Rapid Determination of RNA Modifications in Consensus Motifs by Nuclease Protection with Ion-Tagged Oligonucleotide Probes and Matrix-Assisted Laser Desorption Ionization Mass Spectrometry
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Ion-Tagged Oligonucleotide Synthesis
2.3. Nuclease Protection and Purification of Target RNA
2.4. Matrix-Assisted Laser Desorption Ionization Mass Spectrometry
2.5. Data Analysis
3. Results and Discussion
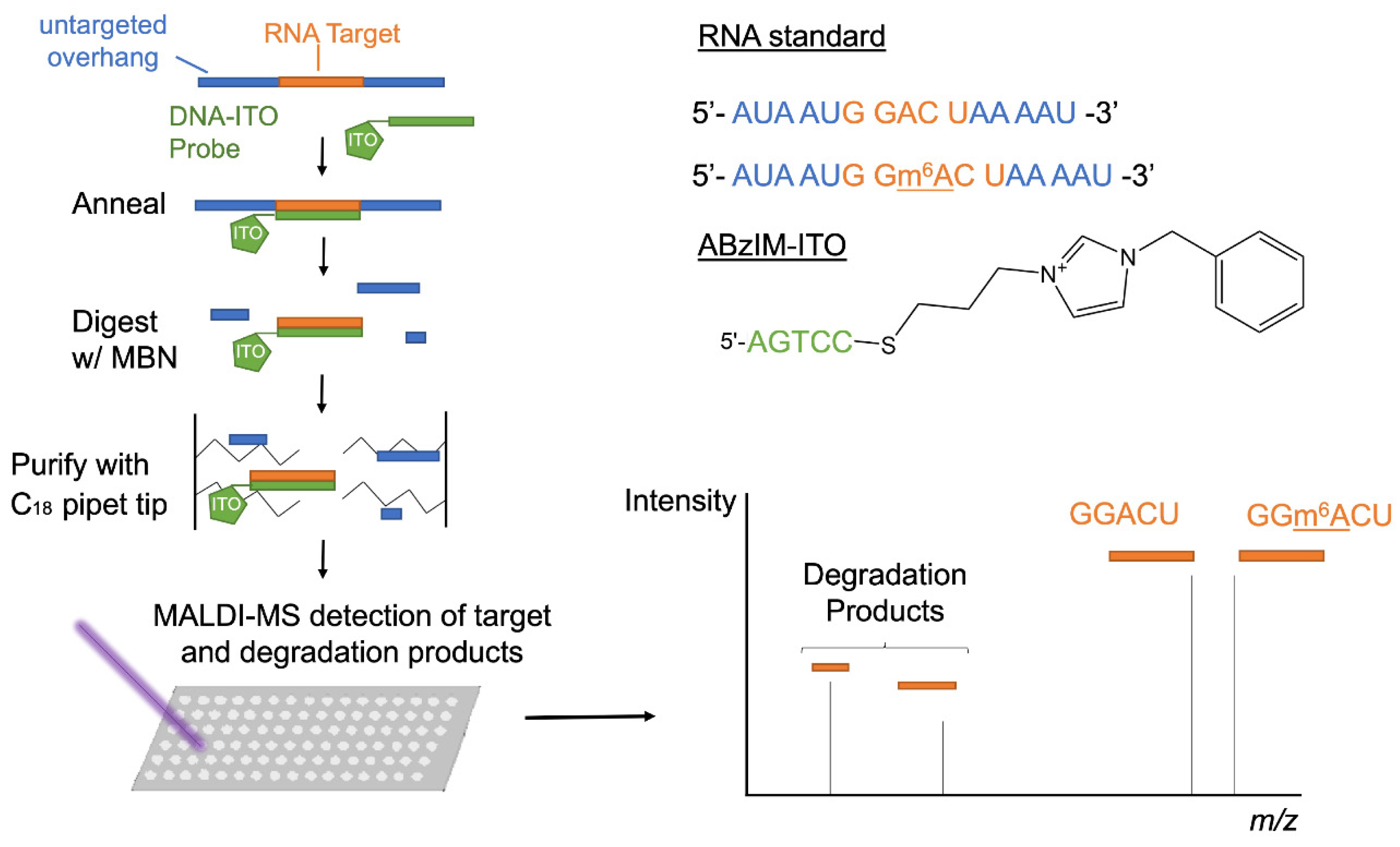
3.1. ITO-Facilitated Nuclease Protection Coupled with MALDI-MS for the Characterization of RNA Consensus Motifs
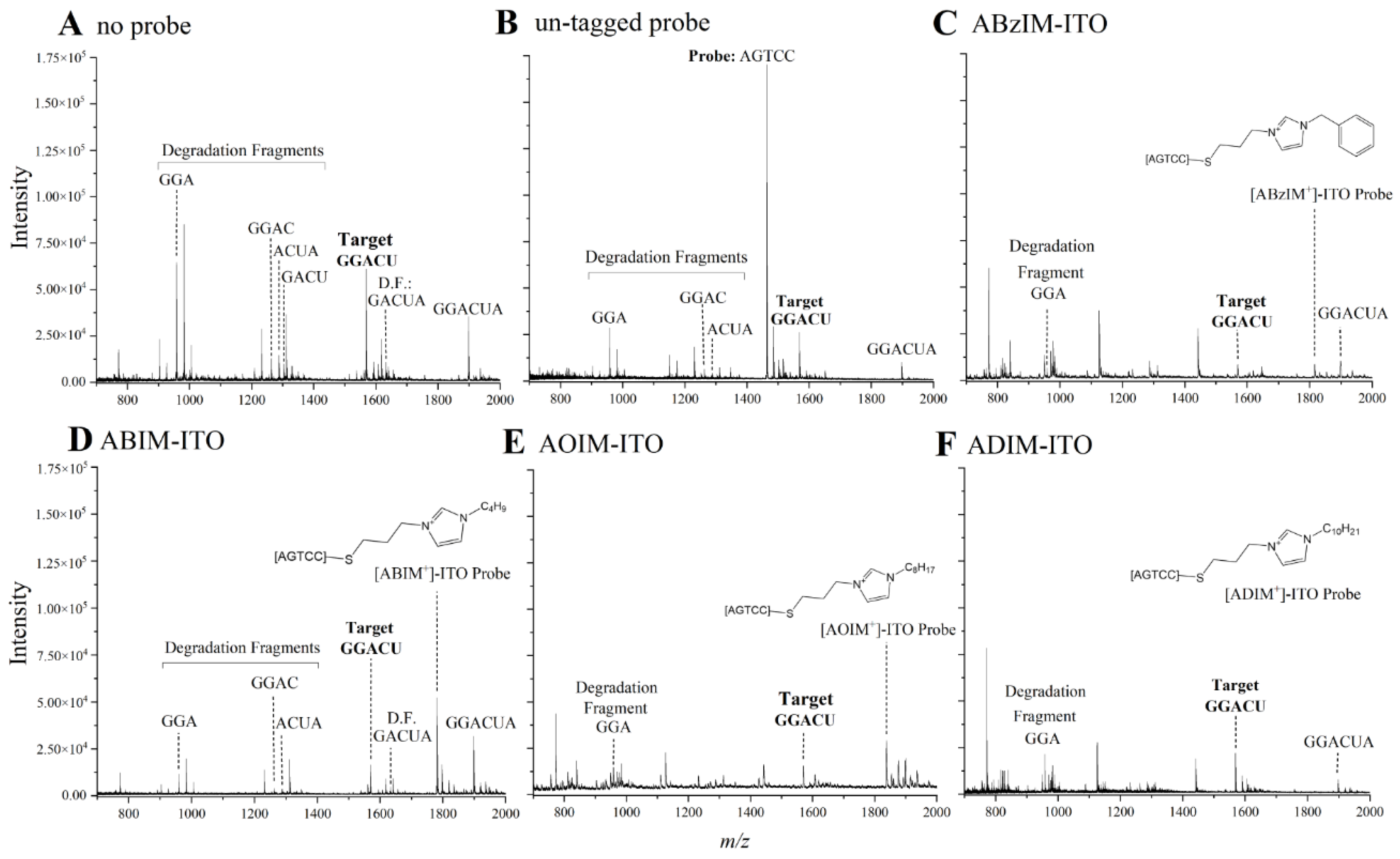
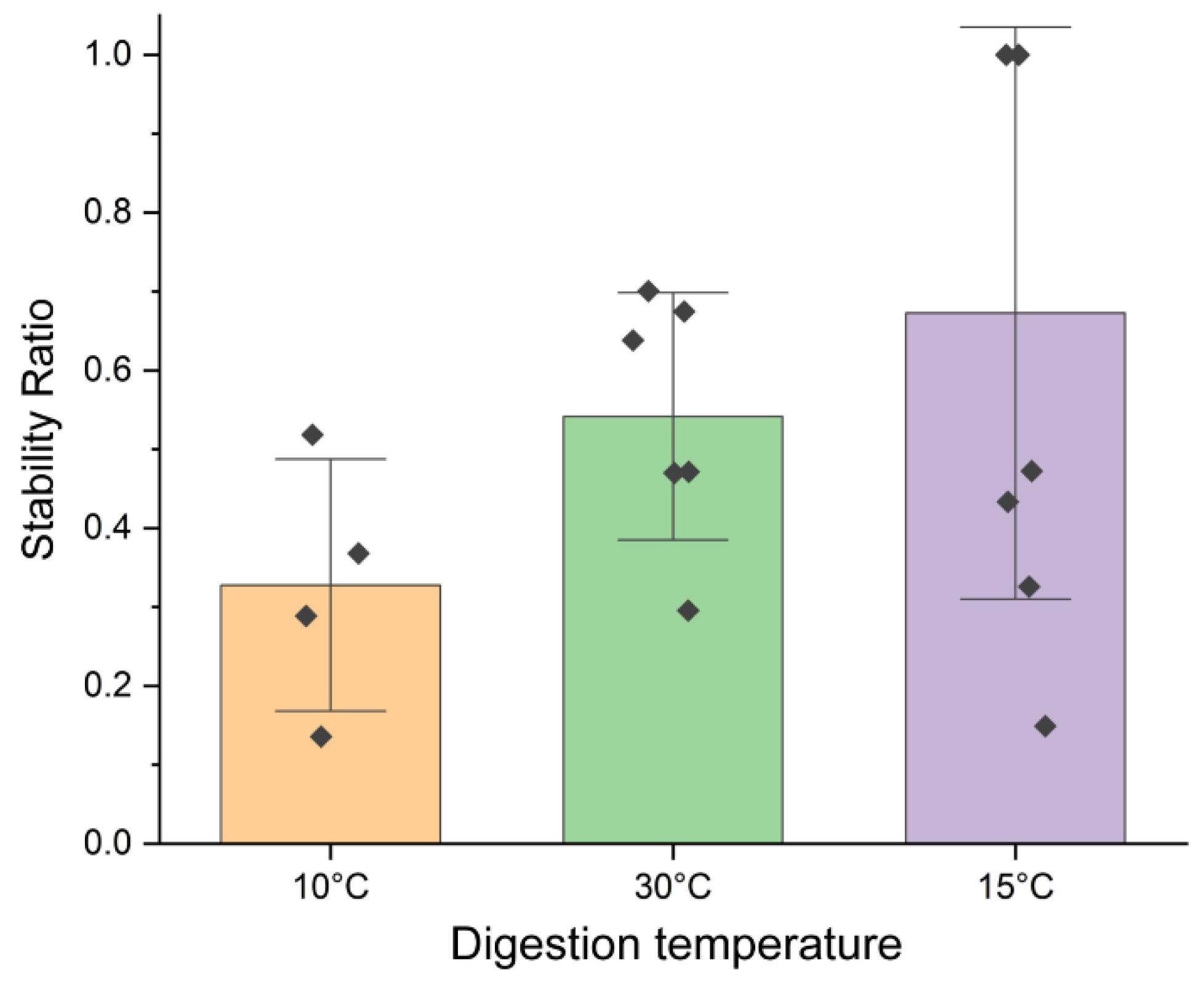
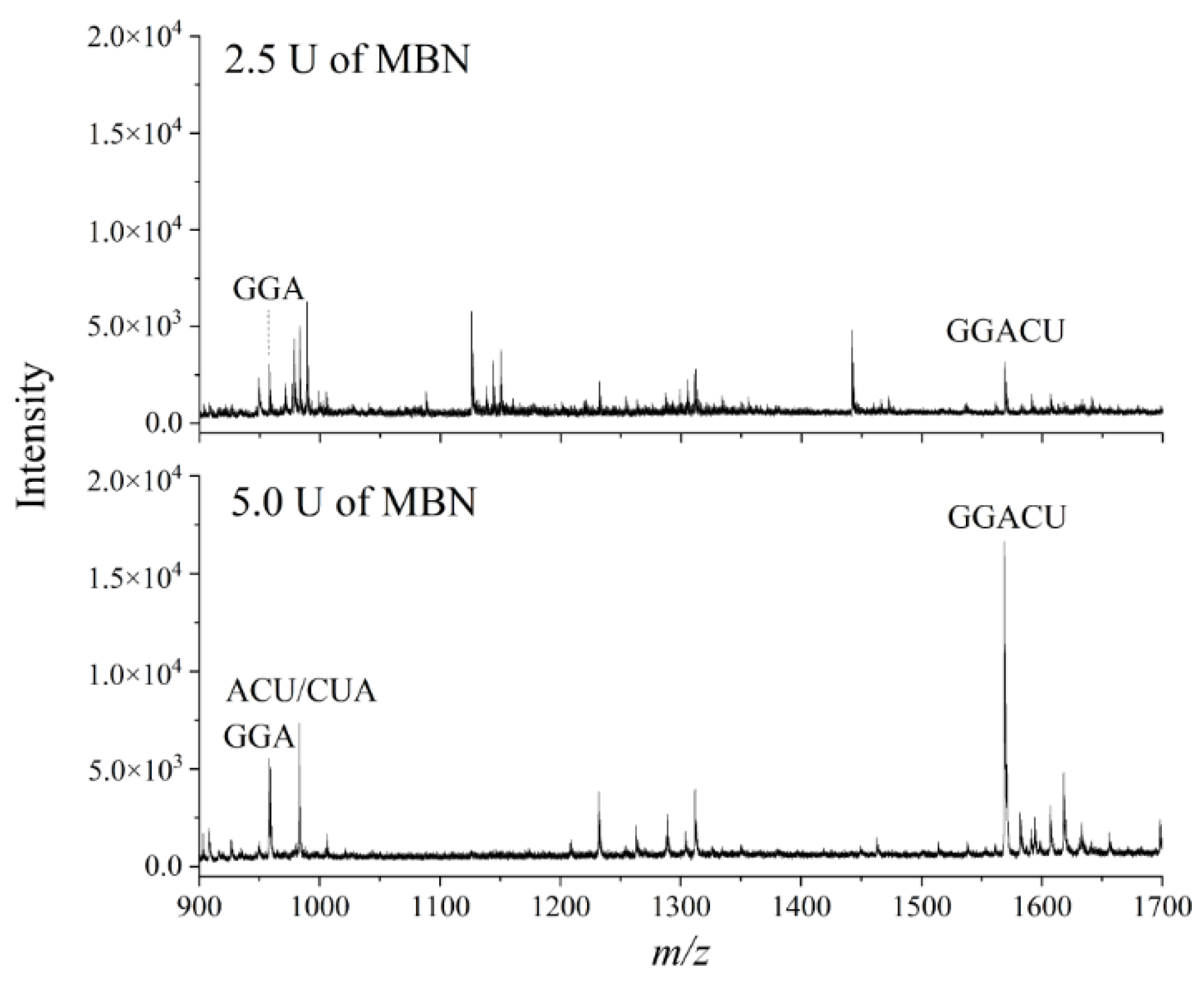
3.2. Optimization of ITO-Based Nuclease Protection Conditions
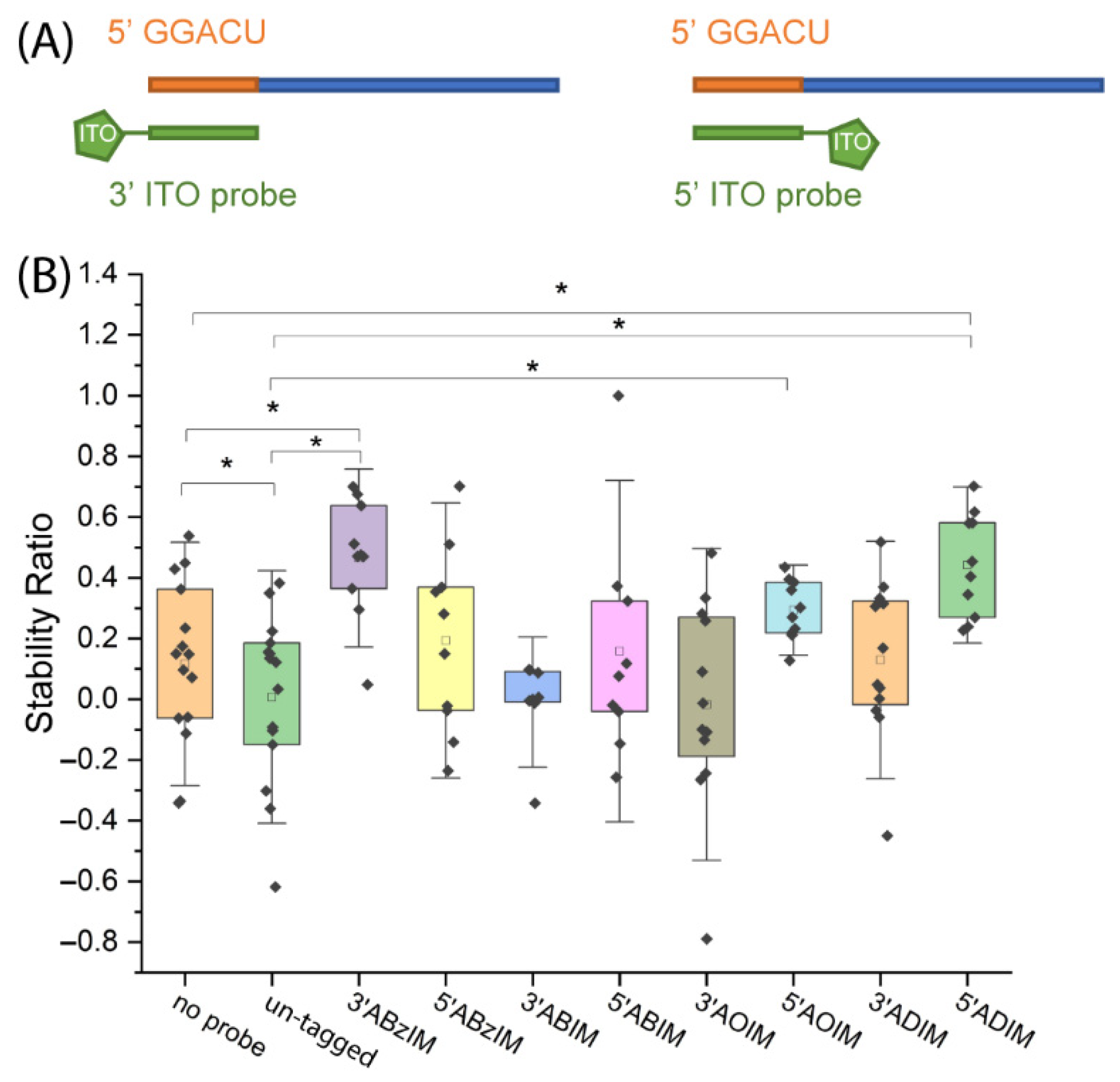
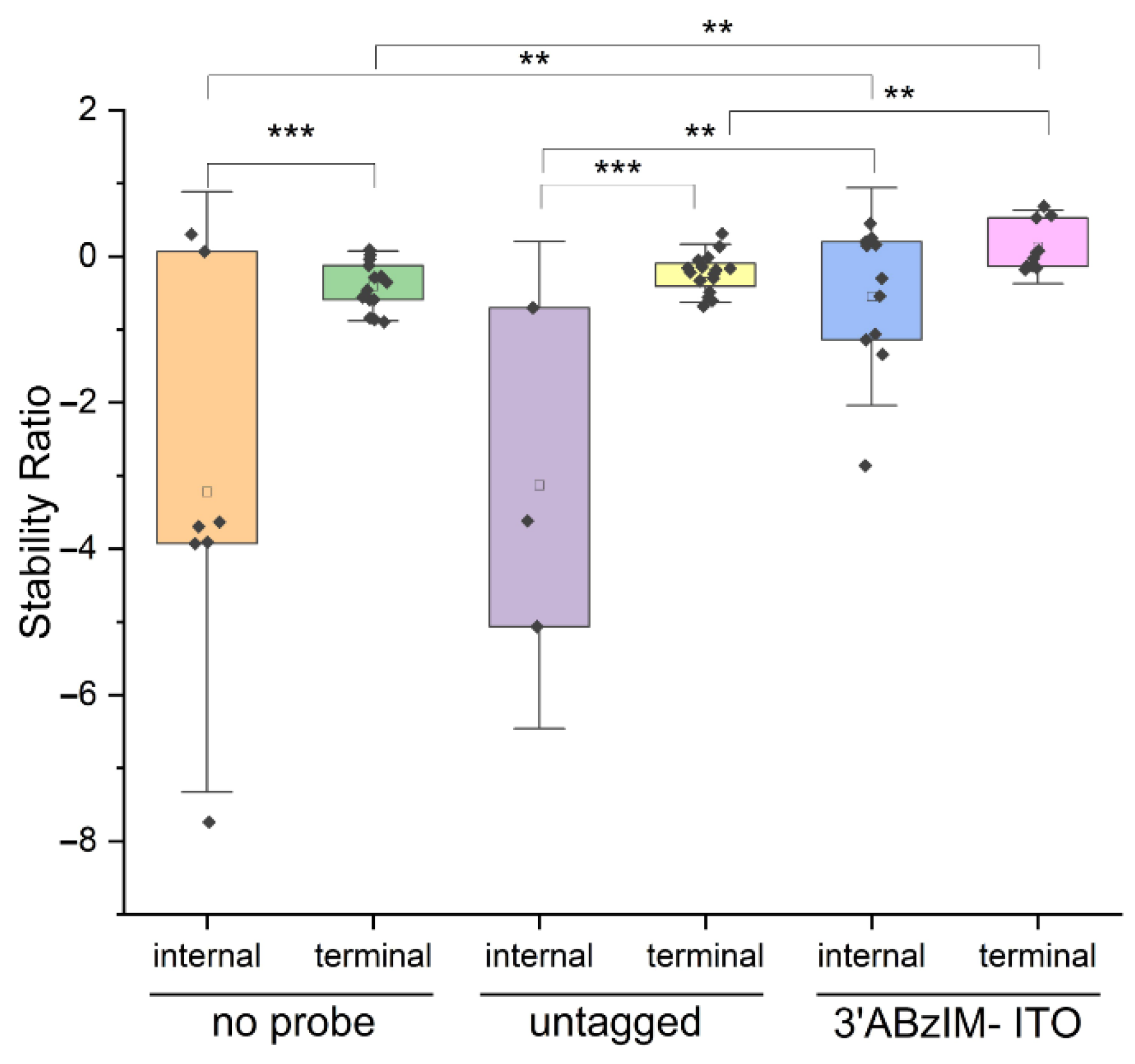
3.3. Comparison of 3′ and 5′ Ion Tag Structures and Sequence Context of Consensus Motifs on Nuclease Protection
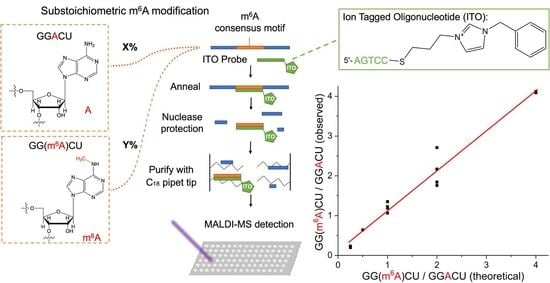
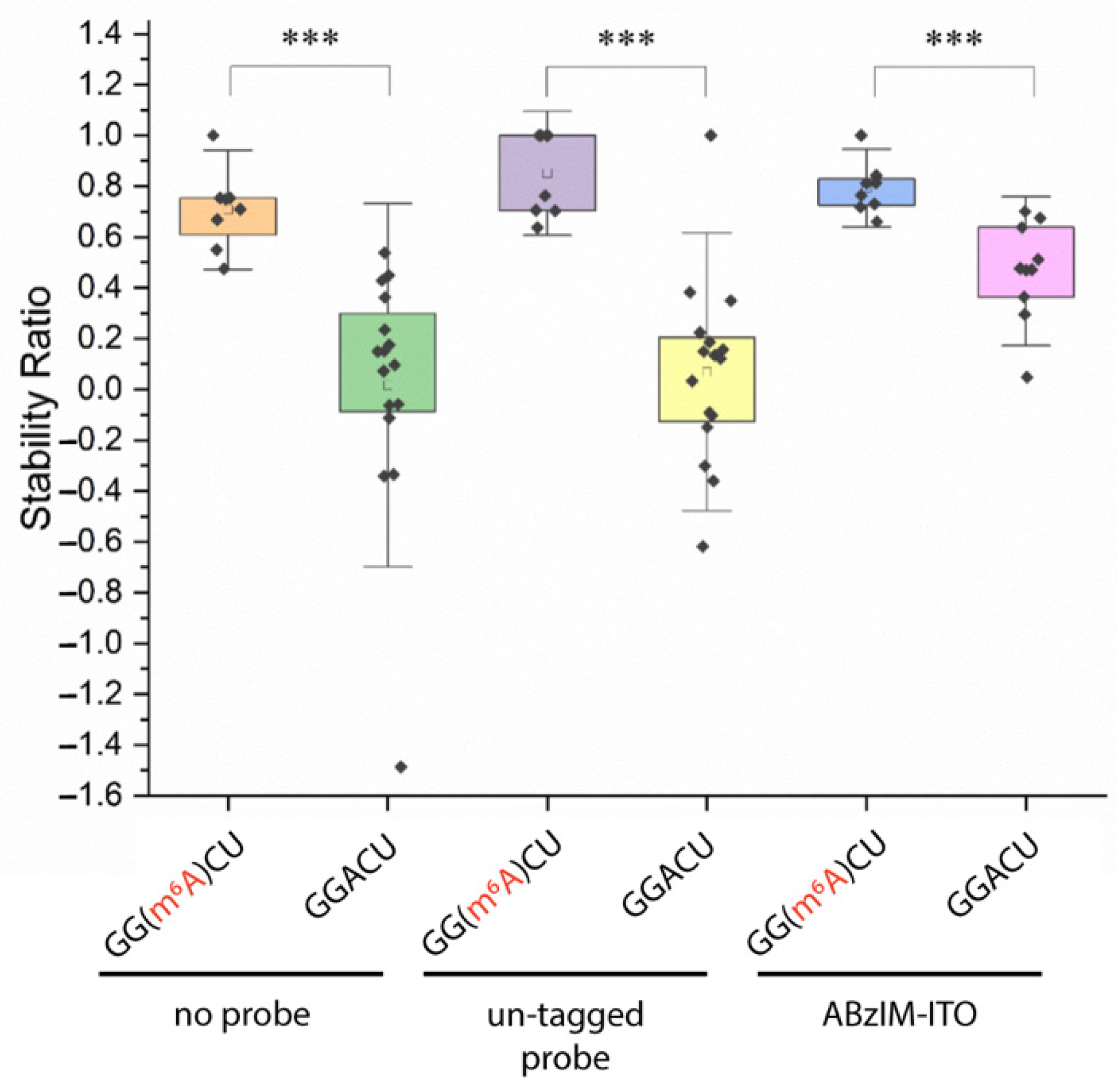
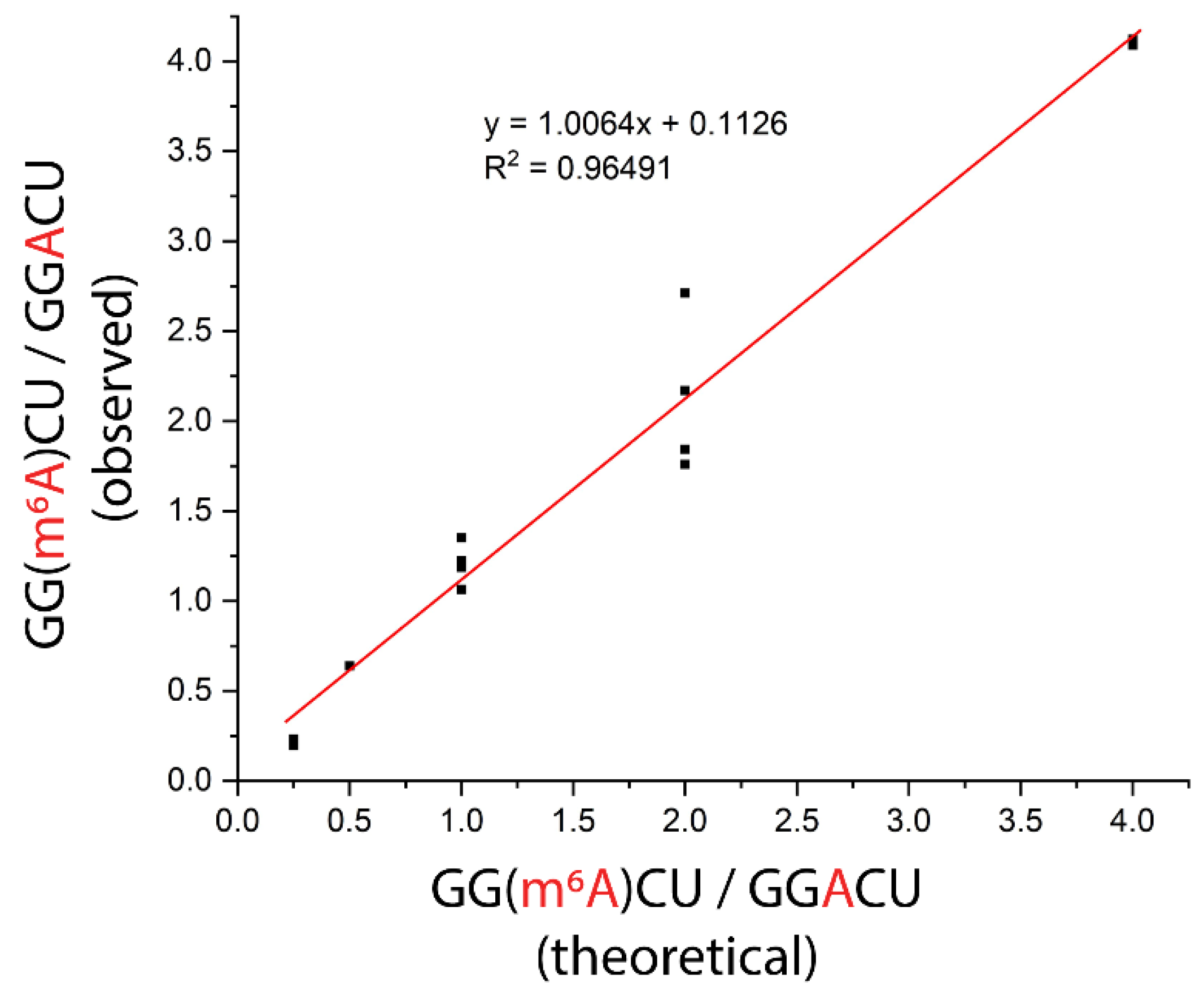
3.4. Evaluation of m6A Stoichiometry in the GGACU Consensus Motif Using ITO-Based Nuclease Protection and MALDI-MS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frye, M.; Harada, B.T.; Behm, M.; He, C. RNA Modifications Modulate Gene Expression during Development. Science 2018, 361, 1346–1349. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic Transcriptomic m 6 A Decoration: Writers, Erasers, Readers and Functions in RNA Metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Lirussi, L.; Demir, Ö.; You, P.; Sarno, A.; Amaro, R.E.; Nilsen, H. RNA Metabolism Guided by RNA Modifications: The Role of SMUG1 in rRNA Quality Control. Biomolecules 2021, 11, 76. [Google Scholar] [CrossRef]
