Genetic Correlation, Pleiotropy, and Molar Morphology in a Longitudinal Sample of Australian Twins and Families
Abstract
1. Introduction
Research Aims and Hypotheses
- 1.
- Paired deciduous traits are genetically independent of one another:dtrait1–dtrait2 ρG = 0; dtrait1–dtrait2 ρG ≠ 1 (no pleiotropy).
- 2.
- Paired permanent traits are genetically independent of one another:PTrait1–PTrait2 ρG = 0; PTrait1–PTrait2 ρG ≠ 1 (no pleiotropy).
- 3.
- Paired deciduous/permanent traits are genetically independent of one another:dtrait1–PTrait1 ρG = 0; dtrait1–PTrait1 ρG ≠ 1 (no pleiotropy).
2. Materials and Methods
2.1. Data Collection Methods
2.2. Analytical Methods
2.2.1. Observer Error
2.2.2. Quantitative Genetic Modeling
3. Results
3.1. Observer Error Results
3.2. Genetic Correlation Results
4. Discussion
4.1. Morphology Versus Size
4.2. Antimeric Correlations and Bilateral Symmetry
4.3. Within-Dentition Correlations (Deciduous–Deciduous and Permanent–Permanent)
4.4. Between-Dentition Correlations (Deciduous–Permanent)
4.5. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Ungar, P.S. Mammal Teeth: Origin, Evolution, and Diversity; John Hopkins University Press: Baltimore, MD, USA, 2010. [Google Scholar] [CrossRef]
- Ungar, P.S. Origins and functions of teeth: From “toothed” worms to mammals. In A Companion to Dental Anthropology; Irish, J.D., Scott, G.R., Eds.; John Wiley & Sons, Inc.: Chichester, UK, 2016; Chapter 3; pp. 21–36. [Google Scholar] [CrossRef]
- Lumsden, A.G.S.; Osborn, J.W. The evolution of chewing: A dentist’s view of palaeontology. J. Dent. 1977, 5, 269–287. [Google Scholar] [CrossRef]
- Luo, Z.-X. Transformation and diversification in early mammal evolution. Nature 2007, 450, 1011–1019. [Google Scholar] [CrossRef]
- Kielan-Jaworowska, Z.; Cifelli, R.L.; Luo, Z.-Z. Mammals from the Age of Dinosaurs: Origins, Evolution, and Structure; Columbia University Press: New York, NY, USA, 2004. [Google Scholar]
- Luo, Z.-X.; Kielan-Jaworowska, Z.; Cifelli, R.L. Evolution of dental replacement in mammals. Bull. Carnegie Mus. Nat. Hist. 2004, 36, 159–175. [Google Scholar] [CrossRef]
- Tucker, A.S.; Fraser, G.J. Evolution and developmental diversity of tooth regeneration. Semin. Cell Dev. Biol. 2014, 25–26, 71–80. [Google Scholar] [CrossRef]
- Buchtová, M.; Stembírek, J.; Glocová, K.; Matalová, E.; Tucker, A.S. Early regression of the dental lamina underlies the development of diphyodont dentitions. J. Dent. Res. 2012, 91, 491–498. [Google Scholar] [CrossRef]
- Cobourne, M.T.; Sharpe, P.T. Making up the numbers: The molecular control of mammalian dental formula. Semin. Cell Dev. Biol. 2010, 21, 314–324. [Google Scholar] [CrossRef]
- Jernvall, J.; Thesleff, I. Tooth shape formation and tooth renewal: Evolving with the same signals. Development 2012, 139, 3487–3497. [Google Scholar] [CrossRef]
- Townsend, G.; Harris, E.F.; Lesot, H.; Clauss, F.; Brook, A. Morphogenetic fields within the human dentition: A new, clinically relevant synthesis of an old concept. Arch. Oral Biol. 2009, 54, S43–S44. [Google Scholar] [CrossRef]
- Tucker, A.S.; Matthews, K.; Sharpe, P.T. Transformation of tooth type by inhibition of BMP signalling. Science 1998, 282, 1136–1138. [Google Scholar] [CrossRef]
- Weiss, K.M.; Stock, D.W.; Zhao, Z. Dynamic interactions and the evolutionary genetics of dental patterning. Crit. Rev. Oral Biol. Med. 1998, 9, 369–398. [Google Scholar] [CrossRef]
- Biggerstaff, R.H. Heritability of the Carabelli’s cusp in twins. J. Dent. Res. 1973, 52, 40–44. [Google Scholar] [CrossRef]
- Dempsey, P.J.; Townsend, G.C. Genetic and environmental contributions to variation in human tooth size. Heredity 2001, 86, 685–693. [Google Scholar] [CrossRef]
- Hardin, A.M. Genetic contributions to dental dimensions in brown-mantled tamarins (Saguinus fuscicollis) and rhesus macaques (Macaca mulatta). Am. J. Phys. Anthropol. 2019, 168, 292–302. [Google Scholar] [CrossRef]
