Abstract
As the basis of animal reproductive activity, normal spermatogenesis directly determines the efficiency of livestock production. An in-depth understanding of spermatogenesis will greatly facilitate animal breeding efforts and male infertility treatment. With the continuous development and application of gene editing technologies, they have become valuable tools to study the mechanism of spermatogenesis. Gene editing technologies have provided us with a better understanding of the functions and potential mechanisms of action of factors that regulate spermatogenesis. This review summarizes the applications of gene editing technologies, especially CRISPR/Cas9, in deepening our understanding of the function of spermatogenesis-related genes and disease treatment. The problems of gene editing technologies in the field of spermatogenesis research are also discussed.
1. Introduction
The world population is gradually expanding. Already, projections have been made that the world population will increase to 9–10 billion by 2050, and the future demand for cereals and animal products will increase to unprecedented levels [1,2]. The demand for animal products is closely related to improvements in animal breeds. With the improvements in people’s living standards and changes in diet structure, the quality of animal products will also be directly related to economic development and people’s quality of life. In recent years, the importance of animal husbandry has been reconsidered by people all over the world, and the impact of animal husbandry on the quality of food and the health of people has been increasingly emphasized. Accordingly, people have put forward higher requirements for livestock breeds. It is well known that the proportion of livestock production value in the total agricultural output objectively reflects the social development and economic development of a country or region. The genetic quality of livestock breeds or populations plays a dominant role in many factors affecting livestock production efficiency. As a result, animal breeding has once again entered the limelight in a high-profile way. Through innovative efforts to improve livestock farming production efficiency, sustainability, and product quality and profitability, animal breeding will greatly contribute to economic and consumer benefits. Recently, significant progress has been made in livestock production through reproductive biotechnology, as seen in simultaneous estrus, semen cryopreservation, and artificial insemination [3]. The rapid developments in molecular biology technology have made it possible to manipulate the genetic material of animals at the molecular level, which in turn has laid the foundation for the molecular breeding of animals.
As a necessary part of sexual reproduction, gametogenesis is the basic guarantee for the continuation of life, reproduction and the completion of evolution. Infertility affects approximately 15% of human couples globally [4]. Effective animal breeding efforts and the expansion of good breeding stock also depend greatly on the quantity and quality of gametes. For decades, one of the main avenues for breeding efforts has been the selective use of sperm from desirable male animals [5]. Precise knowledge of the potential regulatory factors and mechanisms of various aspects of spermatogenesis is important for innovations in animal breeding technology. The extensive application of functional data analysis techniques such as genomics, epigenomics, transcriptomics, proteomics and metabolomics has allowed researchers to better understand the physiological mechanisms that underlie the differences in male fertility [6]. Technological innovations are driving researchers to understand spermatogenesis in a stepwise manner, and the complex molecular mechanisms of spermatogenesis are constantly being explored.
Gene editing technologies can already be described as one of the most important technological advances of the 21st century. Technological innovations from transcription activator-like effector nuclease (TALEN) to CRISPR/Cas9 to prime editing have greatly advanced the development of gene editing technologies and research in the field of gene editing. Although there are still biosafety and ethical issues, it must be said that the emergence of transgenic farm animals and gene-edited animal models has further improved animal production and human health [7,8]. Animal transgenic technology and gene editing technology have broad application prospects in improving the breeding efficiency and disease resistance of livestock and manufacturing bioreactors [9,10,11]. The development and widespread application of these technologies are helping us to better understand the molecular mechanisms of spermatogenesis and thus to better develop new animal breeds and conserve excellent germplasm resources.
2. Spermatogenesis in Brief
Spermatogenesis is a complex biological process based on the cellular transformation of stem cells [12]. Spermatogenesis can be divided into three main stages: the mitotic proliferation of spermatogonia, the meiotic replication of spermatocytes, and the transformation of spermatocytes into spermatozoa [13]. The entire process of spermatogenesis is accomplished in the seminiferous tubules (STs). As the functional unit of the testis, STs consist of a combination of basement membrane, Sertoli cells (SCs) and germ cells (GCs) in various stages of maturation. Spermatogonia begin meiosis and transition to spermatocytes at the basement membrane. Among them, the spermatogonial stem cells (SSCs) located on the basement membrane can both self-renew to maintain a constant number of themselves and differentiate directionally to produce spermatocytes [14]. Because of their pluripotency, the quantity and quality of SSCs are directly related to the health and stability of the entire GC lineage. During the differentiation process of the next spermatogenic stage, GCs are transferred from the basement membrane into the lumen of the STs.
Within STs, there is a complex intercellular communication dialog between GCs and SCs, which together constitute the seminiferous epithelium [15]. SCs, a special group of nondividing cells, are active during the reproductive lifespan of animals and periodically change in terms of morphology and gene expression. The widely known spermatogenesis epithelial cycle is initiated by SCs and maintained by SC–GC cooperation [16]. In addition, the number of SCs directly affects sperm production [17,18]. Therefore, SCs have been considered indispensable conductors of spermatogenesis [19].
The STs are surrounded by a large amount of peritubular tissue, consisting of peritubular myoid cells (PTCs), fibrocyte-like adventitial cells and collagen matrix [20]. A large amount of interstitial tissue fills in the space between the STs in the testis. This interstitial tissue is a loose connective tissue rich in blood vessels and nerves and contains Leydig cells (LCs), macrophages, various immune cells, and some fibroblasts. In addition, a great deal of cellular communication occurs between LCs and SCs or other cells [21,22]. Together, the structural integrity of these tissues and the interplay of cells ensure that spermatogenesis occurs properly. In addition, the regulation of spermatogenesis requires the participation of hormones, paracrine factors, transcription factors, epigenetic regulators, and other substances together with multiple cells [20]. This process requires each factor to send the appropriate regulatory signals to GCs correctly and punctually at each stage of GC development.
3. A Brief Overview of CRISPR/Cas Technology
The defense mechanism of cutting foreign invading viral nucleic acids in clustered regularly interspaced short palindromic repeats (CRISPR) showed researchers its potential to edit genes after being first discovered in 1987 [23]. As a result, CRISPR/Cas, a gene editing technology derived from bacterial or archaeal acquired immunity, was created.
CRISPR/Cas is a technology for RNA-directed modification of target sequences by Cas proteins, consisting mainly of CRISPR clusters, leading sequences (leaders), repeating sequence regions (tracers), and a set of conserved CRISPR-associated genes (Cas genes) [24]. In previous gene editing processes, the construction of DNA-binding structural domains of artificial nuclease-mediated zinc finger nucleases (ZFN) and TALEN required the execution of a protein fusion process. CRISPR/Cas technology utilizes sequence-specific small guide RNA (sgRNA) designed to act as DNA-binding structural domains instead of the protein fusion process. Therefore, CRISPR/Cas, known as the third-generation gene editing technology, is more efficient, simple, accessible, and widely used compared to ZFN and TALEN.