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A Database of RNA Modification Pathways. 2021 Update. Nucleic Acids Res. 2022, 50, D231–D235. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic M6A MRNA Methylation Directs Translational Control of Heat Shock Response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef]
- Chan, C.T.Y.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A Quantitative Systems Approach Reveals Dynamic Control of TRNA Modifications during Cellular Stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef]
- Heiss, M.; Hagelskamp, F.; Marchand, V.; Motorin, Y.; Kellner, S. Cell Culture NAIL-MS Allows Insight into Human TRNA and rRNA Modification Dynamics in Vivo. Nat. Commun. 2021, 12, 389. [Google Scholar] [CrossRef]
- Widagdo, J.; Zhao, Q.-Y.; Kempen, M.-J.; Tan, M.C.; Ratnu, V.S.; Wei, W.; Leighton, L.; Spadaro, P.A.; Edson, J.; Anggono, V.; et al. Experience-Dependent Accumulation of N6-Methyladenosine in the Prefrontal Cortex Is Associated with Memory Processes in Mice. J. Neurosci. 2016, 36, 6771–6777. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, M.; Xie, D.; Huang, Z.; Zhang, L.; Yang, Y.; Ma, D.; Li, W.; Zhou, Q.; Yang, Y.-G.; et al. METTL3-Mediated N 6 -Methyladenosine MRNA Modification Enhances Long-Term Memory Consolidation. Cell Res. 2018, 28, 1050–1061. [Google Scholar] [CrossRef]
- Clark, K.D.; Lee, C.; Gillette, R.; Sweedler, J.V. Characterization of Neuronal RNA Modifications during Non-Associative Learning in Aplysia Reveals Key Roles for TRNAs in Behavioral Sensitization. ACS Cent. Sci. 2021, 7, 1183–1190. [Google Scholar] [CrossRef]
- Clark, K.D.; Philip, M.C.; Tan, Y.; Sweedler, J.V. Biphasic Liquid Microjunction Extraction for Profiling Neuronal RNA Modifications by Liquid Chromatography–Tandem Mass Spectrometry. Anal. Chem. 2020, 92, 12647–12655. [Google Scholar] [CrossRef]
- Clark, K.D.; Rubakhin, S.S.; Sweedler, J.V. Single-Neuron RNA Modification Analysis by Mass Spectrometry: Characterizing RNA Modification Patterns and Dynamics with Single-Cell Resolution. Anal. Chem. 2021, 93, 14537–14544. [Google Scholar] [CrossRef]