- Hlusko, L.J.; Mahaney, M.C. Genetic contributions to expression of the baboon cingular remnant. Arch. Oral Biol. 2003, 48, 663–672. [Google Scholar] [CrossRef]
- Lawrence, J.; Stojanowski, C.M.; Paul, K.S.; Seidel, A.C.; Guatelli-Steinberg, D. Heterogeneous frailty and the expression of linear enamel hypoplasia in a genealogical population. Am. J. Phys. Anthropol. 2021, 176, 638–651. [Google Scholar] [CrossRef]
- Paul, K.S.; Stojanowski, C.M.; Hughes, T.E.; Brook, A.H.; Townsend, G.C. Patterns of heritability across the human diphyodont dental complex: Crown morphology of Australian twins and families. Am. J. Phys. Anthropol. 2020, 172, 447–461. [Google Scholar] [CrossRef]
- Townsend, G.C. Heritability of deciduous tooth size in Australian aboriginals. Am. J. Phys. Anthropol. 1980, 53, 297–300. [Google Scholar] [CrossRef]
- Townsend, G.C.; Brown, T. Heritability of permanent tooth size. Am. J. Phys. Anthropol. 1978, 49, 497–504. [Google Scholar] [CrossRef]
- Hardin, A.M. Genetic correlations in the dental dimensions of Saguinus fuscicollis. Am. J. Phys. Anthropol. 2019, 169, 557–566. [Google Scholar] [CrossRef]
- Hardin, A.M. Genetic correlations in the rhesus macaque dentition. J. Hum. Evol. 2020, 148, 102873. [Google Scholar] [CrossRef]
- Hlusko, L.J.; Mahaney, M.C. Quantitative genetics, pleiotropy, and morphological integration in the dentition of Papio hamadryas. Evol. Biol. 2009, 36, 5–18. [Google Scholar] [CrossRef][Green Version]
- Hlusko, L.J.; Sage, R.D.; Mahaney, M.C. Modularity in the mammalian dentition: Mice and monkeys share a common dental genetic architecture. J. Exp. Zool. Part B Mol. Dev. Evol. 2011, 316, 21–49. [Google Scholar] [CrossRef]
- Stojanowski, C.M.; Paul, K.S.; Seidel, A.C.; Duncan, W.N.; Guatelli-Steinberg, D. Heritability and genetic integration of tooth size in the South Carolina Gullah. Am. J. Phys. Anthropol. 2017, 164, 505–521. [Google Scholar] [CrossRef]
- Grieco, T.M.; Rizk, O.T.; Hlusko, L.J. A modular framework characterized micro- and macroevolution of Old World monkey dentitions. Evolution 2012, 67, 241–259. [Google Scholar] [CrossRef]
- Hlusko, L.J.; Do, N.; Mahaney, M.C. Genetic correlations between mandibular molar cusp areas in baboons. Am. J. Phys. Anthropol. 2007, 132, 445–454. [Google Scholar] [CrossRef]
- Hlusko, L.J.; Maas, M.-L.; Mahaney, M.C. Statistical genetics of molar cusp patterning in pedigreed baboons: Implications for primate dental development and evolution. J. Exp. Zool. Part B Mol. Dev. Evol. 2004, 302, 268–283. [Google Scholar] [CrossRef]
- Koh, C.; Bates, E.; Broughton, E.; Do, N.T.; Fletcher, Z.; Mahaney, M.C.; Hlusko, L.J. Genetic integration of molar cusp size variation in baboons. Am. J. Phys. Anthropol. 2010, 142, 246–260. [Google Scholar] [CrossRef]
- Stojanowski, C.M.; Paul, K.S.; Seidel, A.C.; Duncan, W.N.; Guatelli-Steinberg, D. Heritability and genetic integration of anterior tooth crown variants in the South Carolina Gullah. Am. J. Phys. Anthropol. 2018, 167, 124–143. [Google Scholar] [CrossRef]
- Stojanowski, C.M.; Paul, K.S.; Seidel, A.C.; Duncan, W.N.; Guatelli-Steinberg, D. Quantitative genetic analyses of postcanine morphological crown variation. Am. J. Phys. Anthropol. 2019, 168, 606–631. [Google Scholar] [CrossRef]
- Butler, P.M. Studies of the mammalian dentition, differentiation of the post-canine dentition. Differentiation of the post-canine dentition. Proc. Zool. Soc. Lond. 1939, B109, 1–36. [Google Scholar] [CrossRef]
- Dahlberg, A.A. The changing dentition of man. J. Am. Dent. Assoc. 1945, 32, 676–690. [Google Scholar] [CrossRef]
- Dahlberg, A.A. The dentition of the American Indian. In The Physical Anthropology of the American Indian; Laughlin, W.S., Ed.; Viking Fund Inc.: New York, NY, USA, 1951; pp. 138–176. [Google Scholar]