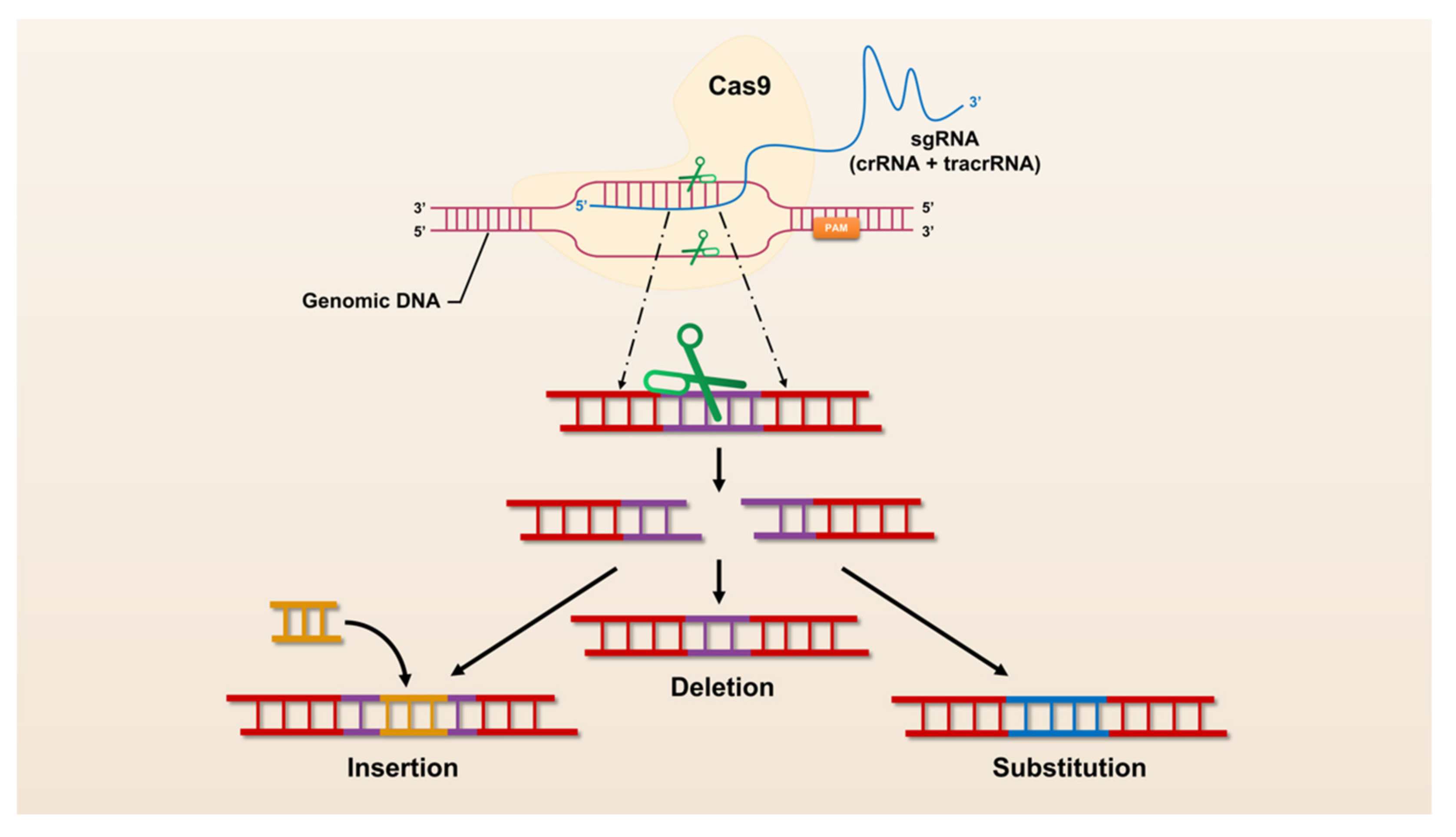
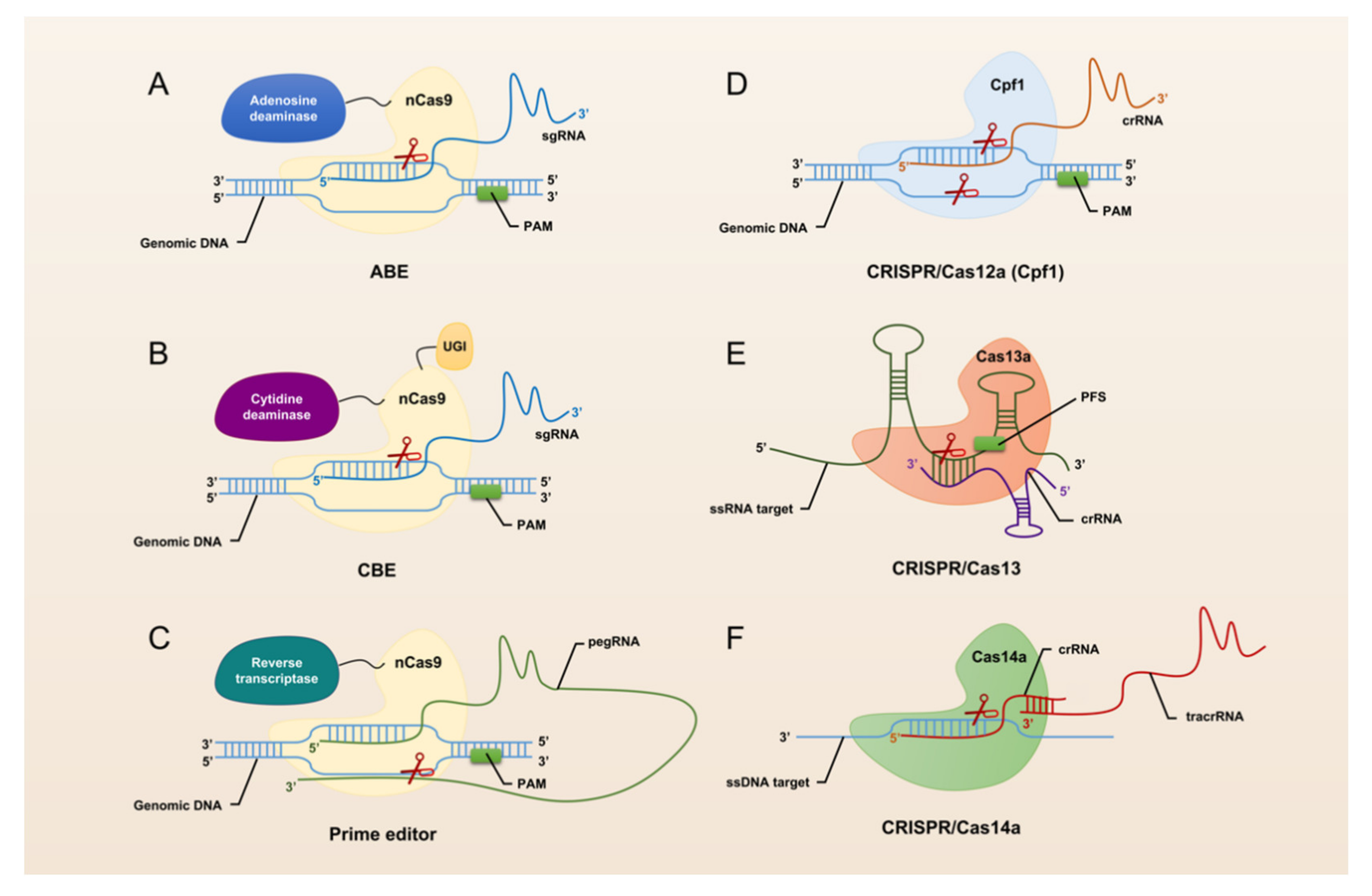
The revolutionary CRISPR/Cas system, CRISPR/Cas9, was universally recognized and widely used by researchers immediately after its advent in 2013. CRISPR/Cas9 has opened the door to a large number of applications for manipulation in almost all organisms, making it easier to achieve gene deletion, insertion, and substitution (Figure 1). However, CRISPR/Cas9 still suffers from defects and limitations such as too large components and high off-target rates. Therefore, a large number of Cas9 homologs have been mined and CRISPR/Cas9 itself is being upgraded (Figure 2).
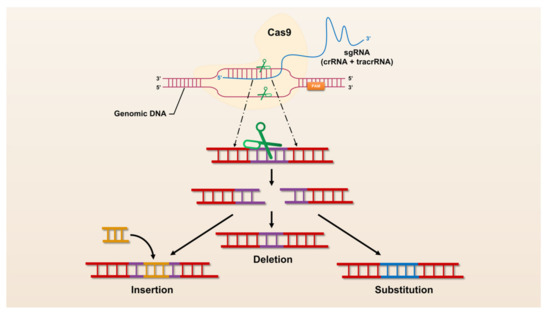
Figure 1.
The mechanism of CRISPR/Cas9 mediated genome engineering.
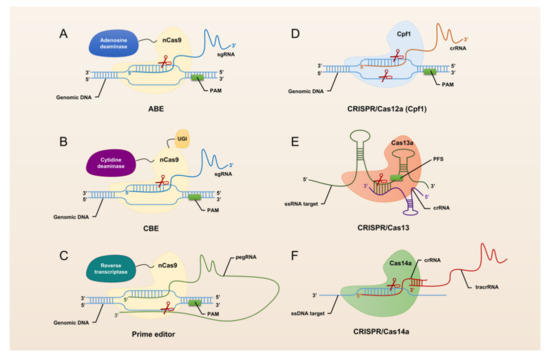
Figure 2.
Schematic summary of CRISPR/Cas systems used for genome editing.
The currently known CRISPR-Cas systems can be divided into two broad classes [25]. One class of CRISPR-Cas systems (types I, III, and IV) function with multi-subunit effector complexes. The other class of CRISPR-Cas systems (types II, V, VI) function using only a single multidomain effector protein.
The hallmark protein of the type I system is Cas3. It has a nuclease and a decapping enzyme structural domain that plays an important role in degrading exogenous DNA recognized by the multi-protein-crRNA complex cascade. The hallmark protein of the type II system is Cas9. Heterologous recombination of CRISPR/Cas9 system in mammalian cells can effectively accomplish gene editing [26]. Through the continuous exploration by researchers, CRISPR/Cas9 has been gradually upgraded from the classical system to base editing (Figure 2A,B) and prime editing (Figure 2C). This makes the application of CRISPR/Cas9 more extensive and the operations that can be realized more abundant and precise. The hallmark protein of the type III system is Cas10, which assembles into a cascade-like interference complex for target search and destruction. The hallmark protein of the type IV system is Csf1, an uncharacterized protein that has been proposed to form part of a cascade-like complex [27]. The hallmark protein of the type V system is Cpf1 (Figure 2D). Proteins such as C2c1 or C2c3 that contain an endonuclease structural domain similar to Cpf1 are also classified as effector proteins of the type V system [28]. In addition, the CRISPR/Cas14 system discovered in 2018 also falls into this type (Figure 2F). With the advent of the CRISPR/Cas14 system, cleavage of targeted single-stranded DNA (ssDNA) became a reality. The type VI system is a Cas13-based RNA targeting system (Figure 2E). Abudayyeh et al. have confirmed the high efficiency and specificity of single CRISPR RNA (crRNA)-guided Cas13-targeted specific RNA knockdown in mammalian cells [29].
