Involvement of Phospholipase C in Photosynthesis and Growth of Maize Seedlings
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth, Handling and Sampling
2.2. RNA-Seq Analysis and qRT-PCR Validation
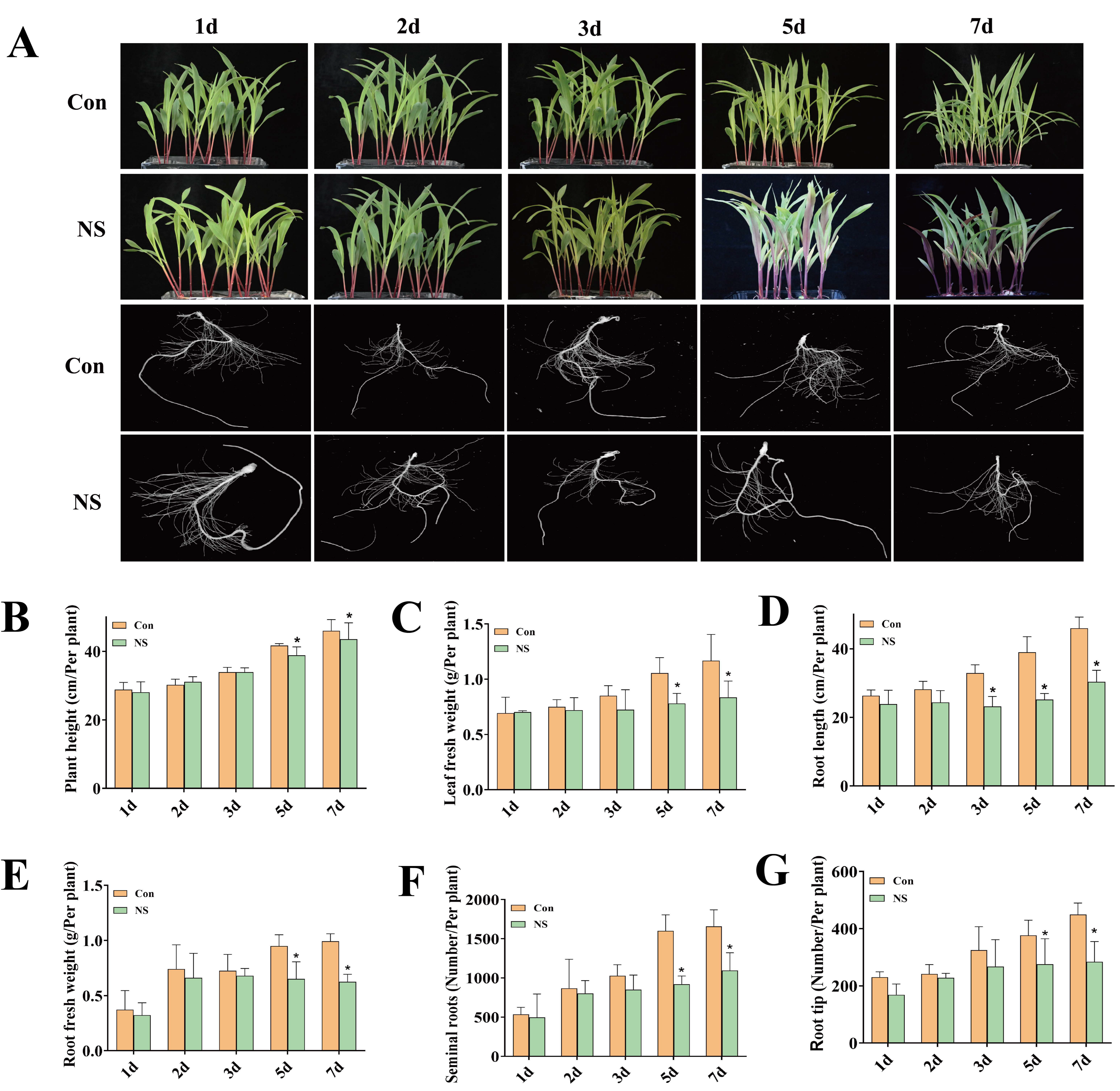
2.3. Phenotypic Analysis of Maize Seedlings under NS Treatment
2.4. Analysis of Photosynthetic Parameters and Chlorophyll Content
2.5. Statistic Analysis
3. Results
3.1. Inhibition of Phospholipase C Activity Affects the Growth and Development of Maize Seedlings
3.2. Transcriptome Analysis of Maize Seedlings under Phospholipase C Inhibition
3.3. Inhibition of Phospholipase C Affects Photosynthesis in Maize Seedlings
3.4. Inhibition of Phospholipase C Affects Photosynthetic Pigment Accumulation in Maize Seedlings
3.5. Inhibition of Phospholipase C Affects Carbon Metabolism in Leaves of Maize Seedlings
3.6. Inhibition of Phospholipase C Activity Affects the Stability of Cell Membranes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Z.; Hubbard, K.G.; Lin, X.; Yang, X. Negative effects of climate warming on maize yield are reversed by the changing of sowing date and cultivar selection in Northeast China. Glob. Change Biol. 2013, 19, 3481–3492. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, B.M. Diversity in global maize germplasm: Characterization and utilization. J. Biosci. 2012, 37, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Devlin, E.B.T.M. Textbook of Biochemistry. Indian Med. Gaz. 1947, 82, 501. [Google Scholar]
- Kadamur, G.; Ross, E.M. Mammalian phospholipase C. Annu. Rev. Physiol. 2013, 75, 127–154. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Bhatnagar, N.; Pandey, A.; Pandey, G.K. Plant phospholipase C family: Regulation and functional role in lipid signaling. Cell Calcium 2015, 58, 139–146. [Google Scholar] [CrossRef]
- Pokotylo, I.; Pejchar, P.; Potocký, M.; Kocourková, D.; Krčková, Z.; Ruelland, E.; Kravets, V.; Martinec, J. The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling. Prog. Lipid Res. 2013, 52, 62–79. [Google Scholar] [CrossRef]
- Pokotylo, I.; Kolesnikov, Y.; Kravets, V.; Zachowski, A.; Ruelland, E. Plant phosphoinositide-dependent phospholipases C: Variations around a canonical theme. Biochimie 2014, 96, 144–157. [Google Scholar] [CrossRef]
- Nakamura, Y.; Awai, K.; Masuda, T.; Yoshioka, Y.; Takamiya, K.; Ohta, H. A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J. Biol. Chem. 2005, 280, 7469–7476. [Google Scholar] [CrossRef] [Green Version]
- Tasma, I.M.; Brendel, V.; Whitham, S.A.; Bhattacharyya, M.K. Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana. Plant Physiol. Biochem. PPB 2008, 46, 627–637. [Google Scholar] [CrossRef]
- Chen, X.; Li, L.; Xu, B.; Zhao, S.; Lu, P.; He, Y.; Ye, T.; Feng, Y.Q.; Wu, Y. Phosphatidylinositol-specific phospholipase C2 functions in auxin-modulated root development. Plant Cell Environ. 2019, 42, 1441–1457. [Google Scholar] [CrossRef]