- Paul, K.S.; Stojanowski, C.M.; Hughes, T.E.; Brook, A.H.; Townsend, G.C. The genetic architecture of anterior tooth morphology in a longitudinal sample of Australian twins and families. Arch. Oral Biol. 2021, 129, 105168. [Google Scholar] [CrossRef]
- AlQahtani, S.J.; Hector, M.P.; Liversidge, H.M. Brief communication: The London atlas of human tooth development and eruption. Am. J. Phys. Anthropol. 2010, 142, 481–490. [Google Scholar] [CrossRef]
- Schour, I.; Massler, M. The development of the human dentition. J. Am. Dent. Assoc. 1941, 28, 1153–1160. [Google Scholar]
- Smith, B.H. Standards of human tooth formation and dental age assessment. In Advances in Dental Anthropology; Kelley, M.A., Larsen, C.S., Eds.; Wiley-Liss: New York, NY, USA, 1991; pp. 143–168. [Google Scholar]
- Bailey, S.E.; Benazzi, S.; Hublin, J.-J. Allometry, merism, and tooth shape of the upper deciduous and permanent M1. Am. J. Phys. Anthropol. 2014, 154, 104–114. [Google Scholar] [CrossRef]
- Paul, K.S.; Astorino, C.M.; Bailey, S.E. The Patterning Cascade Model and Carabelli’s trait expression in metameres of the mixed human dentition: Exploring a morphogenetic model. Am. J. Phys. Anthropol. 2017, 162, 3–18. [Google Scholar] [CrossRef]
- Saunders, S.R.; Mayhall, J.T. Developmental patterns of human dental morphological traits. Arch. Oral Biol. 1982, 27, 45–49. [Google Scholar] [CrossRef]
- Berkovitz, B.K.B.; Holland, G.R.; Moxham, B.J. Oral Anatomy, Embryology, and Histology, 4th ed.; Mosby International Ltd.: Edinburgh, UK, 2009. [Google Scholar]
- Bailey, S.E.; Benazzi, S.; Buti, L.; Hublin, J.-J. Allometry, merism, and tooth shape of the lower second deciduous and first permanent molar. Am. J. Phys. Anthropol. 2016, 159, 93–105. [Google Scholar] [CrossRef]
- Juuri, E.; Jussila, M.; Seidel, K.; Holmes, S.; Wu, P.; Richman, J.; Heikinheimo, K.; Chuong, C.-M.; Arnold, K.; Hochedlinger, K.; et al. Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development 2013, 140, 1424–1432. [Google Scholar] [CrossRef]
- Olley, R.C.; Xavier, G.M.; Seppala, M.; Volponi, A.A.; Geoghegan, F.; Sharpe, P.T.; Cobourne, M.T. Expression analysis of candidate genes regulating successional tooth formation in the human embryo. Front. Physiol. 2014, 5, 445. [Google Scholar] [CrossRef]
- Townsend, G.C.; Pinkerton, S.K.; Rogers, J.R.; Bockmann, M.R.; Hughes, T.E. Twin Studies: Research in Genes, Teeth and Faces; University of Adelaide Press: Adelaide, Australia, 2015. [Google Scholar] [CrossRef]
- Hughes, T.; Bockmann, M.; Mihailidis, S.; Bennett, C.; Harris, A.; Seow, W.K.; Lekkas, D.; Ranjitkar, S.; Rupinskas, L.; Pinkerton, S.; et al. Genetic, epigenetic, and environmental influences on dentofacial structures and oral health: Ongoing studies of Australian twins and their families. Twin Res. Hum. Genet. 2013, 16, 43–51. [Google Scholar] [CrossRef]
- Hughes, T.E.; Townsend, G.C.; Pinkerton, S.K.; Bockmann, M.R.; Seow, W.K.; Brook, A.H.; Richards, L.C.; Mihailidis, S.; Ranjitkar, S.; Lekkas, D. The teeth and faces of twins: Providing insights into dentofacial development and oral health for practising oral health professionals. Aust. Dent. J. 2014, 59, S101–S116. [Google Scholar] [CrossRef]
- Scott, G.R.; Irish, J.D. Human Tooth Crown and Root Morphology: The Arizona State Dental Anthropology System; Cambridge University Press: Cambridge, UK, 2017. [Google Scholar] [CrossRef]
- Turner, C.G., II; Nichol, C.R.; Scott, G.R. Scoring procedures for key morphological traits of the permanent dentition: The Arizona State University Dental Anthropology System. In Advances in Dental Anthropology; Kelly, M.A., Larsen, C.S., Eds.; Wiley-Liss: New York, NY, USA, 1991; pp. 13–31. [Google Scholar]
