Circulating miRNAs as Potential Biomarkers for Patient Stratification in Bipolar Disorder: A Combined Review and Data Mining Approach
Abstract
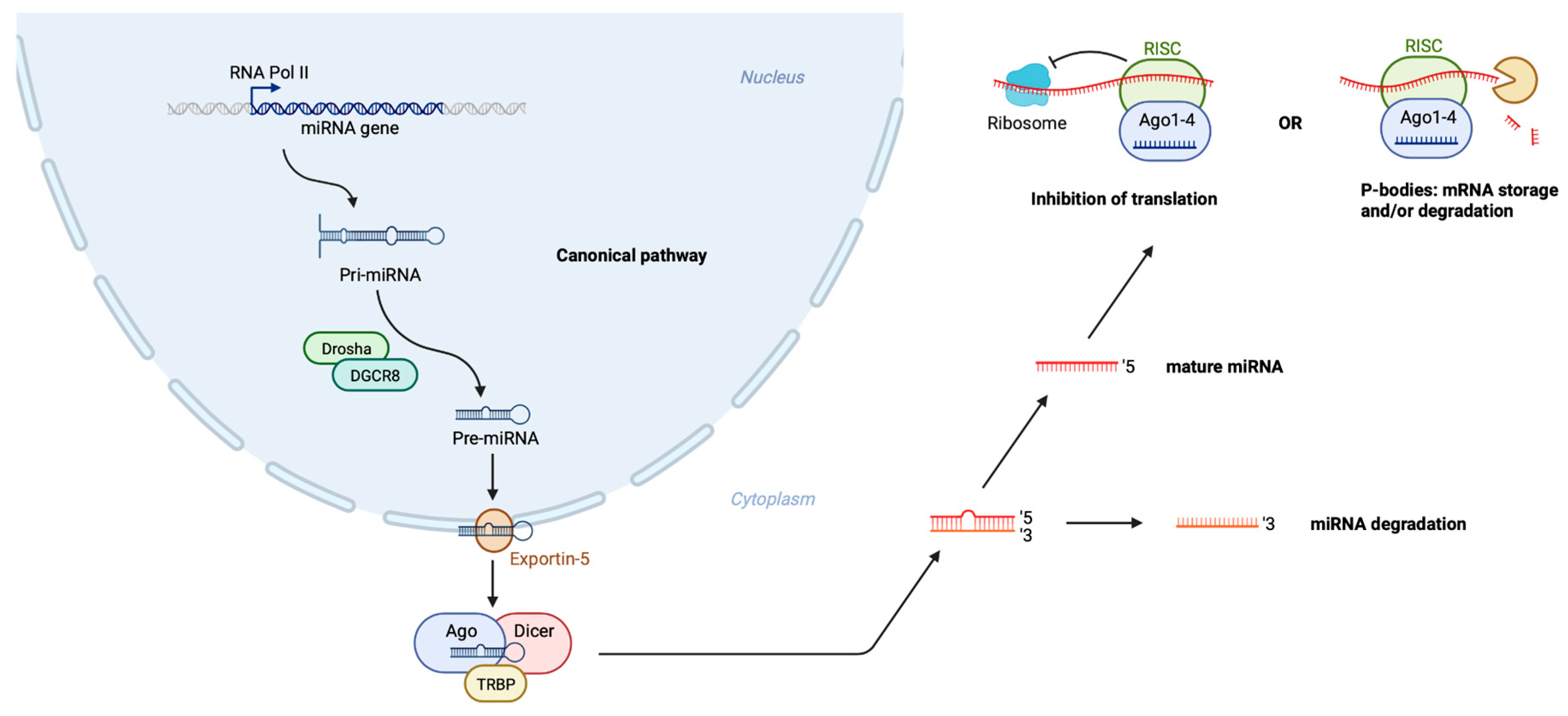
:1. Introduction
2. Materials and Methods
2.1. Literature Review
- (1)
- Search on PubMed was performed with string 1: (‘bipolar disorder’[MeSH]) AND (‘microRNA’[MeSH]). Whereas on EMBASE, string 2: (‘bipolar disorder’[Broad Search]) AND (‘microRNA’[Emtree]) AND (´biological marker´[Broad Search]) was combined with string 3: (‘bipolar disorder’[Broad Search]) AND (‘microRNA’[Emtree]) AND (‘blood’[Broad Search]).
