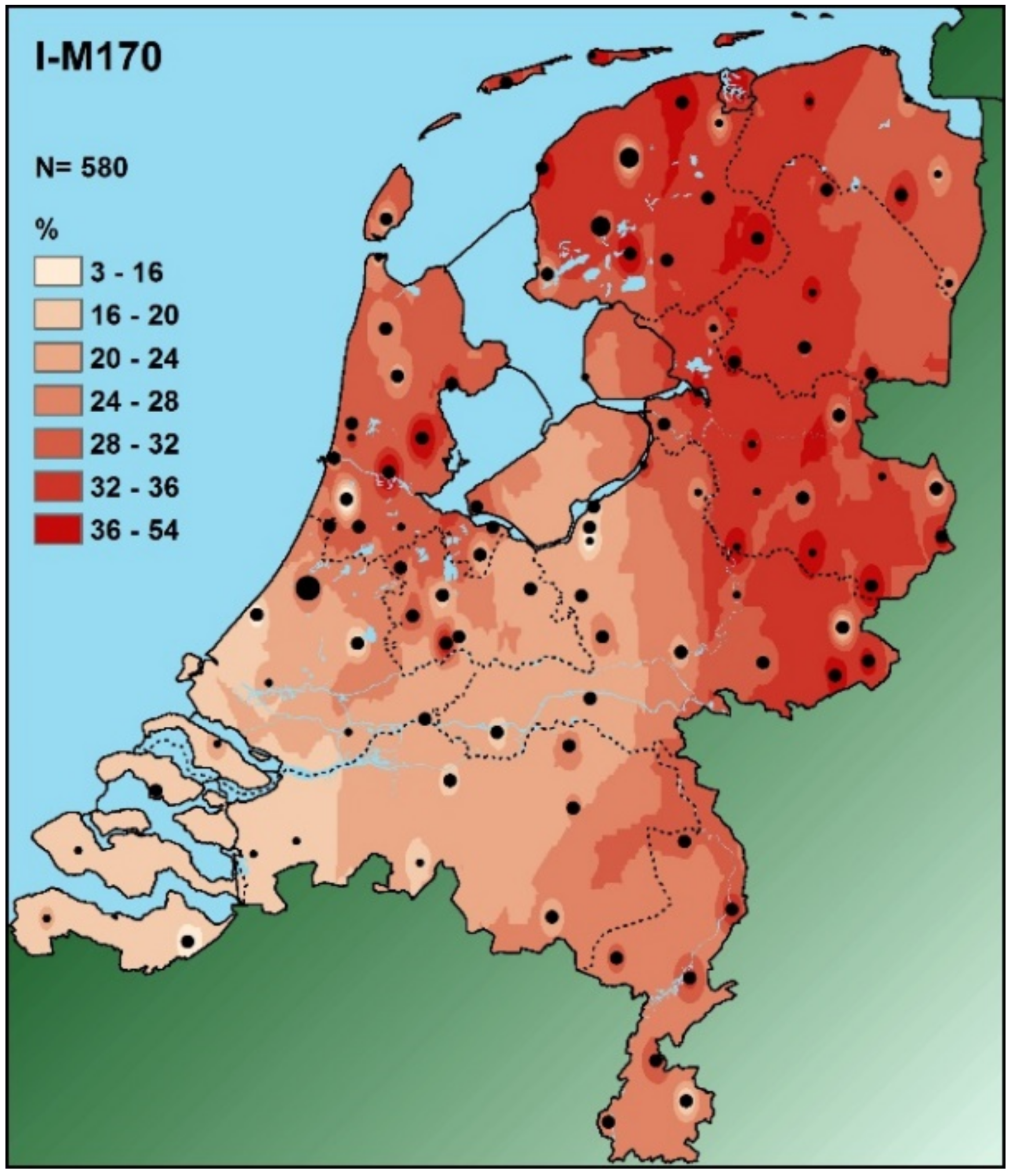
- Altena, E.; Smeding, R.; van der Gaag, K.J.; Larmuseau, M.H.D.; Decorte, R.; Lao, O.; Kayser, M.; Kraaijenbrink, T.; de Knijff, P. The Dutch Y-Chromosomal Landscape. Eur. J. Hum. Genet. 2020, 28, 287–299. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Heap, A.D. A Review of Comparative Studies of Spatial Interpolation Methods in Environmental Sciences: Performance and Impact Factors. Ecol. Inform. 2011, 6, 228–241. [Google Scholar] [CrossRef]
- Pati, D.; Reich, B.J.; Dunson, D.B. Bayesian Geostatistical Modelling with Informative Sampling Locations. Biometrika 2011, 98, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Piel, F.B.; Patil, A.P.; Howes, R.E.; Nyangiri, O.A.; Gething, P.W.; Williams, T.N.; Weatherall, D.J.; Hay, S.I. Global Distribution of the Sickle Cell Gene and Geographical Confirmation of the Malaria Hypothesis. Nat. Commun. 2010, 1, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz-Lacava, A.; Walier, M.; Willuweit, S.; Wienker, T.F.; Fimmers, R.; Baur, M.P.; Roewer, L. Geostatistical Inference of Main Y-STR-Haplotype Groups in Europe. For. Sci. Int. Genet. 2011, 5, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.F.; Weiss, D.A.; Richards, M.; Thomas, M.G.; Bradman, N.; Goldstein, D.B. Genetic Evidence for Different Male and Female Roles during Cultural Transitions in the British Isles. Proc. Natl. Acad. Sci. USA 2001, 98, 5078–5083. [Google Scholar] [CrossRef] [Green Version]
- Zerjal, T.; Xue, Y.; Bertorelle, G.; Wells, R.S.; Bao, W.; Zhu, S.; Qamar, R.; Ayub, Q.; Mohyuddin, A.; Fu, S.; et al. The Genetic Legacy of the Mongols. Am. J. Hum. Genet. 2003, 72, 717–721. [Google Scholar] [CrossRef] [Green Version]
- Balaresque, P.; Poulet, N.; Cussat-Blanc, S.; Gerard, P.; Quintana-Murci, L.; Heyer, E.; Jobling, M.A. Y-Chromosome Descent Clusters and Male Differential Reproductive Success: Young Lineage Expansions Dominate Asian Pastoral Nomadic Populations. Eur. J. Hum. Genet. 2015, 23, 1413–1422. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Zerjal, T.; Bao, W.; Zhu, S.; Lim, S.-K.; Shu, Q.; Xu, J.; Du, R.; Fu, S.; Li, P.; et al. Recent Spread of a Y-Chromosomal Lineage in Northern China and Mongolia. Am. J. Hum. Genet. 2005, 77, 1112–1116. [Google Scholar] [CrossRef] [Green Version]
- Moore, L.T.; McEvoy, B.; Cape, E.; Simms, K.; Bradley, D.G. A Y-Chromosome Signature of Hegemony in Gaelic Ireland. Am. J. Hum. Genet. 2006, 78, 334–338. [Google Scholar] [CrossRef] [Green Version]
- King, T.E.; Jobling, M.A. What’s in a Name? Y Chromosomes, Surnames and the Genetic Genealogy Revolution. Trends Genet. 2009, 25, 351–360. [Google Scholar] [CrossRef] [Green Version]