- Nichol, C.R.; Turner, C.G. Intra- and interobserver concordance in classifying dental morphology. Am. J. Phys. Anthropol. 1986, 69, 229–315. [Google Scholar] [CrossRef]
- Hopper, J.L.; Mathews, J.D. Extensions to multivariate normal models for pedigree analysis. Ann. Hum. Genet. 1982, 46, 373–383. [Google Scholar] [CrossRef]
- Lange, K.; Boehnke, M.; Opitz, J.M. Extensions to pedigree analysis. IV. Covariance components models for multivariate traits. Am. J. Med. Genet. 1983, 14, 513–524. [Google Scholar] [CrossRef]
- Almasy, L.; Blangero, J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998, 62, 1198–1211. [Google Scholar] [CrossRef]
- Blangero, J.; Lange, K.; Almasy, L.; Dyer, T.; Göring, H.; Williams, J.; Peterson, C. SOLAR: Sequential Oligogenic Linkage Analysis Routine; Version 8.1.1; Southwest Foundation for Biomedical Research: San Antonio, TX, USA, 1995–2016. [Google Scholar]
- Harris, E.F. Anthropologic and Genetic Aspects of the Dental Morphology of Solomon Islanders, Melanesia. Ph.D. Thesis, Arizona State University, Tempe, AZ, USA, 1977. [Google Scholar]
- Scott, G.R. Dental Morphology: A Genetic Study of American White Families and Variation in Living Southwest Indians. Ph.D. Thesis, Arizona State University, Tempe, AZ, USA, 1973. [Google Scholar]
- Pilloud, M.A.; Edgar, H.J.H.; George, R.; Scott, G.R. Dental morphology in biodistance analysis. In Biological Distance Analysis; Pilloud, M.A., Hefner, J.T., Eds.; Academic Press: London, UK, 2016; pp. 109–133. [Google Scholar] [CrossRef]
- Scott, G.R.; Pilloud, M.A. Dental morphology. In Biological Anthropology of the Human Skeleton, 3rd ed.; Katzenberg, A., Grauer, A.L., Eds.; Wiley-Liss: New York, NY, USA, 2018; pp. 257–292. [Google Scholar] [CrossRef]
- Cheverud, J.M. Genetic and Environmental Morphological Variation among Social Groups of Rhesus Monkeys (Macaca mulatta) on Cayo Santiago. Ph.D. Thesis, University of Wisconsin, Madison, WI, USA, 1979. [Google Scholar]
- Falconer, D.S. Introduction to Quantitative Genetics; Oliver & Boyd: Edinburgh, UK, 1960. [Google Scholar]
- Falconer, D.S. The inheritance of liability to certain diseases from the incidence among relatives. Ann. Hum. Genet. 1965, 29, 51–76. [Google Scholar] [CrossRef]
- Grüneberg, H. The Pathology of Development; Wiley: New York, NY, USA, 1963. [Google Scholar]
- Brook, A.H. A unifying aetiological explanation for anomalies of tooth number and size in humans. Arch. Oral Biol. 1984, 29, 373–378. [Google Scholar] [CrossRef]
- Brook, A.H.; O’Donnell, M.; Hone, A.; Hart, E.; Hughes, T.; Smith, R.N.; Townsend, G.C. General and craniofacial development are complex adaptive processes influenced by diversity. Aust. Dent. J. 2014, 59, 13–22. [Google Scholar] [CrossRef]
- Gómez-Robles, A.; Polly, P.D. Hominin dentition: Evolutionary, developmental, and functional factors. Evolution 2012, 66, 1024–1043. [Google Scholar] [CrossRef]
- Kieser, J.A.; Groeneveld, H.T. Tooth size and arcadal length correlated in man. Int. J. Anthropol. 1987, 2, 37–46. [Google Scholar] [CrossRef]
- Evans, A.R.; Sanson, G.D. The tooth of perfection: Functional and spatial constraints on mammalian tooth shape. Biol. J. Linn. Soc. 2003, 78, 173–191. [Google Scholar] [CrossRef]
- Scott, G.R.; Maier, C.; Heim, K. Identifying and recording key morphological (nonmetric) crown and root traits. In A Companion to Dental Anthropology; Irish, J.D., Scott, G.R., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 245–264. [Google Scholar] [CrossRef]
- Hanihara, K. Crown characters of the deciduous dentition of the Japanese-American hybrids. In Dental Anthropology; Brothwell, D.R., Ed.; Pergamon Press: London, UK, 1963; pp. 105–124. [Google Scholar] [CrossRef]
- Sciulli, P.W. Evolution of the dentition in prehistoric Ohio Valley Native Americans II. Morphology of the deciduous dentition. Am. J. Phys. Anthropol. 1998, 106, 189–205. [Google Scholar] [CrossRef]