- Huang, W.; Qi, C.-B.; Lv, S.-W.; Xie, M.; Feng, Y.-Q.; Huang, W.-H.; Yuan, B.-F. Determination of DNA and RNA Methylation in Circulating Tumor Cells by Mass Spectrometry. Anal. Chem. 2016, 88, 1378–1384. [Google Scholar] [CrossRef]
- Tegowski, M.; Flamand, M.N.; Meyer, K.D. ScDART-Seq Reveals Distinct M6A Signatures and MRNA Methylation Heterogeneity in Single Cells. Mol. Cell 2022, 82, 868–878.e10. [Google Scholar] [CrossRef]
- Purchal, M.K.; Eyler, D.E.; Tardu, M.; Franco, M.K.; Korn, M.M.; Khan, T.; McNassor, R.; Giles, R.; Lev, K.; Sharma, H.; et al. Pseudouridine Synthase 7 Is an Opportunistic Enzyme That Binds and Modifies Substrates with Diverse Sequences and Structures. Proc. Natl. Acad. Sci. USA 2022, 119, e2109708119. [Google Scholar] [CrossRef]
- Delatte, B.; Wang, F.; Ngoc, L.V.; Collignon, E.; Bonvin, E.; Deplus, R.; Calonne, E.; Hassabi, B.; Putmans, P.; Awe, S.; et al. Transcriptome-Wide Distribution and Function of RNA Hydroxymethylcytosine. Science 2016, 351, 282–285. [Google Scholar] [CrossRef]
- Li, X.; Xiong, X.; Zhang, M.; Wang, K.; Chen, Y.; Zhou, J.; Mao, Y.; Lv, J.; Yi, D.; Chen, X.-W.; et al. Base-Resolution Mapping Reveals Distinct M1A Methylome in Nuclear- and Mitochondrial-Encoded Transcripts. Mol. Cell 2017, 68, 993–1005.e9. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3–METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of MRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Hu, L.; Liu, S.; Peng, Y.; Ge, R.; Su, R.; Senevirathne, C.; Harada, B.T.; Dai, Q.; Wei, J.; Zhang, L.; et al. m6A RNA Modifications Are Measured at Single-Base Resolution across the Mammalian Transcriptome. Nat. Biotechnol. 2022, 1–10. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene Expression Regulation Mediated through Reversible m6A RNA Methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Li, Y.; Wu, K.; Quan, W.; Yu, L.; Chen, S.; Cheng, C.; Wu, Q.; Zhao, S.; Zhang, Y.; Zhou, L. The Dynamics of FTO Binding and Demethylation from the M6A Motifs. RNA Biol. 2019, 16, 1179–1189. [Google Scholar] [CrossRef]