- Claerhout, S.; Vandenbosch, M.; Nivelle, K.; Gruyters, L.; Peeters, A.; Larmuseau, M.H.D.; Decorte, R. Determining Y-STR Mutation Rates in Deep-Routing Genealogies: Identification of Haplogroup Differences. For. Sci. Int. Genet. 2018, 34, 1–10. [Google Scholar] [CrossRef]
- Larmuseau, M.H.D.; Van Geystelen, A.; van Oven, M.; Decorte, R. Genetic Genealogy Comes of Age: Perspectives on the Use of Deep-Rooted Pedigrees in Human Population Genetics. Am. J. Phys. Anthropol. 2013, 150, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Calafell, F.; Larmuseau, M.H.D. The Y Chromosome as the Most Popular Marker in Genetic Genealogy Benefits Interdisciplinary Research. Hum. Genet. 2017, 136, 559–573. [Google Scholar] [CrossRef]
- Claerhout, S.; Roelens, J.; Van der Haegen, M.; Verstraete, P.; Larmuseau, M.H.D.; Decorte, R. Ysurnames? The Patrilineal Y-Chromosome and Surname Correlation for DNA Kinship Research. For. Sci. Int. Genet. 2020, 44, 102204. [Google Scholar] [CrossRef] [Green Version]
- Crow, J.F.; Mange, A.P. Measurement of Inbreeding from the Frequency of Marriages between Persons of the Same Surname. Eugen. Quart. 1965, 12, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Manni, F.; Toupance, B.; Sabbagh, A.; Heyer, E. New Method for Surname Studies of Ancient Patrilineal Population Structures, and Possible Application to Improvement of Y-Chromosome Sampling. Am. J. Phys. Anthropol. 2005, 126, 214–228. [Google Scholar] [CrossRef] [PubMed]
- Manni, F.; Heeringa, W.; Toupance, B.; Nerbonne, J. Do Surname Differences Mirror Dialect Variation. Hum. Biol. 2008, 80, 41–64. [Google Scholar] [CrossRef] [Green Version]
- Kayser, M.; de Knijff, P. Improving Human Forensics through Advances in Genetics, Genomics and Molecular Biology. Nat. Rev. Genet. 2011, 12, 179–192. [Google Scholar] [CrossRef]
- Visage Consortium. Available online: https://www.visage-h2020.eu/#about (accessed on 10 October 2021).
- Jong, L.; M’Charek, A. The High-Profile Case as ‘Fire Object’: Following the Marianne Vaatstra Murder Case through the Media. Crime Media Cult. 2018, 14, 347–363. [Google Scholar] [CrossRef] [Green Version]
- Bartram, I.; Plümecke, T.; Schultz, S. Genetic Racial Profiling: Extended DNA Analyses and Entangled Processes of Discrimination. Sci. Technol. Stud. 2022. Available online: https://sciencetechnologystudies.journal.fi/article/download/101384/65867/213339 (accessed on 1 April 2022).


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Knijff, P. On the Forensic Use of Y-Chromosome Polymorphisms. Genes 2022, 13, 898. https://doi.org/10.3390/genes13050898
de Knijff P. On the Forensic Use of Y-Chromosome Polymorphisms. Genes. 2022; 13(5):898. https://doi.org/10.3390/genes13050898
Chicago/Turabian Stylede Knijff, Peter. 2022. "On the Forensic Use of Y-Chromosome Polymorphisms" Genes 13, no. 5: 898. https://doi.org/10.3390/genes13050898