- Lesot, H.; Brook, A.H. Epithelial histogenesis during tooth development. Arch. Oral Biol. 2009, 54, S25–S33. [Google Scholar] [CrossRef]
- Balic, A.; Thesleff, I. Chapter Seven—Tissue interactions regulating tooth development and renewal. Curr. Top. Dev. Biol. 2015, 115, 157–186. [Google Scholar] [CrossRef]
- Bei, M. Molecular genetics of tooth development. Curr. Opin. Genet. Dev. 2009, 19, 504–510. [Google Scholar] [CrossRef]
- Brook, A.H. Multilevel complex interactions between genetic, epigenetic and environmental factors in the aetiology of anomalies of dental development. Arch. Oral Biol. 2009, 54, S3–S17. [Google Scholar] [CrossRef]
- Jernvall, J.; Thesleff, I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech. Dev. 2000, 92, 19–29. [Google Scholar] [CrossRef]
- Sharpe, P.T. Neural crest and tooth morphogenesis. Adv. Dent. Res. 2001, 15, 4–7. [Google Scholar] [CrossRef]
- Edgar, H.J.H.; Daly, E.S. Dental Morphology for Anthropology: An Illustrated Manual; Routledge: New York, NY, USA, 2017. [Google Scholar] [CrossRef]
- Bunn, J.M.; Boyer, D.M.; Lipman, Y.; St. Clair, E.; Jernvall, J.; Daubechies, I. Comparing Dirichlet normal surface energy of tooth crowns, a new technique of molar shape quantification for dietary inference, with previous methods in isolation and combination. Am. J. Phys. Anthropol. 2011, 145, 247–261. [Google Scholar] [CrossRef]
- Gaboutchian, A.V.; Knyaz, V.A.; Novikov, M.M.; Vasilyev, S.V.; Leybova, N.A.; Korost, D.V.; Cherebylo, S.A.; Kudaev, A.A. Automated digital odontometry: Measurement data analyses in cases of complicated dental morphology. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, XLIII, B2. [Google Scholar] [CrossRef]
- Kullmer, O.; Benazzi, S.; Fiorenza, L.; Schulz, D.; Bacso, S.; Winzen, O. Technical note: Occlusal fingerprint analysis: Quantification of tooth wear patterns. Am. J. Phys. Anthropol. 2009, 139, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Kullmer, O.; Benazzi, S.; Schulz, D.; Gunz, P.; Kordos, L.; Begun, D.R. Dental arch restoration using tooth macrowear patterns with application to Rudapithecus hungaricus, from the late Miocene of Rudabánya, Hungary. J. Hum. Evol. 2013, 64, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Pampush, J.D.; Winchester, J.M.; Morse, P.E.; Vining, A.Q.; Boyer, D.M.; Kay, R.F. Introducing molar: A new R package for quantitative topographic analysis of teeth (and other topographic surfaces). J. Mamm. Evol. 2016, 23, 397–412. [Google Scholar] [CrossRef]
- Silvester, C.M.; Hillson, S. A critical assessment of the potential for structure-from-motion photogrammetry to produce high fidelity 3D dental models. Am. J. Phys. Anthropol. 2020, 173, 381–392. [Google Scholar] [CrossRef]
- Skinner, M.M.; Evans, A.; Smith, T.; Jernvall, J.; Tafforeau, P.; Kupczik, K.; Olejniczak, A.J.; Rosas, A.; Radovčić, J.; Thackery, J.F.; et al. Brief communication: Contributions of enamel-dentine junction shape and enamel deposition to primate molar crown complexity. Am. J. Phys. Anthropol. 2010, 142, 157–163. [Google Scholar] [CrossRef]
- Winchester, J.M. MorphoTester: An open source application for morphological topographic analysis. PLoS ONE 2016, 11, e0147649. [Google Scholar] [CrossRef] [PubMed]
- Yong, R.; Ranjitkar, S.; Townsend, G.C.; Smith, R.N.; Evans, A.R.; Hughes, T.E.; Lekkas, D.; Brook, A.H. Dental phenomics: Advancing genotype to phenotype correlations in craniofacial research. Aust. Dent. J. 2014, 59, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Jernvall, J. Linking development with generation of novelty in mammalian teeth. Proc. Natl. Acad. Sci. USA 2000, 97, 2641–2645. [Google Scholar] [CrossRef]
- Jernvall, J.; Jung, H.-S. Genotype, phenotype, and developmental biology of molar tooth characters. Yearb. Phys. Anthropol. 2000, 43, 171–190. [Google Scholar] [CrossRef]