- Weng, Y.-L.; Wang, X.; An, R.; Cassin, J.; Vissers, C.; Liu, Y.; Liu, Y.; Xu, T.; Wang, X.; Wong, S.Z.H.; et al. Epitranscriptomic M6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron 2018, 97, 313–325.e6. [Google Scholar] [CrossRef]
- Liu, J.; Eckert, M.A.; Harada, B.T.; Liu, S.-M.; Lu, Z.; Yu, K.; Tienda, S.M.; Chryplewicz, A.; Zhu, A.C.; Yang, Y.; et al. M6A MRNA Methylation Regulates AKT Activity to Promote the Proliferation and Tumorigenicity of Endometrial Cancer. Nat. Cell Biol 2018, 20, 1074–1083. [Google Scholar] [CrossRef]
- Zhao, F.; Xu, Y.; Gao, S.; Qin, L.; Austria, Q.; Siedlak, S.L.; Pajdzik, K.; Dai, Q.; He, C.; Wang, W.; et al. METTL3-Dependent RNA M6A Dysregulation Contributes to Neurodegeneration in Alzheimer’s Disease through Aberrant Cell Cycle Events. Mol. Neurodegener. 2021, 16, 70. [Google Scholar] [CrossRef]
- Hess, M.E.; Hess, S.; Meyer, K.D.; Verhagen, L.A.W.; Koch, L.; Brönneke, H.S.; Dietrich, M.O.; Jordan, S.D.; Saletore, Y.; Elemento, O.; et al. The Fat Mass and Obesity Associated Gene (Fto) Regulates Activity of the Dopaminergic Midbrain Circuitry. Nat. Neurosci. 2013, 16, 1042–1048. [Google Scholar] [CrossRef]
- Krogh, N.; Nielsen, H. Sequencing-Based Methods for Detection and Quantitation of Ribose Methylations in RNA. Methods 2019, 156, 5–15. [Google Scholar] [CrossRef]
- Helm, M.; Motorin, Y. Detecting RNA Modifications in the Epitranscriptome: Predict and Validate. Nat. Rev. Genet. 2017, 18, 275–291. [Google Scholar] [CrossRef]
- Limbach, P.A.; Paulines, M.J. Going Global: The New Era of Mapping Modifications in RNA. WIREs RNA 2017, 8, e1367. [Google Scholar] [CrossRef]
- Hartstock, K.; Ovcharenko, A.; Kueck, N.A.; Spacek, P.; Cornelissen, N.V.; Hüwel, S.; Dieterich, C.; Rentmeister, A. MePMe-Seq: Antibody-Free Simultaneous M6A and M5C Mapping in MRNA by Metabolic Propargyl Labeling and Sequencing. bioRxiv 2022. [Google Scholar] [CrossRef]