With the successive discovery of Cas proteins, the CRISPR/Cas system can edit DNA, but also has more functions such as editing RNA and single- and double-stranded nucleotides. This extends the editing scope of the CRISPR/Cas system while extending its applications in biomedicine, agriculture, forestry or other fields [30,31]. CRISPR/Cas technology allows us to see more possibilities in all fields of scientific research.
4. CRISPR/Cas9: An In-Depth Exploration of Functional Genes for Spermatogenesis
With development and upgrading, gene editing technologies have become very intuitive in helping us understand more about key genes and proteins in spermatogenesis through the easy construction of cellular or animal models. The advent of a large number of animal models, particularly mouse models, has led to a renewed understanding of the role of these genes and proteins in spermatogenesis. A significant number of genes have been found to be critical for spermatogenesis and male fertility, but many genes that we thought were associated with spermatogenesis have been shown to be dispensable.
The most widely used gene editing technology in the field of spermatogenesis research is currently the CRISPR/Cas9 system. A database search revealed that more than 100 related studies have been reported since 2015, and 55 genes have been further confirmed to be relevant to male GC proliferation, sperm head and tail formation, or sperm motility (Table 1).

Table 1.
Genes related to spermatogenesis unearthed by gene editing technology.
4.1. Spermatogenesis Associated 16 (Spata16)
The protein SPATA16, encoded by Spata16, is also known as NYD-SP12. SPATA16 was first identified and characterized as a novel testis-specific protein in 2003. Min Xu et al. found that Spata16 mRNA expression levels were 30-fold higher in human adult testes than in fetal testes, and they speculated that SPATA16 may be involved in spermatogenesis through its role in the Golgi apparatus [89]. Subsequent studies have confirmed that Spata16 is closely associated with acrosome formation and that its pathogenic mutations can cause globozoospermia and male infertility [90,91,92].
Based on a mutation (851G→A, R284Q) localized in the fourth exon of Spata16 found in patients with globozoospermia, Yoshitaka Fujihara et al. successfully constructed the corresponding point mutation mouse model Spata16pm/pm by CRISPR/Cas9 [72]. Interestingly, this mutant mouse has normal reproductive capacity, which may be because the mutation does not affect the splicing of the fourth intron in Spata16pm/pm mice. However, the sperm of Spata16−781/−781 mutant mice stalled and exhibited sterility after the deletion of 781 bp around the fourth exon of mouse Spata16 with CRISPR/Cas9. This suggests that the C-terminus of the fourth exon of Spata16 encoding the TPR structural domain is essential for male fertility. Furthermore, the Spata16 mutation produces a different phenotype in humans and mice. The same mutation causes globozoospermia in humans but spermatogenic arrest in mice. Thus, the mechanism of Spata16 in spermatogenesis requires further studies for elucidation.
4.2. Doublesex and Mab-3 Related Transcription Factor 1 (Dmrt1)
As the first sex differentiation gene identified and the only one that is evolutionarily conserved among mammalian species, Dmrt1 plays a key role in sex determination, differentiation and development by controlling testicular development and male GC proliferation [93,94,95]. Dmrt1 is specifically expressed in testes and is very dynamically expressed in somatic cells and GCs [96,97]. Human Dmrt1 is linked to sex determination because of chromosome 9, where it is located. The deletion of the distal short arm of chromosome 9 was found to be associated with 46, XY gonadal hypoplasia and XY sex reversal in a large number of clinical cases [98,99,100]. The findings of these clinical trials and studies support in a stepwise manner that Dmrt1 deficiency is directly associated with disorders of sexual development. Subsequently, Shinseog Kim et al. identified the different functions of Dmrt1 in GCs versus supporting cells by conditional gene targeting. Their study confirmed the multiple roles of Dmrt1 in controlling the remodeling and differentiation of the juvenile testis [101]. It can be said that Dmrt1 is required for the establishment of postnatal spermatogenesis and the maintenance of the pool of progenitor cells that participate in adult spermatogenesis. In addition, Dmrt1 is involved in regulating the self-renewal of SSCs and maintaining their pluripotency [102,103,104]. The progressive exploration of Dmrt1 function has made researchers more certain that understanding Dmrt1 will likely help enable artificial manipulation of spermatogenesis.
Gene editing technology has made it possible to determine the function of Dmrt1 in more detail. CRISPR/Cas9-mediated Dmrt1 mutation animal models were first implemented in tilapia. A distinct phenotype was observed for the G0 generation of Dmrt1 mutant tilapia constructed by Minghui Li et al., and this phenotype was consistent with the gonadal phenotype induced by TALENs [105]. Qiaohong Lin et al. successfully generated Dmrt1 mutant zebrafish by CRISPR/Cas9 [33]. The deletion of Dmrt1 caused defective testicular development. The differentiation of GCs of all types was severely impaired, and their number was drastically reduced. GC-associated and testicular somatic-cell-associated genes were also differentially dysregulated due to Dmrt1 deletion. All these results support the hypothesis that Dmrt1 is involved in regulating testicular development and male GC proliferation. By constructing a joint study in Amh mutant zebrafish, Qiaohong Lin et al. also found that Amh and Dmrt1 synergistically maintain spermatogenesis by regulating male GC self-renewal and differentiation [33]. The construction of other Dmrt1-deficient models, such as mouse and chicken models, has allowed us to further define the critical role of Dmrt1 in sex determination and spermatogenesis [106].
4.3. Dpy-19-like 2 (Dpy19l2)
Similar to Spata16 and Pick1, Dpy19l2 is the third gene in which defects have been identified to be closely associated with globozoospermia [107]. Dpy19l2, located in the inner nuclear membrane, actively participates in the attachment process of the acrosome to the nuclear envelope [108,109]. A homozygous deletion of Dpy19l2 blocks sperm head elongation and acrosome formation, leading to male infertility [110]. Afterward, the discovery of various novel point mutations, nonsense mutations and missense mutations of Dpy19l2 further deepened the understanding of Dpy19l2 mutation-induced globozoospermia [111,112,113]. Yueshuai Guo et al. revealed a large number of differentially expressed proteins between the sperm of Dpy19l2-deficient human globozoospermia and those of normozoospermia by tandem mass (TMT) quantitative proteomics analysis [114]. This finding implies that pathogenic mutations in Dpy19l2 induce aberrant expression of many unknown factors. There are also many mechanisms that we do not yet understand that work together to cause globozoospermia. With the discovery of FAM209, the first protein that interacts with DPY19L2, our knowledge of the in vivo mechanism of action of DPY19L2 has become even richer. Studies have confirmed that the FAM209-DPY19L2 complex maintains normal acrosome biogenesis and spermatogenesis [115]. In addition, the use of intracytoplasmic sperm injection (ICSI) and calcium carrier assisted oocyte activation (AOA) has made it possible to cure infertility caused by Dpy19l2 dysfunction [116,117].
4.4. Testis Specific 10 (Tsga10)
Tsga10 was first identified and characterized in 2001 by M H Modarressi et al. [118]. Tsga10 is a testis-specific expressed gene consisting of 19 exons. Modarressi et al. then found that the Tsga10-encoded sperm cell protein is processed into fibrous sheath protein in mature sperm [119]. Therefore, Tsga10 is considered to be an important regulatory gene for the formation of the fibrous sheath of the sperm tail. As research on acephalic spermatozoa syndrome continues to advance, Tsga10 has been identified as one of the candidate genes for the syndrome [120]. Loss-of-function mutations or deletions in Tsga10 directly cause acephalic spermatozoa syndrome [121,122,123]. To further analyze the function of Tsga10, Geng Luo et al. constructed Tsga10+/− mice by CRISPR/Cas9 [80]. Tsga10+/− mice showed disturbed mitochondrial sheath formation, significantly low sperm motility, and male sterility. This finding further defines the role of Tsga10 in spermatogenesis. Rezvan Asgari et al. further found that abnormal spermatogenesis due to the deletion of Tsga10 may be associated with autophagy [124]. Reduced Tsga10 expression attenuated its inhibition of HIF-1, leading to diminished autophagy and the overproduction of reactive oxygen species (ROS) and resulting in impaired sperm maturation.