- Jernvall, J.; Åberg, T.; Kettunen, P.; Keränan, S.; Thesleff, I. The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouth tooth enamel knot. Development 1998, 125, 161–169. [Google Scholar] [CrossRef]
- Jernvall, J.; Kettunen, P.; Karavanova, I.; Martin, L.B.; Thesleff, I. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: Non-dividing cells express growth stimulating Fgf-4 gene. Int. J. Dev. Biol. 1994, 38, 463–469. [Google Scholar] [CrossRef]
- Keränen, S.; Åberg, T.; Kettunen, P.; Thesleff, I.; Jernvall, J. Association of developmental regulatory genes with the development of different molar tooth shapes in two species of rodents. Dev. Genes Evol. 1998, 208, 477–486. [Google Scholar] [CrossRef]
- Thesleff, I.; Sharpe, P. Signalling networks regulating dental development. Mech. Dev. 1997, 67, 111–123. [Google Scholar] [CrossRef]
- Zhao, Z.; Weiss, K.M.; Stock, D.W. Development and evolution of dentition patterns and their genetic basis. In Development, Function, and Evolution of Teeth; Teaford, M., Smith, M.M., Ferguson, M., Eds.; Cambridge University Press: Cambridge, UK, 2000; pp. 152–172. [Google Scholar] [CrossRef]
- Butler, P.M. The ontogeny of molar pattern. Biol. Rev. 1956, 31, 30–69. [Google Scholar] [CrossRef]
- Christensen, G.J.; Kraus, B.S. Initial calcification of the human permanent first molar. J. Dent. Res. 1965, 44, 1338–1342. [Google Scholar] [CrossRef]
- Kraus, B.S.; Jordan, R.E. The Human Dentition Before Birth; Lea Febiger: Philadelphia, PA, USA, 1965. [Google Scholar]
- Hunter, J.P.; Guatelli-Steinberg, D.; Weston, T.C.; Durner, R.; Betsinger, T.K. Model of tooth morphogenesis predicts Carabelli cusp expression, size, and symmetry in humans. PLoS ONE 2010, 5, e11844. [Google Scholar] [CrossRef]
- Moormann, S.; Guatelli-Steinberg, D.; Hunter, J. Metamerism, morphogenesis, and the expression of Carabelli and other dental traits in humans. Am. J. Phys. Anthropol. 2013, 150, 400–408. [Google Scholar] [CrossRef]
- Atchley, W.R. Developmental quantitative genetics and the evolution of ontogenies. Evolution 1987, 41, 316–330. [Google Scholar] [CrossRef]
- Bochdanovitz, Z.; de Jong, G. Antagonistic pleiotropy for life-history traits at the gene expression level. Proc. R. Soc. Lond. B. 2004, 271, S75–S78. [Google Scholar] [CrossRef]
- Norry, F.M.; Vilardi, J.C.; Hasson, E. Negative genetic correlation between traits of the Drosphila head, and interspecific divergence in head shape. Heredity 2000, 85, 177–183. [Google Scholar] [CrossRef]
- Riska, B. Some models for development, growth, and morphometric correlation. Evolution 1986, 40, 1301–1311. [Google Scholar] [CrossRef]
- Jernvall, J. Mammalian molar cusp patterns: Developmental mechanisms of diversity. Acta Zool. Fenn. 1995, 198, 1–61. [Google Scholar]
- Weiss, K.M. Duplication with variation: Metameric logic in evolution from genes to morphology. Yearb. Phys. Anthropol. 1990, 33, 1–23. [Google Scholar] [CrossRef]
- Dahlberg, A.A. The evolutionary significance of the protostylid. Am. J. Phys. Anthropol. 1950, 8, 15–25. [Google Scholar] [CrossRef]
- Bateson, W. Materials for the Study of Variation: Treated with Special Regard to Discontinuity in the Origin of Species; Macmillan and Company: London, UK, 1894. [Google Scholar] [CrossRef][Green Version]
- Butler, P.M. Comparison of the development of the second deciduous molar and first permanent molar in man. Arch. Oral Biol. 1967, 12, 1245–1260. [Google Scholar] [CrossRef]
- Edgar, H.J.H.; Lease, L.R. Correlations between deciduous and permanent tooth morphology in a European American sample. Am. J. Phys. Anthropol. 2007, 133, 726–734. [Google Scholar] [CrossRef]
- Farmer, V.; Townsend, G. Crown size variability in the deciduous dentition of South Australian children. Am. J. Hum. Biol 1993, 5, 681–690. [Google Scholar] [CrossRef]