- Werner, S.; Galliot, A.; Pichot, F.; Kemmer, T.; Marchand, V.; Sednev, M.V.; Lence, T.; Roignant, J.-Y.; König, J.; Höbartner, C.; et al. NOseq: Amplicon Sequencing Evaluation Method for RNA M6A Sites after Chemical Deamination. Nucleic Acids Res. 2021, 49, e23. [Google Scholar] [CrossRef]
- Lobue, P.A.; Jora, M.; Addepalli, B.; Limbach, P.A. Oligonucleotide Analysis by Hydrophilic Interaction Liquid Chromatography-Mass Spectrometry in the Absence of Ion-Pair Reagents. J. Chromatogr. A 2019, 1595, 39–48. [Google Scholar] [CrossRef]
- Hagelskamp, F.; Borland, K.; Ramos, J.; Hendrick, A.G.; Fu, D.; Kellner, S. Broadly Applicable Oligonucleotide Mass Spectrometry for the Analysis of RNA Writers and Erasers in Vitro. Nucleic Acids Res. 2020, 48, e41. [Google Scholar] [CrossRef]
- Yu, N.; Lobue, P.A.; Cao, X.; Limbach, P.A. RNAModMapper: RNA Modification Mapping Software for Analysis of Liquid Chromatography Tandem Mass Spectrometry Data. Anal. Chem. 2017, 89, 10744–10752. [Google Scholar] [CrossRef]
- Nakayama, H.; Akiyama, M.; Taoka, M.; Yamauchi, Y.; Nobe, Y.; Ishikawa, H.; Takahashi, N.; Isobe, T. Ariadne: A Database Search Engine for Identification and Chemical Analysis of RNA Using Tandem Mass Spectrometry Data. Nucleic Acids Res. 2009, 37, e47. [Google Scholar] [CrossRef]
- Zhang, N.; Shi, S.; Jia, T.Z.; Ziegler, A.; Yoo, B.; Yuan, X.; Li, W.; Zhang, S. A General LC-MS-Based RNA Sequencing Method for Direct Analysis of Multiple-Base Modifications in RNA Mixtures. Nucleic Acids Res. 2019, 47, e125. [Google Scholar] [CrossRef]
- Douthwaite, S.; Kirpekar, F. Identifying Modifications in RNA by MALDI Mass Spectrometry. In Methods in Enzymology; RNA Modification; Academic Press: Cambridge, MA, USA, 2007; Volume 425, pp. 1–20. [Google Scholar] [CrossRef]