5. CRISPR/Cas9: Potential New Tools for Treating Abnormal Spermatogenesis and Male Infertility
The successful gene editing of mouse SSCs using TALEN or CRISPR/Cas9 indicates that gene editing technologies will become an important tool for studying spermatogenesis and revealing the mechanism of the development of spermatogenesis abnormalities. In addition to advancing basic research, the combination of gene editing technologies and SSC transplantation technology makes it possible to produce transgenic animals more rapidly and to better achieve the conservation of good animal breeds. However, the complexity and inefficiency of traditional gene editing technologies have limited the success rate of editing SSCs. The creation of CRISPR/Cas9 has greatly solved this problem.
Yuxuan Wu et al. provided the first theoretical justification for the use of CRISPR/Cas9 to correct genetic defects in 2013 [125]. Two years later, they once again used CRISPR/Cas9 to efficiently edit Crygc in mouse SSCs and successfully completed the repair and correction of genetic defects in mice [126]. The correction of SSCs by CRISPR/Cas9-mediated homology-directed repair (HDR) has led to a new therapeutic direction for male infertility caused by GC genetic defects. The point mutation in the SSCs of Kitw/Kitwv mice was subsequently corrected by Xiaoyu Li et al. via CRISPR/Cas9 in vitro [127]. After being transplanted back into the testis, the repaired SSCs successfully restored the natural fertility of the mice. Xianyu Zhang et al. also attempted to establish a SSC transplant recipient mouse model to achieve more effective SSC transplantation [128]. Although the Etv5−/− mice that were successfully constructed could adopt and support foreign SSCs and produce donor-derived sperm, the efficiency of this process was very low. The quantity and quality of sperm produced by Etv5−/− mice were not ideal. It can thus be seen that research is still needed to further refine the protocol of SSC transplantation to construct transgenic animals. There is no denying that the application of gene editing technologies has allowed researchers to delve further into the complex and mysterious process of spermatogenesis. Gene editing technologies have also opened up new avenues of development in the medical, biomedical and agricultural fields.
6. Perspectives
As an interdisciplinary field involving histology, embryology, molecular biology, genetics, etc., the study of spermatogenesis is one of the research hotspots in the field of reproductive biology. The regulatory roles of DNA and RNA methylation modifications, histone modifications, noncoding RNAs, exosomes, hormones, and various testicular somatic cells during spermatogenesis have been discovered in a stepwise manner [129,130,131,132,133]. With the rapid innovation and optimization of gene editing technology, more key factors in the process of spermatogenesis have been identified. From the initial systemic knockout to the target-specific knockout, researchers can better understand the spermatogenesis process. The successive emergence of novel Cas proteins such as Cas12, Cas13, Cas14, etc. has also expanded and extended the editing scope and practical applications of CRISPR/Cas technology. This will allow CRISPR/Cas technology to play a greater driving role in the field of spermatogenesis. However, most of the existing research on spermatogenesis is still carried out by gene knockout using the CRISPR/Cas9 system to construct animal models. Studies evaluating gene function by phenotypes such as fecundity, sperm count, and sperm motility in knockout animals still account for a disproportionate number of studies. At the same time, many mechanisms involved in the process of spermatogenesis are still unknown, which also leads to the fact that, although the current gene editing technology has promoted research in spermatogenesis, the exploration of deeper molecular mechanisms of spermatogenesis is still lacking.
In spermatogenesis studies, most of the existing animal models are fish or mouse knockout models, which may be due to the easier handling and lower cost of fish and mice. However, there is a lack of an accepted and complete system of phenotypic evaluation indicators for the models. Single-gene knockout animal models exploring the presence or absence of reproductive disorders have limitations for our deeper understanding of spermatogenesis. Morphologically normal sperm produced by knockout animals may also have some recessive abnormalities leading to infertility. Therefore, visual indicators such as sperm viability, sperm count, and testicular histomorphology alone can no longer meet the needs of existing research. The need for a more in-depth evaluation of animal models constructed by gene editing is urging researchers to explore the underlying causes of spermatogenesis rather than making the crude assumption that a particular gene or protein is important or unimportant.
Although the use of mice as research subjects can reveal universal mechanisms in mammals, these studies are still at a stage where they can provide only a theoretical basis for practical animal breeding work. At this point in time, very little research has been carried out on livestock and poultry. This fact suggests that the mechanisms of spermatogenesis are not as clear-cut as we thought and that there are still many problems that need to be solved in the practical application of gene editing technology in production. Therefore, more research in livestock and poultry is needed in the future. A comprehensive and systematic analysis of the mechanisms underlying the various biological processes involved in spermatogenesis will facilitate the development of animal breeding, as well as developmental and reproductive biology.
In addition, the combination of gene editing and SSC transplantation offers new ideas for the treatment of male infertility, but further research is needed to establish the optimum transplantation process [128]. However, in the absence of extensive basic research and clinical trials, this therapeutic idea can be only a new direction. There is still a long way to go until gene editing becomes a safe, effective and controllable tool for treatment. Ethical issues also limit the use of gene editing technologies to some extent. In conclusion, there is still much space to be explored in both gene editing and spermatogenesis research. However, it is undeniable that gene editing already holds great promise for the research and treatment of human infertility and the acceleration of animal breeding processes. Meanwhile, in-depth research on spermatogenesis will provide new strategies for the diagnosis and treatment of male reproductive system diseases and the conservation of rare and endangered animal germplasm resources.
Author Contributions
Conceptualization, F.G. and W.-Z.R.; writing—original draft preparation, H.-Q.W. and T.W.; writing—review and editing, H.-Q.W. and T.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, 31972570.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Statens Veterinärmedicinska Anstalt. Food and agriculture organization of the United Nations. 2013. [Google Scholar]
- Steinfeld, H.; Gerber, P.; Wassenaar, T.D.; Castel, V.; Rosales, M.; Rosales, M.; de Haan, C. Livestock’s Long Shadow: Environmental issues and Options; Food & Agriculture Organization: Rome, Italy, 2006. [Google Scholar]