- Zhang, Q.; van Wijk, R.; Zarza, X.; Shahbaz, M.; van Hooren, M.; Guardia, A.; Scuffi, D.; García-Mata, C.; Van den Ende, W.; Hoffmann-Benning, S.; et al. Knock-Down of Arabidopsis PLC5 Reduces Primary Root Growth and Secondary Root Formation While Overexpression Improves Drought Tolerance and Causes Stunted Root Hair Growth. Plant Cell Physiol. 2018, 59, 2004–2019. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, Z.; Barton, D.; Armour, W.J.; Li, M.Y.; Liao, L.F.; McKellar, H.L.; Pethybridge, K.A.; Marc, J. Inhibition of phospholipase C disrupts cytoskeletal organization and gravitropic growth in Arabidopsis roots. Planta 2010, 232, 1263–1279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; van Wijk, R.; Shahbaz, M.; Roels, W.; Schooten, B.V.; Vermeer, J.E.M.; Zarza, X.; Guardia, A.; Scuffi, D.; García-Mata, C.; et al. Arabidopsis Phospholipase C3 is Involved in Lateral Root Initiation and ABA Responses in Seed Germination and Stomatal Closure. Plant Cell Physiol. 2018, 59, 469–486. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wei, Y.; Zhang, J.; Yang, L.; Liu, X.; Zhang, H.; Shao, W.; He, L.; Li, Z.; Zhang, Y.; et al. Membrane Lipids’ Metabolism and Transcriptional Regulation in Maize Roots under Cold Stress. Front. Plant Sci. 2021, 12, 639132. [Google Scholar] [CrossRef] [PubMed]
- Bogorad, L.J.M.I.E. Porphyrin synthesis. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1962; Volume 5, pp. 885–895. [Google Scholar]
- Das, A.; Guyer, L.; Hörtensteiner, S. Chlorophyll and Chlorophyll Catabolite Analysis by HPLC. Methods Mol. Biol. 2018, 1744, 223–235. [Google Scholar] [CrossRef] [Green Version]
- Silva, S.; Costa, E.M.; Calhau, C.; Morais, R.M.; Pintado, M.E. Anthocyanin extraction from plant tissues: A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 3072–3083. [Google Scholar] [CrossRef]
- Mamode Cassim, A.; Gouguet, P.; Gronnier, J.; Laurent, N.; Germain, V.; Grison, M.; Boutté, Y.; Gerbeau-Pissot, P.; Simon-Plas, F.; Mongrand, S. Plant lipids: Key players of plasma membrane organization and function. Prog. Lipid Res. 2019, 73, 1–27. [Google Scholar] [CrossRef]
- Grison, M.S.; Brocard, L.; Fouillen, L.; Nicolas, W.; Wewer, V.; Dörmann, P.; Nacir, H.; Benitez-Alfonso, Y.; Claverol, S.; Germain, V.; et al. Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell 2015, 27, 1228–1250. [Google Scholar] [CrossRef] [Green Version]
- Markham, J.E.; Jaworski, J.G. Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. RCM 2007, 21, 1304–1314. [Google Scholar] [CrossRef]
- Markham, J.E.; Li, J.; Cahoon, E.B.; Jaworski, J.G. Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 2006, 281, 22684–22694. [Google Scholar] [CrossRef] [Green Version]