- Liversidge, H.M.; Molleson, T.I. Deciduous tooth size and morphogenetic field in the children from Christ Church, Spitalfields. Arch. Oral Biol. 1999, 44, 7–13. [Google Scholar] [CrossRef]
- Bockmann, M.R.; Hughes, T.E.; Townsend, G.C. Genetic modeling of primary tooth emergence: A study of Australian twins. Twin Res. Hum. Genet. 2010, 13, 573–581. [Google Scholar] [CrossRef]
- Edgar, H.J.H.; Ousley, S.D. Dominance in dental morphological traits: Implications for biological distance studies. In Biological Distance Analysis; Pilloud, M.A., Hefner, J.T., Eds.; Academic Press: London, UK, 2016; pp. 317–332. [Google Scholar] [CrossRef]
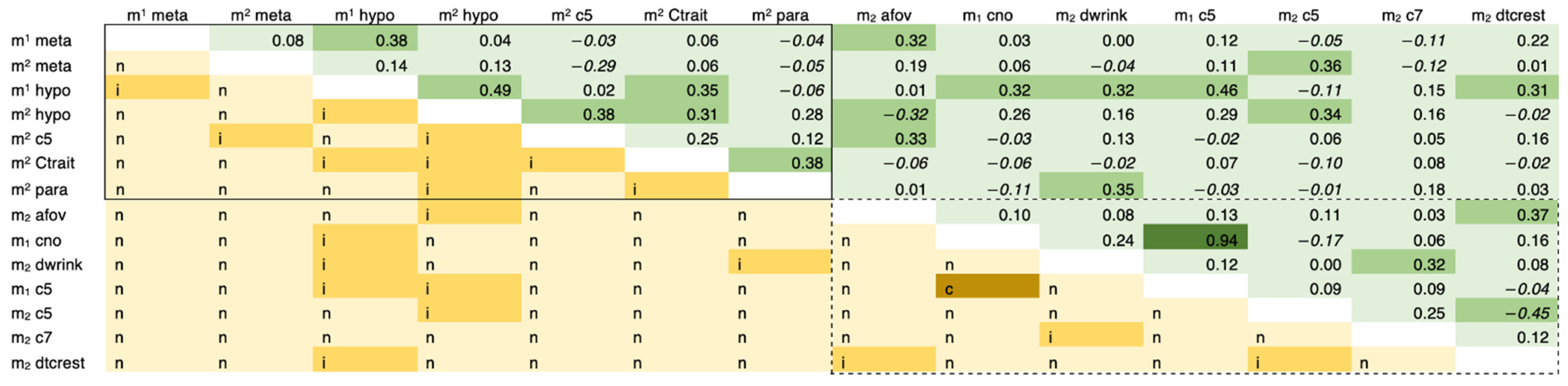
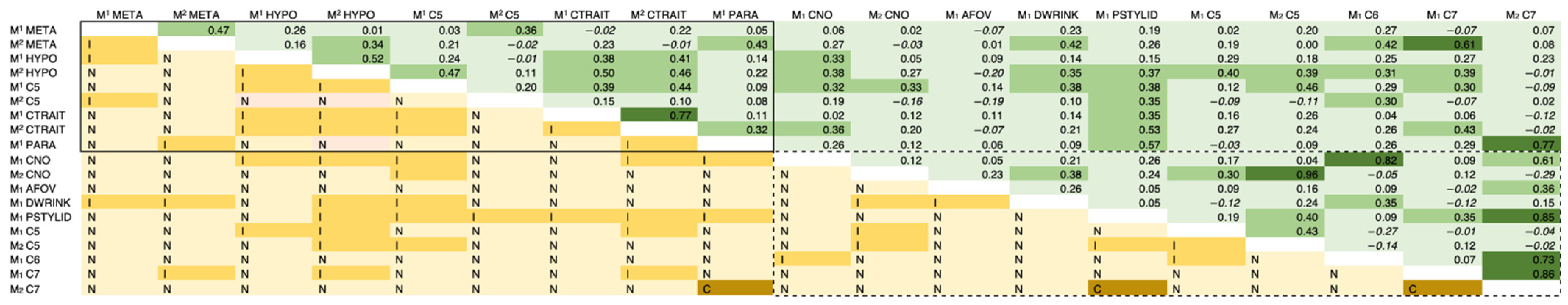
| Morphological Trait 1 | Abbreviation | Dental Elements Scored |
|---|---|---|
| Metacone | META | m1, m2, M1, M2, M3 |
| Hypocone | HYPO | m1, m2, M1, M2, M3 |
| Cusp 5 (Metaconule) 2 | C5 | m1, m2, M1, M2, M3 |
| Carabelli’s Trait 2 | CTRAIT | m1, m2, M1, M2, M3 |
| Parastyle 2 | PARA | m1, m2, M1, M2, M3 |
| Anterior Fovea 2 | AFOV | m1, m2, M1 |
| Deflecting Wrinkle | DWRINK | m2, M1 |
| Cusp 5 (Hypoconulid) | C5 | m1, m2, M1, M2, M3 |
| Cusp 6 | C6 | m1, m2, M1, M2, M3 |
| Cusp 7 2 | C7 | m1, m2, M1, M2, M3 |
| Cusp Number | CNO | m1, m2, M1, M2, M3 |
| Groove Pattern | GROOVE | m2, M1, M2, M3 |
| Protostylid 2 | PSTYLID | m1, m2, M1, M2, M3 |
| Distal Trigonid Crest | DTCREST | m2, M1, M2, M3 |
| Genetic | Environmental | Phenotypic | |||||
|---|---|---|---|---|---|---|---|
| Trait 1 | N/Cov 2 | ρG 3 | P(ρG = 0) 4 | P(|ρG| = 1) 4 | ρE 5 | P (ρE = 0) 4 | ρP 6 |
| DECIDUOUS (l−r) | |||||||
| m1 meta e | 249 | 0.978 ± 0.077 ** | <0.001 | 0.387 | −0.144 ± 0.131 | 0.283 | 0.597 |
| m2 meta | 278/a | 0.852 ± 0.094 ** | <0.001 | 0.057 | −0.144 ± 0.111 | 0.208 | 0.434 |
| m1 hypo w | 251 | 1.000 ± − ** | <0.001 | − | 0.114 ± 0.106 | 0.282 | 0.642 |
| m2 hypo | 279 | 0.992 ± 0.056 ** | <0.001 | 0.445 | 0.005 ± 0.112 | 0.967 | 0.648 |
| m2 c5 | 272/s | 0.868 ± 0.087 ** | <0.001 | 0.070 | 0.132 ± 0.140 | 0.345 | 0.625 |
| m2 ctrait w | 276 | 1.000 ± − ** | <0.001 | − | 0.227 ± 0.112 | 0.046 | 0.758 |
| m2 para | 278 | 1.000 ± − ** | <0.001 | − | 0.263 ± 0.117 | 0.021 | 0.638 |
| m2 afov | 268 | 0.753 ± 0.139 ** | <0.001 | 0.053 | 0.182 ± 0.120 | 0.134 | 0.446 |
| m2 dwrink | 261 | 1.000 ± − ** | <0.001 | − | −0.152 ± 0.118 | 0.228 | 0.607 |
| m1 c5 | 239/s | 1.000 ± − ** | <0.001 | − | −0.085 ± 0.125 | 0.501 | 0.578 |
| m2 c5 | 281/a, a*s | 0.875 ± 0.176 ** | 0.001 | 0.249 | 0.173 ± 0.120 | 0.161 | 0.412 |
| m1 c6 w | − | − | − | − | − | − | − |
| m2 c6 cf | − | − | − | − | − | − | − |
| m2 c7 | 283 | 0.938 ± 0.054 ** | <0.001 | 0.120 | 0.107 ± 0.121 | 0.374 | 0.687 |
| m1 cno w | 239/s | 1.000 ± − ** | <0.001 | − | 0.020 ± 0.126 | 0.871 | 0.591 |
| m2 cno cf | − | − | − | − | − | − | − |
| m2 dtcrest w | 253/a | 1.000 ± − ** | <0.001 | − | 0.269 ± 0.185 | 0.179 | 0.492 |
| PERMANENT (L−R) | |||||||
| M1 META E | 327 | 1.000 ± − ** | <0.001 | − | 0.221 ± 0.095 | 0.028 | 0.555 |
| M2 META | 151/S, A*S | 0.990 ± 0.232 ** | 0.002 | 0.483 | 0.544 ± 0.127 | <0.001 | 0.668 |
| M1 HYPO | 319/S | 1.000 ± − ** | <0.001 | − | −0.206 ± 0.108 | 0.073 | 0.620 |
| M2 HYPO | 112 | 1.000 ± − ** | <0.001 | − | −0.523 ± 0.312 | 0.276 | 0.878 |
| M1 C5 | 292/A, A*S | 0.968 ± 0.051 ** | <0.001 | 0.263 | −0.085 ± 0.143 | 0.558 | 0.675 |
| M2 C5 W | 117 | 1.000 ± − ** | <0.001 | − | −0.498 ± 0.220 | 0.095 | 0.596 |
| M1 CTRAIT | 302/A, A*S | 0.965 ± 0.032 ** | <0.001 | 0.119 | 0.438 ± 0.100 | <0.001 | 0.801 |
| M2 CTRAIT | 135/A, A*S | 0.792 ± 0.100 * | <0.001 | 0.023 | 0.056 ± 0.257 | 0.828 | 0.641 |
| M1 PARA E | 314 | 0.886 ± 0.110 ** | <0.001 | 0.148 | −0.429 ± 0.118 | 0.001 | 0.306 |
| M2 PARA W | 154 | 0.093 ± − | 1.000 | 0.500 | 0.440 ± − | <0.001 | 0.439 |