- Bourdon, G.; Cadoret, V.; Charpigny, G.; Couturier-Tarrade, A.; Dalbies-Tran, R.; Flores, M.J.; Froment, P.; Raliou, M.; Reynaud, K.; Saint-Dizier, M.; et al. Progress and challenges in developing organoids in farm animal species for the study of reproduction and their applications to reproductive biotechnologies. Vet. Res. 2021, 52, 42. [Google Scholar] [CrossRef]
- Saha, S.; Roy, P.; Corbitt, C.; Kakar, S.S. Application of Stem Cell Therapy for Infertility. Cells 2021, 10, 1613. [Google Scholar] [CrossRef] [PubMed]
- Giassetti, M.I.; Ciccarelli, M.; Oatley, J.M. Spermatogonial Stem Cell Transplantation: Insights and Outlook for Domestic Animals. Annu. Rev. Anim. Biosci. 2019, 7, 385–401. [Google Scholar] [CrossRef] [PubMed]
- Ozbek, M.; Hitit, M.; Kaya, A.; Jousan, F.D.; Memili, E. Sperm Functional Genome Associated With Bull Fertility. Front. Vet. Sci. 2021, 8, 610888. [Google Scholar] [CrossRef]
- Houdebine, L.M. Use of transgenic animals to improve human health and animal production. Reprod. Domest. Anim. 2005, 40, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.V.; Tiley, L.S.; Sang, H.M. Developments in transgenic technology: Applications for medicine. Trends Mol. Med. 2005, 11, 293–298. [Google Scholar] [CrossRef]
- Fan, J.; Watanabe, T. Transgenic rabbits as therapeutic protein bioreactors and human disease models. Pharmacol. Ther. 2003, 99, 261–282. [Google Scholar] [CrossRef]
- Lillico, S.G.; McGrew, M.J.; Sherman, A.; Sang, H.M. Transgenic chickens as bioreactors for protein-based drugs. Drug Discov. Today 2005, 10, 191–196. [Google Scholar] [CrossRef]
- Xu, K.; Zhou, Y.; Mu, Y.; Liu, Z.; Hou, S.; Xiong, Y.; Fang, L.; Ge, C.; Wei, Y.; Zhang, X.; et al. CD163 and pAPN double-knockout pigs are resistant to PRRSV and TGEV and exhibit decreased susceptibility to PDCoV while maintaining normal production performance. eLife 2020, 9, e57132. [Google Scholar] [CrossRef]
- De Kretser, D.M.; Loveland, K.L.; Meinhardt, A.; Simorangkir, D.; Wreford, N. Spermatogenesis. Hum. Reprod. 1998, 13 (Suppl. 1), 1–8. [Google Scholar] [CrossRef] [PubMed]
- De Rooij, D.G. The nature and dynamics of spermatogonial stem cells. Development 2017, 144, 3022–3030. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhang, Y.; Qu, R.; He, Y.; Tian, X.; Zeng, W. Spermatogonial stem cells from domestic animals: Progress and prospects. Reproduction 2014, 147, R65–R74. [Google Scholar] [CrossRef] [PubMed]
- Griswold, M.D. Spermatogenesis: The Commitment to Meiosis. Physiol. Rev. 2016, 96, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Griswold, M.D. 50 years of spermatogenesis: Sertoli cells and their interactions with germ cells. Biol. Reprod. 2018, 99, 87–100. [Google Scholar] [CrossRef]
- Griswold, M.D. The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Biol. 1998, 9, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Thompson, T.L.; Berndtson, W.E. Testicular weight, Sertoli cell number, daily sperm production, and sperm output of sexually mature rabbits after neonatal or prepubertal hemicastration. Biol. Reprod. 1993, 48, 952–957. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Russell, L.D.; Ren, H.P.; Sinha Hikim, I.; Schulze, W.; Sinha Hikim, A.P. A comparative study in twelve mammalian species of volume densities, volumes, and numerical densities of selected testis components, emphasizing those related to the Sertoli cell. Am. J. Anat. 1990, 188, 21–30. [Google Scholar] [CrossRef]
- Neto, F.T.; Bach, P.V.; Najari, B.B.; Li, P.S.; Goldstein, M. Spermatogenesis in humans and its affecting factors. Semin. Cell Dev. Biol. 2016, 59, 10–26. [Google Scholar] [CrossRef]
- Saez, J.M.; Avallet, O.; Naville, D.; Perrard-Sapori, M.H.; Chatelain, P.G. Sertoli-Leydig cell communications. Ann. N. Y. Acad. Sci. 1989, 564, 210–231. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Wu, J.; Liu, B.; Jiang, Y.; Chen, W.; Li, J.; He, Q.; He, Z. The roles and mechanisms of Leydig cells and myoid cells in regulating spermatogenesis. Cell Mol. Life Sci. 2019, 76, 2681–2695. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lin, Q.; Jin, S.; Gao, C. The CRISPR-Cas toolbox and gene editing technologies. Mol. Cell 2022, 82, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 2015, 60, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef]
- Strich, J.R.; Chertow, D.S. CRISPR-Cas Biology and Its Application to Infectious Diseases. J. Clin. Microbiol 2019, 57, JCM-01307. [Google Scholar] [CrossRef]
- Fang, X.; Huang, L.L.; Xu, J.; Ma, C.Q.; Chen, Z.H.; Zhang, Z.; Liao, C.H.; Zheng, S.X.; Huang, P.; Xu, W.M.; et al. Proteomics and single-cell RNA analysis of Akap4-knockout mice model confirm indispensable role of Akap4 in spermatogenesis. Dev. Biol. 2019, 454, 118–127. [Google Scholar] [CrossRef]
- Lin, Q.; Mei, J.; Li, Z.; Zhang, X.; Zhou, L.; Gui, J.F. Distinct and Cooperative Roles of amh and dmrt1 in Self-Renewal and Differentiation of Male Germ Cells in Zebrafish. Genetics 2017, 207, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Coutton, C.; Martinez, G.; Kherraf, Z.E.; Amiri-Yekta, A.; Boguenet, M.; Saut, A.; He, X.; Zhang, F.; Cristou-Kent, M.; Escoffier, J.; et al. Bi-allelic Mutations in ARMC2 Lead to Severe Astheno-Teratozoospermia Due to Sperm Flagellum Malformations in Humans and Mice. Am. J. Hum. Genet. 2019, 104, 331–340. [Google Scholar] [CrossRef]
- Shen, C.; Xu, J.; Zhou, Q.; Lin, M.; Lv, J.; Zhang, X.; Wu, Y.; Chen, X.; Yu, J.; Huang, X.; et al. E3 ubiquitin ligase ASB17 is required for spermiation in mice. Transl. Androl. Urol. 2021, 10, 4320–4332. [Google Scholar] [CrossRef]
- Lu, C.; Zhang, Y.; Qin, Y.; Xu, Q.; Zhou, R.; Cui, Y.; Zhu, Y.; Zhang, X.; Zhang, J.; Wei, X.; et al. Human X chromosome exome sequencing identifies BCORL1 as contributor to spermatogenesis. J. Med. Genet. 2021, 58, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, W.; Zhang, P.; Gao, F.; Zhao, X.; Shum, W.W.; Zeng, X. Cabs1 Maintains Structural Integrity of Mouse Sperm Flagella during Epididymal Transit of Sperm. Int. J. Mol. Sci. 2021, 22, 652. [Google Scholar] [CrossRef] [PubMed]
- Young, S.A.; Miyata, H.; Satouh, Y.; Kato, H.; Nozawa, K.; Isotani, A.; Aitken, R.J.; Baker, M.A.; Ikawa, M. CRISPR/Cas9-Mediated Rapid Generation of Multiple Mouse Lines Identified Ccdc63 as Essential for Spermiogenesis. Int. J. Mol. Sci. 2015, 16, 24732–24750. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Tang, W.; Li, H.; Hua, R.; Yuan, Y.; Zhang, Y.; Zhu, Y.; Cui, Y.; Sha, J. T-complex protein 1 subunit zeta-2 (CCT6B) deficiency induces murine teratospermia. PeerJ 2021, 9, e11545. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Zhu, H.; Zhang, A.; Lin, J.; Zhang, G.; Liu, D.; Xiao, Y.; Ye, C.; Sun, D.; Wu, B.; et al. Cdc14a has a role in spermatogenesis, sperm maturation and male fertility. Exp. Cell Res. 2020, 395, 112178. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Miyata, H.; Kaneda, Y.; Castaneda, J.M.; Lu, Y.; Morohoshi, A.; Yu, Z.; Matzuk, M.M.; Ikawa, M. CIB4 is essential for the haploid phase of spermatogenesis in micedagger. Biol. Reprod. 2020, 103, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Liu, X.; Liu, X.; Li, T.; Zhu, P.; Liu, Z.; Xue, H.; Wang, W.; Yang, X.; Liu, J.; et al. Integrated Analyses of Phenotype and Quantitative Proteome of CMTM4 Deficient Mice Reveal Its Association with Male Fertility. Mol. Cell Proteomics 2019, 18, 1070–1084. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, A.G.; Seroussi, U.; Lehrbach, N.J.; Renaud, M.S.; Sundby, A.E.; Molnar, R.I.; Lao, R.X.; Willis, A.R.; Woock, J.R.; Aber, M.J.; et al. Two isoforms of the essential C. elegans Argonaute CSR-1 differentially regulate sperm and oocyte fertility. Nucleic Acids Res. 2021, 49, 8836–8865. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Xiao, H.; Shi, H.; Wang, T.; Sun, L.; Tao, W.; Kocher, T.D.; Li, M.; Wang, D. Loss of Cyp11c1 causes delayed spermatogenesis due to the absence of 11-ketotestosterone. J. Endocrinol. 2020, 244, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Le, H.T.; Hasegawa, Y.; Daitoku, Y.; Kato, K.; Miznuo-Iijima, S.; Dinh, T.T.H.; Kuba, Y.; Osawa, Y.; Mikami, N.; Morimoto, K.; et al. Generation of B6-Ddx4(em1(CreERT2)Utr), a novel CreERT2 knock-in line, for germ cell lineage by CRISPR/Cas9. Genesis 2020, 58, e23367. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, Y.; Xie, S.; Yin, Q.; Tang, C.; Ni, Z.; Fei, J.; Zhang, Y. CRISPR/Cas9-mediated genome editing reveals the synergistic effects of β-defensin family members on sperm maturation in rat epididymis. FASEB J. 2018, 32, 1354–1363. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, H.; Li, M.; Cheng, Y.; Jiang, D.; Sun, L.; Tao, W.; Zhou, L.; Wang, Z.; Wang, D. Isolation of doublesex- and mab-3-related transcription factor 6 and its involvement in spermatogenesis in tilapia. Biol. Reprod. 2014, 91, 136. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ouyang, J.; Li, X.; Xiao, X.; Sun, W.; Li, S.; Zhou, L.; Liao, Y.; Zhang, Q. DNAH17 is essential for rat spermatogenesis and fertility. J. Genet. 2021, 100, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Ma, H.; Khan, T.; Ma, A.; Li, T.; Zhang, H.; Gao, J.; Zhou, J.; Li, Y.; Yu, C.; et al. A DNAH17 missense variant causes flagella destabilization and asthenozoospermia. J. Exp. Med. 2020, 217, e20182365. [Google Scholar] [CrossRef]
- Pierre, V.; Martinez, G.; Coutton, C.; Delaroche, J.; Yassine, S.; Novella, C.; Pernet-Gallay, K.; Hennebicq, S.; Ray, P.F.; Arnoult, C. Absence of Dpy19l2, a new inner nuclear membrane protein, causes globozoospermia in mice by preventing the anchoring of the acrosome to the nucleus. Development 2012, 139, 2955–2965. [Google Scholar] [CrossRef]
- N’Tumba-Byn, T.; Yamada, M.; Seandel, M. Loss of tyrosine kinase receptor Ephb2 impairs proliferation and stem cell activity of spermatogonia in culturedagger. Biol. Reprod. 2020, 102, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Devlin, D.J.; Nozawa, K.; Ikawa, M.; Matzuk, M.M. Knockout of family with sequence similarity 170 member A (Fam170a) causes male subfertility, while Fam170b is dispensable in micedagger. Biol. Reprod. 2020, 103, 205–222. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Gao, Q.; Feng, T.; Zheng, Y.; Guo, J.; Zeng, W. FTO Knockout Causes Chromosome Instability and G2/M Arrest in Mouse GC-1 Cells. Front. Genet. 2018, 9, 732. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ai, N.; Chen, W.; Wong, Q.W.; Ge, W. Loss of Growth Hormone Gene (gh1) in Zebrafish Arrests Folliculogenesis in Females and Delays Spermatogenesis in Males. Endocrinology 2019, 160, 568–586. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhao, W.; Liu, J.; Wang, Y.; Yuan, C.; Zhang, F.; Jin, G.; Qin, Q. Effects of HIF-1alpha on Spermatogenesis of Varicocele Rats by Regulating VEGF/PI3K/Akt Signaling Pathway. Reprod. Sci. 2021, 28, 1161–1174. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, J.; Wang, D.; Wang, Y.; Zhang, F.; Jin, G.; Yuan, C.; Wang, X.; Qin, Q. Effect of silencing HIF-1alpha gene on testicle spermatogenesis function in varicocele rats. Cell Tissue Res. 2019, 378, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Saju, J.M.; Hossain, M.S.; Liew, W.C.; Pradhan, A.; Thevasagayam, N.M.; Tan, L.S.E.; Anand, A.; Olsson, P.E.; Orban, L. Heat Shock Factor 5 Is Essential for Spermatogenesis in Zebrafish. Cell Rep. 2018, 25, 3252–3261.e4. [Google Scholar] [CrossRef] [PubMed]
- Oura, S.; Miyata, H.; Noda, T.; Shimada, K.; Matsumura, T.; Morohoshi, A.; Isotani, A.; Ikawa, M. Chimeric analysis with newly established EGFP/DsRed2-tagged ES cells identify HYDIN as essential for spermiogenesis in mice. Exp. Anim. 2019, 68, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, X.; Dai, S.; Xiao, H.; Qi, S.; Li, Y.; Zheng, Q.; Jie, M.; Cheng, C.H.K.; Wang, D. Regulation of spermatogenesis and reproductive capacity by Igf3 in tilapia. Cell Mol. Life Sci. 2020, 77, 4921–4938. [Google Scholar] [CrossRef]
- Wen, Z.; Liu, D.; Zhu, H.; Sun, X.; Xiao, Y.; Lin, Z.; Zhang, A.; Ye, C.; Gao, J. Deficiency for Lcn8 causes epididymal sperm maturation defects in mice. Biochem. Biophys. Res. Commun. 2021, 548, 7–13. [Google Scholar] [CrossRef]
- Gong, Q.Q.; Dou, Z.L.; Wang, X.; Zhang, K.Y.; Chen, H.; Gao, J.G.; Sun, X.Y. Epididymal initial segment-specific Cre recombinase activity in Lcn8-Cre knock-in mice. Mol. Biol. Rep. 2021, 48, 6015–6023. [Google Scholar] [CrossRef]
- Bae, H.S.; Jin, Y.K.; Ham, S.; Kim, H.K.; Shin, H.; Cho, G.B.; Lee, K.J.; Lee, H.; Kim, K.M.; Koo, O.J.; et al. CRISRP/Cas9-mediated knockout of Mct8 reveals a functional involvement of Mct8 in testis and sperm development in a rat. Sci. Rep. 2020, 10, 11148. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Huang, Q.; Zhang, L.; Liu, H.; Zhang, D.; Yuan, S.; Yap, Y.; Qu, W.; Shiang, R.; Song, S.; et al. A single amino acid mutation in the mouse MEIG1 protein disrupts a cargo transport system necessary for sperm formation. J. Biol. Chem. 2021, 297, 101312. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.; Kam, C.; Shen, C.; Jin, W.; Wang, J.; Lee, K.M.; Jiang, L.; Xia, J. PICK1 deficiency causes male infertility in mice by disrupting acrosome formation. J. Clin. Invest. 2009, 119, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Xu, J.; Chen, K.; Liu, Y.; Yang, X.; Tang, L.; Luo, X.; Liu, Z.; Li, M.; Walters, J.R.; et al. BmPMFBP1 regulates the development of eupyrene sperm in the silkworm, Bombyx mori. PLoS Genet. 2022, 18, e1010131. [Google Scholar] [CrossRef]
- Kobayashi, K.; Endo, T.; Matsumura, T.; Lu, Y.; Yu, Z.; Matzuk, M.M.; Ikawa, M. Prss55 but not Prss51 is required for male fertility in micedagger. Biol. Reprod. 2020, 103, 223–234. [Google Scholar] [CrossRef]
- Yang, L.; Li, Y.; Wu, Y.; Sun, S.; Song, Q.; Wei, J.; Sun, L.; Li, M.; Wang, D.; Zhou, L. Rln3a is a prerequisite for spermatogenesis and fertility in male fish. J. Steroid. Biochem. Mol. Biol. 2020, 197, 105517. [Google Scholar] [CrossRef]
- Li, D.; Li, F.; Meng, L.; Wei, H.; Zhang, Q.; Jiang, F.; Chen, D.N.; Li, W.; Tan, Y.Q.; Li, J.D. RNF216 regulates meiosis and PKA stability in the testes. FASEB J. 2021, 35, e21460. [Google Scholar] [CrossRef]
- Melnick, A.F.; Gao, Y.; Liu, J.; Ding, D.; Predom, A.; Kelly, C.; Hess, R.A.; Chen, C. RNF216 is essential for spermatogenesis and male fertilitydagger. Biol. Reprod. 2019, 100, 1132–1134. [Google Scholar] [CrossRef]
- Wei, L.; Tang, Y.; Zeng, X.; Li, Y.; Zhang, S.; Deng, L.; Wang, L.; Wang, D. The transcription factor Sox30 is involved in Nile tilapia spermatogenesis. J. Genet. Genomics 2021. [Google Scholar] [CrossRef]
- Feng, C.A.; Spiller, C.; Merriner, D.J.; O’Bryan, M.K.; Bowles, J.; Koopman, P. SOX30 is required for male fertility in mice. Sci. Rep. 2017, 7, 17619. [Google Scholar] [CrossRef]
- Fujihara, Y.; Oji, A.; Larasati, T.; Kojima-Kita, K.; Ikawa, M. Human Globozoospermia-Related Gene Spata16 Is Required for Sperm Formation Revealed by CRISPR/Cas9-Mediated Mouse Models. Int. J. Mol. Sci 2017, 18, 2208. [Google Scholar] [CrossRef]