- Madsen, C.T.; Mengel-Jørgensen, J.; Kirpekar, F.; Douthwaite, S. Identifying the Methyltransferases for M5U747 and M5U1939 in 23S rRNA Using MALDI Mass Spectrometry. Nucleic Acids Res. 2003, 31, 4738–4746. [Google Scholar] [CrossRef][Green Version]
- Kirpekar, F.; Douthwaite, S.; Roepstorff, P. Mapping Posttranscriptional Modifications in 5S Ribosomal RNA by MALDI Mass Spectrometry. RNA 2000, 6, 296–306. [Google Scholar] [CrossRef]
- Kellersberger, K.A.; Yu, E.T.; Merenbloom, S.I.; Fabris, D. Atmospheric Pressure MALDI-FTMS of Normal and Chemically Modified RNA. J. Am. Soc. Mass Spectrom. 2005, 16, 199–207. [Google Scholar] [CrossRef]
- Berkenkamp, S.; Kirpekar, F.; Hillenkamp, F. Infrared MALDI Mass Spectrometry of Large Nucleic Acids. Science 1998, 281, 260–262. [Google Scholar] [CrossRef]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.-L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of TRNAAsp by the DNA Methyltransferase Homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef]
- Hossain, M.; Limbach, P.A. Mass Spectrometry-Based Detection of Transfer RNAs by Their Signature Endonuclease Digestion Products. RNA 2007, 13, 295–303. [Google Scholar] [CrossRef]
- Andersen, T.E.; Porse, B.T.; Kirpekar, F. A Novel Partial Modification at C2501 in Escherichia Coli 23S Ribosomal RNA. RNA 2004, 10, 907–913. [Google Scholar] [CrossRef][Green Version]
- Thomsen, R.; Nielsen, P.S.; Jensen, T.H. Dramatically Improved RNA in Situ Hybridization Signals Using LNA-Modified Probes. RNA 2005, 11, 1745–1748. [Google Scholar] [CrossRef]
- Clark, K.D.; Varona, M.; Anderson, J.L. Ion-Tagged Oligonucleotides Coupled with a Magnetic Liquid Support for the Sequence-Specific Capture of DNA. Angew. Chem. Int. Ed. 2017, 56, 7630–7633. [Google Scholar] [CrossRef]
- Clark, K.D.; Zhu, C.; Anderson, J.L. Maximizing Ion-Tagged Oligonucleotide Loading on Magnetic Ionic Liquid Supports for the Sequence-Specific Extraction of Nucleic Acids. Anal. Chem. 2019, 91, 5945–5952. [Google Scholar] [CrossRef]