| M1 AFOV E | 294/A, A*S | 1.000 ± − ** | <0.001 | − | −0.098 ± 0.155 | 0.528 | 0.655 |
| M1 DWRINK | 301 | 0.973 ± 0.059 ** | <0.001 | 0.324 | −0.278 ± 0.112 | 0.023 | 0.580 |
| M1 PSTYLID | 293/S | 0.916 ± 0.100 ** | <0.001 | 0.207 | 0.282 ± 0.158 | 0.103 | 0.605 |
| M1 C5 | 280/ALL | 0.935 ± 0.046 * | <0.001 | 0.062 | −0.149 ± 0.131 | 0.275 | 0.696 |
| M2 C5 W | 145/S | 1.000 ± − ** | <0.001 | − | 0.290 ± 0.249 | 0.283 | 0.784 |
| M1 C6 | 281/A*S | 1.000 ± − ** | <0.001 | − | 0.152 ± 0.114 | 0.183 | 0.512 |
| M2 C6 W | 144 | −0.075 ± − | 1.000 | 0.500 | −0.018 ± − | 0.878 | −0.018 |
| M1 C7 | 330/S | 1.000 ± − ** | <0.001 | − | 0.144 ± 0.134 | 0.263 | 0.694 |
| M2 C7 E | 187 | 1.000 ± − ** | 0.047 | − | 0.182 ± 0.152 | 0.269 | 0.368 |
| M1 CNO | 293 | 1.000 ± − ** | <0.001 | − | 0.233 ± 0.111 | 0.039 | 0.588 |
| M2 CNO | 140 | 0.991 ± 0.092 ** | <0.001 | 0.462 | −0.144 ± 0.308 | 0.653 | 0.695 |
| M1 DTCREST W | 300/S | 0.327 ± − * | <0.001 | <0.001 | 1.000 ± − | <0.001 | 0.338 |
| M2 DTCREST W | 182 | 0.900 ± − | 1.000 | 1.000 | 0.920 ± − | 1.000 | 0.920 |
| Genetic | Environmental | Phenotypic | |||||
|---|---|---|---|---|---|---|---|
| Trait 1 | N/Cov 2 | ρG 3 | P (ρG = 0) 4 | P (|ρG| = 1) 4 | ρE 5 | P (ρE = 0) 4 | ρP 6 |
| Meta (m2−M1) E | 355/Y | 0.189 ± 0.158 | 0.230 | <0.001 | 0.099 ± 0.148 | 0.502 | 0.147 |
| Meta (m2−M2) | 344/Y | 0.183 ± 0.186 | 0.315 | <0.001 | −0.239 ± 0.155 | 0.142 | 0.003 |
| Hypo (m2−M1) | 352/N | 0.597 ± 0.089 * | <0.001 | <0.001 | 0.126 ± 0.140 | 0.370 | 0.460 |
| Hypo (m2−M2) | 332/N | 0.445 ± 0.143 * | 0.006 | <0.001 | −0.128 ± 0.441 | 0.776 | 0.346 |
| C5 (m2−M1) | 349/Y | 0.547 ± 0.132 * | <0.001 | <0.001 | −0.323 ± 0.133 | 0.032 | 0.234 |
| C5 (m2−M2) | 326/Y | −0.157 ± 0.175 | 0.372 | <0.001 | 0.483 ± 0.181 | 0.034 | −0.062 |
| CTrait (m2−M1) | 354/Y | 0.635 ± 0.075 * | <0.001 | <0.001 | 0.040 ± 0.131 | 0.760 | 0.491 |
| Ctrait (m2−M2) | 337/N | 0.368 ± 0.147 * | 0.019 | <0.001 | −0.340 ± 0.186 | 0.099 | 0.253 |
| Para (m2−M1) E | 356/Y | 0.239 ± 0.122 | 0.057 | <0.001 | 0.265 ± 0.124 | 0.043 | 0.243 |
| AFov (m2−M1)E | 329/Y | 0.691 ± 0.139 * | <0.001 | 0.032 | −0.077 ± 0.138 | 0.583 | 0.353 |
| DWrink (m2−M1) | 332/N | 0.520 ± 0.078 * | <0.001 | <0.001 | −0.016 ± 0.158 | 0.920 | 0.440 |
| Pstylid (m2−M1) E | 340/Y | 0.659 ± 0.128 * | <0.001 | 0.009 | −0.300 ± 0.132 | 0.038 | 0.300 |
| C5 (m2−M1) | 343/Y | 0.168 ± 0.165 | 0.329 | 0.008 | 0.195 ± 0.138 | 0.170 | 0.146 |
| C5 (m2−M2) | 335/Y | −0.391 ± 0.240 | 0.109 | 0.018 | 0.502 ± 0.202 | 0.050 | 0.042 |
| C7 (m2−M1) | 352/Y | 0.649 ± 0.154 * | <0.001 | 0.027 | −0.234 ± 0.136 | 0.107 | 0.265 |
| C7 (m2−M2) E | 345/Y | 0.455 ± 0.240 | 0.056 | 0.071 | −0.096 ± 0.214 | 0.657 | 0.180 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paul, K.S.; Stojanowski, C.M.; Hughes, T.; Brook, A.H.; Townsend, G.C. Genetic Correlation, Pleiotropy, and Molar Morphology in a Longitudinal Sample of Australian Twins and Families. Genes 2022, 13, 996. https://doi.org/10.3390/genes13060996
Paul KS, Stojanowski CM, Hughes T, Brook AH, Townsend GC. Genetic Correlation, Pleiotropy, and Molar Morphology in a Longitudinal Sample of Australian Twins and Families. Genes. 2022; 13(6):996. https://doi.org/10.3390/genes13060996
Chicago/Turabian StylePaul, Kathleen S., Christopher M. Stojanowski, Toby Hughes, Alan H. Brook, and Grant C. Townsend. 2022. "Genetic Correlation, Pleiotropy, and Molar Morphology in a Longitudinal Sample of Australian Twins and Families" Genes 13, no. 6: 996. https://doi.org/10.3390/genes13060996
APA StylePaul, K. S., Stojanowski, C. M., Hughes, T., Brook, A. H., & Townsend, G. C. (2022). Genetic Correlation, Pleiotropy, and Molar Morphology in a Longitudinal Sample of Australian Twins and Families. Genes, 13(6), 996. https://doi.org/10.3390/genes13060996