- Girault, M.S.; Dupuis, S.; Ialy-Radio, C.; Stouvenel, L.; Viollet, C.; Pierre, R.; Favier, M.; Ziyyat, A.; Barbaux, S. Deletion of the Spata3 Gene Induces Sperm Alterations and In Vitro Hypofertility in Mice. Int. J. Mol. Sci. 2021, 22, 1959. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kwon, J.T.; Jeong, J.; Kim, J.; Hong, S.H.; Kim, J.; Park, Z.Y.; Chung, K.H.; Eddy, E.M.; Cho, C. SPATC1L maintains the integrity of the sperm head-tail junction. EMBO. Rep. 2018, 19, e45991. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, K.; Zhang, Q.; Miyata, H.; Devlin, D.J.; Yu, Z.; Oura, S.; Koyano, T.; Matsuyama, M.; Ikawa, M.; Matzuk, M.M. Knockout of serine-rich single-pass membrane protein 1 (Ssmem1) causes globozoospermia and sterility in male micedagger. Biol. Reprod. 2020, 103, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Khan, R.; Yu, C.; Alsheimer, M.; Jiang, X.; Ma, H.; Shi, Q. The testis-specific LINC component SUN3 is essential for sperm head shaping during mouse spermiogenesis. J. Biol. Chem. 2020, 295, 6289–6298. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, Y.; Qin, D.; Gui, Y.; Wang, S.; Du, G.; Yang, F.; Li, L.; Yuan, S.; Wang, M.; et al. Transcription factor-like 5 is a potential DNA- and RNA-binding protein essential for maintaining male fertility in mice. J. Cell Sci. 2022, 135, jcs259036. [Google Scholar] [CrossRef]
- Feng, M.; Bai, Y.; Chen, Y.; Wang, K. Knockout of the Transducin-Like Enhancer of Split 6 Gene Affects the Proliferation and Cell Cycle Process of Mouse Spermatogonia. Int. J. Mol. Sci. 2020, 21, 5827. [Google Scholar] [CrossRef]
- Larasati, T.; Noda, T.; Fujihara, Y.; Shimada, K.; Tobita, T.; Yu, Z.; Matzuk, M.M.; Ikawa, M. Tmprss12 is required for sperm motility and uterotubal junction migration in micedagger. Biol. Reprod. 2020, 103, 254–263. [Google Scholar] [CrossRef]
- Luo, G.; Hou, M.; Wang, B.; Liu, Z.; Liu, W.; Han, T.; Zhang, D.; Zhou, X.; Jia, W.; Tan, Y.; et al. Tsga10 is essential for arrangement of mitochondrial sheath and male fertility in mice. Andrology 2021, 9, 368–375. [Google Scholar] [CrossRef]
- Nayyab, S.; Gervasi, M.G.; Tourzani, D.A.; Caraballo, D.A.; Jha, K.N.; Teves, M.E.; Cui, W.; Georg, G.I.; Visconti, P.E.; Salicioni, A.M. TSSK3, a novel target for male contraception, is required for spermiogenesis. Mol. Reprod. Dev. 2021, 88, 718–730. [Google Scholar] [CrossRef]
- Liu, W.; He, X.; Yang, S.; Zouari, R.; Wang, J.; Wu, H.; Kherraf, Z.E.; Liu, C.; Coutton, C.; Zhao, R.; et al. Bi-allelic Mutations in TTC21A Induce Asthenoteratospermia in Humans and Mice. Am. J. Hum. Genet. 2019, 104, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Liu, Z.; Zheng, Y.; Feng, T.; Gao, Q.; Zeng, W. YTHDF2 promotes spermagonial adhesion through modulating MMPs decay via m(6)A/mRNA pathway. Cell Death Dis. 2020, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, E.A.; Seymour, K.A.; Sigrist, K.; Rooij, D.; Freiman, R.N. ZFP628 Is a TAF4b-Interacting Transcription Factor Required for Mouse Spermiogenesis. Mol. Cell Biol. 2020, 40, e00228-19. [Google Scholar] [CrossRef] [PubMed]
- Nakasuji, T.; Ogonuki, N.; Chiba, T.; Kato, T.; Shiozawa, K.; Yamatoya, K.; Tanaka, H.; Kondo, T.; Miyado, K.; Miyasaka, N.; et al. Complementary Critical Functions of Zfy1 and Zfy2 in Mouse Spermatogenesis and Reproduction. PLoS Genet. 2017, 13, e1006578. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Matsumura, T.; Bakse, J.; Holmlund, H.; Blanchet, G.; Carrot, E.; Ikawa, M.; Ward, M.A. Loss of mouse Y chromosome gene Zfy1 and Zfy2 leads to spermatogenesis impairment, sperm defects, and infertility. Biol. Reprod. 2022, ioac057. [Google Scholar] [CrossRef]
- Hu, X.; Shen, B.; Liao, S.; Ning, Y.; Ma, L.; Chen, J.; Lin, X.; Zhang, D.; Li, Z.; Zheng, C.; et al. Gene knockout of Zmym3 in mice arrests spermatogenesis at meiotic metaphase with defects in spindle assembly checkpoint. Cell Death Dis. 2017, 8, e2910. [Google Scholar] [CrossRef]
- Wu, X.L.; Yun, D.M.; Gao, S.; Liang, A.J.; Duan, Z.Z.; Wang, H.S.; Wang, G.S.; Sun, F. The testis-specific gene 1700102P08Rik is essential for male fertility. Mol. Reprod. Dev. 2020, 87, 231–240. [Google Scholar] [CrossRef]
- Xu, M.; Xiao, J.; Chen, J.; Li, J.; Yin, L.; Zhu, H.; Zhou, Z.; Sha, J. Identification and characterization of a novel human testis-specific Golgi protein, NYD-SP12. Mol. Hum. Reprod. 2003, 9, 9–17. [Google Scholar] [CrossRef]
- Dam, A.H.; Koscinski, I.; Kremer, J.A.; Moutou, C.; Jaeger, A.S.; Oudakker, A.R.; Tournaye, H.; Charlet, N.; Lagier-Tourenne, C.; van Bokhoven, H.; et al. Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am. J. Hum. Genet. 2007, 81, 813–820. [Google Scholar] [CrossRef]
- ElInati, E.; Fossard, C.; Okutman, O.; Ghedir, H.; Ibala-Romdhane, S.; Ray, P.F.; Saad, A.; Hennebicq, S.; Viville, S. A new mutation identified in SPATA16 in two globozoospermic patients. J. Assist. Reprod. Genet. 2016, 33, 815–820. [Google Scholar] [CrossRef]
- Behvarz, M.; Rahmani, S.A.; Siasi Torbati, E.; Danaei Mehrabad, S.; Bikhof Torbati, M. Association of CATSPER1, SPATA16 and TEX11 genes polymorphism with idiopathic azoospermia and oligospermia risk in Iranian population. BMC Med. Genom. 2022, 15, 47. [Google Scholar] [CrossRef] [PubMed]
- Raymond, C.S.; Shamu, C.E.; Shen, M.M.; Seifert, K.J.; Hirsch, B.; Hodgkin, J.; Zarkower, D. Evidence for evolutionary conservation of sex-determining genes. Nature 1998, 391, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; McClive, P.J.; Western, P.S.; Reed, K.J.; Sinclair, A.H. Conservation of a sex-determining gene. Nature 1999, 402, 601–602. [Google Scholar] [CrossRef] [PubMed]
- Raymond, C.S.; Kettlewell, J.R.; Hirsch, B.; Bardwell, V.J.; Zarkower, D. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev. Biol. 1999, 215, 208–220. [Google Scholar] [CrossRef]
- Moniot, B.; Berta, P.; Scherer, G.; Sudbeck, P.; Poulat, F. Male specific expression suggests role of DMRT1 in human sex determination. Mech. Dev. 2000, 91, 323–325. [Google Scholar] [CrossRef]
- Zhang, T.; Zarkower, D. DMRT proteins and coordination of mammalian spermatogenesis. Stem. Cell Res. 2017, 24, 195–202. [Google Scholar] [CrossRef]
- Ottolenghi, C.; McElreavey, K. Deletions of 9p and the quest for a conserved mechanism of sex determination. Mol. Genet. Metab. 2000, 71, 397–404. [Google Scholar] [CrossRef]
- Raymond, C.S.; Parker, E.D.; Kettlewell, J.R.; Brown, L.G.; Page, D.C.; Kusz, K.; Jaruzelska, J.; Reinberg, Y.; Flejter, W.L.; Bardwell, V.J.; et al. A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Hum. Mol. Genet. 1999, 8, 989–996. [Google Scholar] [CrossRef]
- Muroya, K.; Okuyama, T.; Goishi, K.; Ogiso, Y.; Fukuda, S.; Kameyama, J.; Sato, H.; Suzuki, Y.; Terasaki, H.; Gomyo, H.; et al. Sex-determining gene(s) on distal 9p: Clinical and molecular studies in six cases. J. Clin. Endocrinol Metab. 2000, 85, 3094–3100. [Google Scholar] [CrossRef]
- Kim, S.; Bardwell, V.J.; Zarkower, D. Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev. Biol. 2007, 307, 314–327. [Google Scholar] [CrossRef]
- Takashima, S.; Hirose, M.; Ogonuki, N.; Ebisuya, M.; Inoue, K.; Kanatsu-Shinohara, M.; Tanaka, T.; Nishida, E.; Ogura, A.; Shinohara, T. Regulation of pluripotency in male germline stem cells by Dmrt1. Genes Dev. 2013, 27, 1949–1958. [Google Scholar] [CrossRef] [PubMed]