- Emaus, M.N.; Varona, M.; Anderson, J.L. Sequence-Specific Preconcentration of a Mutation Prone KRAS Fragment from Plasma Using Ion-Tagged Oligonucleotides Coupled to qPCR Compatible Magnetic Ionic Liquid Solvents. Anal. Chim. Acta 2019, 1068, 1–10. [Google Scholar] [CrossRef]
- Emaus, M.N.; Zhu, C.; Anderson, J.L. Selective Hybridization and Capture of KRAS DNA from Plasma and Blood Using Ion-Tagged Oligonucleotide Probes Coupled to Magnetic Ionic Liquids. Anal. Chim. Acta 2020, 1094, 1–10. [Google Scholar] [CrossRef]
- Marengo, A.; Emaus, M.N.; Bertea, C.M.; Bicchi, C.; Rubiolo, P.; Cagliero, C.; Anderson, J.L. Arabidopsis Thaliana ITS Sequence-Specific DNA Extraction by Ion-Tagged Oligonucleotides Coupled with a Magnetic Ionic Liquid. Anal. Bioanal. Chem. 2019, 411, 6583–6590. [Google Scholar] [CrossRef]
- Pires de Castro, C.S.; SouzaDe, J.R.; Bloch, C. Mechanism of DNA Cleavage Catalyzed by Mung Bean Nuclease. Inorg. Chim. Acta 2004, 357, 2579–2592. [Google Scholar] [CrossRef]
- Peifer, C.; Sharma, S.; Watzinger, P.; Lamberth, S.; Kötter, P.; Entian, K.-D. Yeast Rrp8p, a Novel Methyltransferase Responsible for M1A 645 Base Modification of 25S rRNA. Nucleic Acids Res. 2013, 41, 1151–1163. [Google Scholar] [CrossRef]
- Moreira, B.G.; You, Y.; Owczarzy, R. Cy3 and Cy5 Dyes Attached to Oligonucleotide Terminus Stabilize DNA Duplexes: Predictive Thermodynamic Model. Biophys. Chem. 2015, 198, 36–44. [Google Scholar] [CrossRef]
- Markham, N.R.; Zuker, M. DINAMelt Web Server for Nucleic Acid Melting Prediction. Nucleic Acids Res. 2005, 33, W577–W581. [Google Scholar] [CrossRef]
- Zhang, L.-S.; Liu, C.; Ma, H.; Dai, Q.; Sun, H.-L.; Luo, G.; Zhang, Z.; Zhang, L.; Hu, L.; Dong, X.; et al. Transcriptome-Wide Mapping of Internal N7-Methylguanosine Methylome in Mammalian MRNA. Mol. Cell 2019, 74, 1304–1316.e8. [Google Scholar] [CrossRef]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.-J.; Chen, Q.; et al. Reversible Methylation of M6Am in the 5′ Cap Controls MRNA Stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef]