- Schrick, K.; Shiva, S.; Arpin, J.C.; Delimont, N.; Isaac, G.; Tamura, P.; Welti, R. Steryl glucoside and acyl steryl glucoside analysis of Arabidopsis seeds by electrospray ionization tandem mass spectrometry. Lipids 2012, 47, 185–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mongrand, S.; Morel, J.; Laroche, J.; Claverol, S.; Carde, J.P.; Hartmann, M.A.; Bonneu, M.; Simon-Plas, F.; Lessire, R.; Bessoule, J.J. Lipid rafts in higher plant cells: Purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 2004, 279, 36277–36286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tejos, R.; Sauer, M.; Vanneste, S.; Palacios-Gomez, M.; Li, H.; Heilmann, M.; van Wijk, R.; Vermeer, J.E.; Heilmann, I.; Munnik, T.; et al. Bipolar Plasma Membrane Distribution of Phosphoinositides and Their Requirement for Auxin-Mediated Cell Polarity and Patterning in Arabidopsis. Plant Cell 2014, 26, 2114–2128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meldrum, E.; Parker, P.J.; Carozzi, A. The PtdIns-PLC superfamily and signal transduction. Biochim. Biophys. Acta 1991, 1092, 49–71. [Google Scholar] [CrossRef]
- Hong, Y.; Zhao, J.; Guo, L.; Kim, S.C.; Deng, X.; Wang, G.; Zhang, G.; Li, M.; Wang, X. Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 2016, 62, 55–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Block, M.A.; Douce, R.; Joyard, J.; Rolland, N. Chloroplast envelope membranes: A dynamic interface between plastids and the cytosol. Photosynth. Res. 2007, 92, 225–244. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Gu, T.; Khan, I.; Zada, A.; Jia, T. Research Progress in the Interconversion, Turnover and Degradation of Chlorophyll. Cells 2021, 10, 3134. [Google Scholar] [CrossRef]
- Rüdiger, W.; Warren, M.J.; Smith, A.G. Regulation of the Late Steps of Chlorophyll Biosynthesis; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Gu, Y.; He, L.; Zhao, C.; Wang, F.; Yan, B.; Gao, Y.; Li, Z.; Yang, K.; Xu, J. Biochemical and Transcriptional Regulation of Membrane Lipid Metabolism in Maize Leaves under Low Temperature. Front. Plant Sci. 2017, 8, 2053. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Shen, Z.; Xu, P.; Zhu, J.; Lu, Q.; Shen, Y.; Wang, Y.; Yao, C.; Li, J.; Wang, Y.; et al. Analysis of photosynthetic pigments and chlorophyll fluorescence characteristics of different strains of Porphyra yezoensis. J. Appl. Phycol. 2012, 24, 881–886. [Google Scholar] [CrossRef]
- Krahmer, J.; Ganpudi, A.; Abbas, A.; Romanowski, A.; Halliday, K.J. Phytochrome, Carbon Sensing, Metabolism, and Plant Growth Plasticity. Plant Physiol. 2018, 176, 1039–1048. [Google Scholar] [CrossRef] [Green Version]