- Cui, N.; Hao, G.; Zhao, Z.; Wang, F.; Cao, J.; Yang, A. MicroRNA-224 regulates self-renewal of mouse spermatogonial stem cells via targeting DMRT1. J. Cell Mol. Med. 2016, 20, 1503–1512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Oatley, J.; Bardwell, V.J.; Zarkower, D. DMRT1 Is Required for Mouse Spermatogonial Stem Cell Maintenance and Replenishment. PLoS Genet. 2016, 12, e1006293. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, H.; Zhao, J.; Fang, L.; Shi, H.; Li, M.; Sun, Y.; Zhang, X.; Jiang, D.; Zhou, L.; et al. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 2014, 197, 591–599. [Google Scholar] [CrossRef]
- Agbor, V.A.; Tao, S.; Lei, N.; Heckert, L.L. A Wt1-Dmrt1 transgene restores DMRT1 to sertoli cells of Dmrt1(-/-) testes: A novel model of DMRT1-deficient germ cells. Biol. Reprod. 2013, 88, 51. [Google Scholar] [CrossRef]
- Koscinski, I.; Elinati, E.; Fossard, C.; Redin, C.; Muller, J.; Velez de la Calle, J.; Schmitt, F.; Ben Khelifa, M.; Ray, P.F.; Kilani, Z.; et al. DPY19L2 deletion as a major cause of globozoospermia. Am. J. Hum. Genet. 2011, 88, 344–350. [Google Scholar] [CrossRef]
- Yassine, S.; Escoffier, J.; Abi Nahed, R.; Pierre, V.; Karaouzene, T.; Ray, P.F.; Arnoult, C. Dynamics of Sun5 localization during spermatogenesis in wild type and Dpy19l2 knock-out mice indicates that Sun5 is not involved in acrosome attachment to the nuclear envelope. PLoS ONE 2015, 10, e0118698. [Google Scholar] [CrossRef]
- Pereira, C.D.; Serrano, J.B.; Martins, F.; da Cruz, E.S.O.A.B.; Rebelo, S. Nuclear envelope dynamics during mammalian spermatogenesis: New insights on male fertility. Biol. Rev. Camb. Philos. Soc. 2019, 94, 1195–1219. [Google Scholar] [CrossRef]
- Harbuz, R.; Zouari, R.; Pierre, V.; Ben Khelifa, M.; Kharouf, M.; Coutton, C.; Merdassi, G.; Abada, F.; Escoffier, J.; Nikas, Y.; et al. A recurrent deletion of DPY19L2 causes infertility in man by blocking sperm head elongation and acrosome formation. Am. J. Hum. Genet. 2011, 88, 351–361. [Google Scholar] [CrossRef]
- Zhu, F.; Gong, F.; Lin, G.; Lu, G. DPY19L2 gene mutations are a major cause of globozoospermia: Identification of three novel point mutations. Mol. Hum. Reprod. 2013, 19, 395–404. [Google Scholar] [CrossRef]
- Coutton, C.; Zouari, R.; Abada, F.; Ben Khelifa, M.; Merdassi, G.; Triki, C.; Escalier, D.; Hesters, L.; Mitchell, V.; Levy, R.; et al. MLPA and sequence analysis of DPY19L2 reveals point mutations causing globozoospermia. Hum. Reprod. 2012, 27, 2549–2558. [Google Scholar] [CrossRef]
- Li, Y.Z.; Wu, R.F.; Zhu, X.S.; Liu, W.S.; Ye, Y.Y.; Lu, Z.X.; Li, N. Identification of a novel deletion mutation in DPY19L2 from an infertile patient with globozoospermia: A case report. Mol. Cytogenet. 2020, 13, 24. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Jiang, J.; Zhang, H.; Wen, Y.; Zhang, H.; Cui, Y.; Tian, J.; Jiang, M.; Liu, X.; Wang, G.; et al. Proteomic Analysis of Dpy19l2-Deficient Human Globozoospermia Reveals Multiple Molecular Defects. Proteomics Clin. Appl. 2019, 13, e1900007. [Google Scholar] [CrossRef] [PubMed]
- Castaneda, J.M.; Shimada, K.; Satouh, Y.; Yu, Z.; Devlin, D.J.; Ikawa, M.; Matzuk, M.M. FAM209 associates with DPY19L2, and is required for sperm acrosome biogenesis and fertility in mice. J. Cell Sci. 2021, 134, jcs259206. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.L.; Zhu, F.X.; Yan, J.; Chen, L.; Tang, W.H.; Xiao, S.; Mo, W.K.; Zhang, Z.G.; He, X.J.; Qiao, J.; et al. Novel DPY19L2 variants in globozoospermic patients and the overcoming this male infertility. Asian J. Androl. 2019, 21, 183–189. [Google Scholar] [CrossRef]
- Kuentz, P.; Vanden Meerschaut, F.; Elinati, E.; Nasr-Esfahani, M.H.; Gurgan, T.; Iqbal, N.; Carre-Pigeon, F.; Brugnon, F.; Gitlin, S.A.; Velez de la Calle, J.; et al. Assisted oocyte activation overcomes fertilization failure in globozoospermic patients regardless of the DPY19L2 status. Hum. Reprod. 2013, 28, 1054–1061. [Google Scholar] [CrossRef]
- Modarressi, M.H.; Cameron, J.; Taylor, K.E.; Wolfe, J. Identification and characterisation of a novel gene, TSGA10, expressed in testis. Gene 2001, 262, 249–255. [Google Scholar] [CrossRef]
- Modarressi, M.H.; Behnam, B.; Cheng, M.; Taylor, K.E.; Wolfe, J.; van der Hoorn, F.A. Tsga10 encodes a 65-kilodalton protein that is processed to the 27-kilodalton fibrous sheath protein. Biol. Reprod. 2004, 70, 608–615. [Google Scholar] [CrossRef]
- Sha, Y.W.; Sha, Y.K.; Ji, Z.Y.; Mei, L.B.; Ding, L.; Zhang, Q.; Qiu, P.P.; Lin, S.B.; Wang, X.; Li, P.; et al. TSGA10 is a novel candidate gene associated with acephalic spermatozoa. Clin. Genet. 2018, 93, 776–783. [Google Scholar] [CrossRef]
- Xiang, M.; Wang, Y.; Xu, W.; Zheng, N.; Zhang, J.; Duan, Z.; Zha, X.; Shi, X.; Wang, F.; Cao, Y.; et al. Pathogenesis of acephalic spermatozoa syndrome caused by splicing mutation and de novo deletion in TSGA10. J. Assist. Reprod. Genet. 2021, 38, 2791–2799. [Google Scholar] [CrossRef]
- Liu, G.; Wang, N.; Zhang, H.; Yin, S.; Dai, H.; Lin, G.; Li, W. Novel mutations in PMFBP1, TSGA10 and SUN5: Expanding the spectrum of mutations that may cause acephalic spermatozoa. Clin. Genet. 2020, 97, 938–939. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Wei, X.; Sha, Y.; Li, N.; Yan, X.; Cheng, L.; Qiao, D.; Zhou, W.; Wu, R.; Liu, Q.; et al. Loss-of-function mutation in TSGA10 causes acephalic spermatozoa phenotype in human. Mol. Genet. Genomic Med. 2020, 8, e1284. [Google Scholar] [CrossRef]
- Asgari, R.; Bakhtiari, M.; Rezazadeh, D.; Yarani, R.; Esmaeili, F.; Mansouri, K. TSGA10 as a Potential Key Factor in the Process of Spermatid Differentiation/Maturation: Deciphering Its Association with Autophagy Pathway. Reprod. Sci. 2021, 28, 3228–3240. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liang, D.; Wang, Y.; Bai, M.; Tang, W.; Bao, S.; Yan, Z.; Li, D.; Li, J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem. Cell 2013, 13, 659–662. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, H.; Fan, X.; Zhang, Y.; Zhang, M.; Wang, Y.; Xie, Z.; Bai, M.; Yin, Q.; Liang, D.; et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015, 25, 67–79. [Google Scholar] [CrossRef]
- Li, X.; Sun, T.; Wang, X.; Tang, J.; Liu, Y. Restore natural fertility of Kit(w)/Kit(wv) mouse with nonobstructive azoospermia through gene editing on SSCs mediated by CRISPR-Cas9. Stem. Cell Res. Ther. 2019, 10, 271. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, X.; Li, G.; Zhang, M.; Xing, P.; Li, Z.; Chen, B.; Yang, H.; Wu, Z. Establishment of Etv5 gene knockout mice as a recipient model for spermatogonial stem cell transplantation. Biol. Open 2021, 10, bio056804. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, Y.; Gao, Y.; Lin, Z.; Yang, S.; Wang, T.; Wang, Q.; Xie, N.; Hua, R.; Liu, M.; et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018, 28, 879–896. [Google Scholar] [CrossRef]
- Wen, L.; Liu, Q.; Xu, J.; Liu, X.; Shi, C.; Yang, Z.; Zhang, Y.; Xu, H.; Liu, J.; Yang, H.; et al. Recent advances in mammalian reproductive biology. Sci. China Life Sci. 2020, 63, 18–58. [Google Scholar] [CrossRef]
- Smith, L.B.; Walker, W.H. The regulation of spermatogenesis by androgens. Semin. Cell Dev. Biol 2014, 30, 2–13. [Google Scholar] [CrossRef]
- Kubota, H.; Brinster, R.L. Spermatogonial stem cells. Biol. Reprod. 2018, 99, 52–74. [Google Scholar] [CrossRef] [PubMed]
- Chocu, S.; Calvel, P.; Rolland, A.D.; Pineau, C. Spermatogenesis in mammals: Proteomic insights. Syst. Biol. Reprod. Med. 2012, 58, 179–190. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).