- Akichika, S.; Hirano, S.; Shichino, Y.; Suzuki, T.; Nishimasu, H.; Ishitani, R.; Sugita, A.; Hirose, Y.; Iwasaki, S.; Nureki, O.; et al. Cap-Specific Terminal N6-Methylation of RNA by an RNA Polymerase II–Associated Methyltransferase. Science 2019, 363, eaav0080. [Google Scholar] [CrossRef]
- Roost, C.; Lynch, S.R.; Batista, P.J.; Qu, K.; Chang, H.Y.; Kool, E.T. Structure and Thermodynamics of N6-Methyladenosine in RNA: A Spring-Loaded Base Modification. J. Am. Chem. Soc. 2015, 137, 2107–2115. [Google Scholar] [CrossRef]
- Meng, Z.; Limbach, P.A. Quantitation of Ribonucleic Acids Using 18O Labeling and Mass Spectrometry. Anal. Chem. 2005, 77, 1891–1895. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melzer, M.E.; Sweedler, J.V.; Clark, K.D. Rapid Determination of RNA Modifications in Consensus Motifs by Nuclease Protection with Ion-Tagged Oligonucleotide Probes and Matrix-Assisted Laser Desorption Ionization Mass Spectrometry. Genes 2022, 13, 1008. https://doi.org/10.3390/genes13061008
Melzer ME, Sweedler JV, Clark KD. Rapid Determination of RNA Modifications in Consensus Motifs by Nuclease Protection with Ion-Tagged Oligonucleotide Probes and Matrix-Assisted Laser Desorption Ionization Mass Spectrometry. Genes. 2022; 13(6):1008. https://doi.org/10.3390/genes13061008
Chicago/Turabian StyleMelzer, Madeline E., Jonathan V. Sweedler, and Kevin D. Clark. 2022. "Rapid Determination of RNA Modifications in Consensus Motifs by Nuclease Protection with Ion-Tagged Oligonucleotide Probes and Matrix-Assisted Laser Desorption Ionization Mass Spectrometry" Genes 13, no. 6: 1008. https://doi.org/10.3390/genes13061008
APA StyleMelzer, M. E., Sweedler, J. V., & Clark, K. D. (2022). Rapid Determination of RNA Modifications in Consensus Motifs by Nuclease Protection with Ion-Tagged Oligonucleotide Probes and Matrix-Assisted Laser Desorption Ionization Mass Spectrometry. Genes, 13(6), 1008. https://doi.org/10.3390/genes13061008