- Sagar, S.; Singh, A. Emerging role of phospholipase C mediated lipid signaling in abiotic stress tolerance and development in plants. Plant Cell Rep. 2021, 40, 2123–2133. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.W.; Lin, Y.T.; Li, H.M. Increased ratio of galactolipid MGDG: DGDG induces jasmonic acid overproduction and changes chloroplast shape. New Phytol. 2020, 228, 1327–1335. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.; Schnarrenberger, C. The evolution of the Calvin cycle from prokaryotic to eukaryotic chromosomes: A case study of functional redundancy in ancient pathways through endosymbiosis. Curr. Genet. 1997, 32, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Massange-Sánchez, J.A.; Casados-Vázquez, L.E.; Juarez-Colunga, S.; Sawers, R.J.H.; Tiessen, A. The Phosphoglycerate Kinase (PGK) Gene Family of Maize (Zea mays var. B73). Plants 2020, 9, 1639. [Google Scholar] [CrossRef] [PubMed]
- Andre, C.; Froehlich, J.E.; Moll, M.R.; Benning, C. A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2006–2022. [Google Scholar] [CrossRef] [Green Version]
- Turner, W.L.; Knowles, V.L.; Plaxton, W.C. Cytosolic pyruvate kinase: Subunit composition, activity, and amount in developing castor and soybean seeds, and biochemical characterization of the purified castor seed enzyme. Planta 2005, 222, 1051–1062. [Google Scholar] [CrossRef]
- Furumoto, T.; Yamaguchi, T.; Ohshima-Ichie, Y.; Nakamura, M.; Tsuchida-Iwata, Y.; Shimamura, M.; Ohnishi, J.; Hata, S.; Gowik, U.; Westhoff, P.; et al. A plastidial sodium-dependent pyruvate transporter. Nature 2011, 476, 472–475. [Google Scholar] [CrossRef] [Green Version]
- Horvath, S.E.; Daum, G. Lipids of mitochondria. Prog. Lipid Res. 2013, 52, 590–614. [Google Scholar] [CrossRef]
- Caiveau, O.; Fortune, D.; Cantrel, C.; Zachowski, A.; Moreau, F. Consequences of omega -6-oleate desaturase deficiency on lipid dynamics and functional properties of mitochondrial membranes of Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 5788–5794. [Google Scholar] [CrossRef] [Green Version]
- Stitt, M.; Fernie, A.R. From measurements of metabolites to metabolomics: An ‘on the fly’ perspective illustrated by recent studies of carbon-nitrogen interactions. Curr. Opin. Biotechnol. 2003, 14, 136–144. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.; Liu, X.; Ge, S.; Zhang, H.; Che, X.; Liu, S.; Liu, D.; Li, H.; Gu, X.; He, L.; et al. Involvement of Phospholipase C in Photosynthesis and Growth of Maize Seedlings. Genes 2022, 13, 1011. https://doi.org/10.3390/genes13061011
Wei Y, Liu X, Ge S, Zhang H, Che X, Liu S, Liu D, Li H, Gu X, He L, et al. Involvement of Phospholipase C in Photosynthesis and Growth of Maize Seedlings. Genes. 2022; 13(6):1011. https://doi.org/10.3390/genes13061011
Chicago/Turabian StyleWei, Yulei, Xinyu Liu, Shengnan Ge, Haiyang Zhang, Xinyang Che, Shiyuan Liu, Debin Liu, Huixin Li, Xinru Gu, Lin He, and et al. 2022. "Involvement of Phospholipase C in Photosynthesis and Growth of Maize Seedlings" Genes 13, no. 6: 1011. https://doi.org/10.3390/genes13061011