- (2)
- The following search strings were applied and combined using PubMed, string 1: (‘bipolar disorder’[MeSH]) AND (‘drug’[All Fields]) AND (‘efficacy’[All Fields]); string 2: (‘bipolar disorder’[MeSH]) AND (‘micrornas’[MeSH] AND (‘antipsychotic agents’[All Fields]); string 3: (‘bipolar disorder’[MeSH Terms]) AND (‘micrornas’[MeSH Terms]) AND (‘lithium’[All Fields]). Using EMBASE the following strings were applied in combination string 4: (‘bipolar disorder’[Broad Search]) AND (‘microRNA’[Emtree]) AND (‘drug efficacy’[Broad Search]); string 5: (‘bipolar disorder’[Broad Search]) AND (‘microRNA’[Emtree]) AND (‘antipsychotic agents’[Broad Search]).
2.2. Data Review
3. Results
3.1. Literature Reviews
3.1.1. Literature Review of Changes in Peripheral Circulating miRNAs in Bipolar Disorder
BD Type I
BD Type II
3.1.2. Literature Review of Exposome-Associated Changes in Peripheral Circulatory miRNAs in Bipolar Disorder
3.2. Data Mining
3.2.1. Genetic Risk in miRNA-Hosting Genomic Loci in Bipolar Disorder
3.2.2. Expression Characteristics of Putative Bipolar Disorder miRNA Biomarkers in the Human Brain
miR-106b
miR-142
miR-652
miR-125a
miR-221
3.2.3. Predicted Regulomes of Putative miRNA Biomarkers and Their Implication in Mental Health
4. Discussion
4.1. Asenapine Exposure
4.2. Lithium Exposure
4.2.1. miR-142
4.2.2. miR-652
4.2.3. miR-125a
4.2.4. miR-221
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO International Statistical Classification of Diseases and Related Health Problems, 11th ed.; WHO: Geneva, Switzerland, 2019.
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders; American Psychiatric Association: Washington, DC, USA, 2013; ISBN 0-89042-555-8. [Google Scholar]
- Duffy, A.; Vandeleur, C.; Heffer, N.; Preisig, M. The Clinical Trajectory of Emerging Bipolar Disorder among the High-Risk Offspring of Bipolar Parents: Current Understanding and Future Considerations. Int. J. Bipolar Disord. 2017, 5, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coryell, W.; Leon, A.C.; Turvey, C.; Akiskal, H.S.; Mueller, T.; Endicott, J. The Significance of Psychotic Features in Manic Episodes: A Report from the NIMH Collaborative Study. J. Affect. Disord. 2001, 67, 79–88. [Google Scholar] [CrossRef]
- Phillips, M.L.; Kupfer, D.J. Bipolar Disorder Diagnosis: Challenges and Future Directions. Lancet 2013, 381, 1663–1671. [Google Scholar] [CrossRef] [Green Version]
- Hirschfeld, R.M.A.; Lewis, L.; Vornik, L.A. Perceptions and Impact of Bipolar Disorder: How Far Have We Really Come? Results of the National Depressive and Manic-Depressive Association 2000 Survey of Individuals with Bipolar Disorder. J. Clin. Psychiatry 2003, 64, 161–174. [Google Scholar] [PubMed]
- Coleman, J.R.I.; Gaspar, H.A.; Bryois, J.; Byrne, E.M.; Forstner, A.J.; Holmans, P.A.; de Leeuw, C.A.; Mattheisen, M.; McQuillin, A.; Whitehead Pavlides, J.M.; et al. The Genetics of the Mood Disorder Spectrum: Genome-Wide Association Analyses of More Than 185,000 Cases and 439,000 Controls. Biol. Psychiatry 2020, 88, 169–184. [Google Scholar] [CrossRef]
- Stahl, E.A.; Breen, G.; Forstner, A.J.; McQuillin, A.; Ripke, S.; Trubetskoy, V.; Mattheisen, M.; Wang, Y.; Coleman, J.R.I.; Gaspar, H.A.; et al. Genome-Wide Association Study Identifies 30 Loci Associated with Bipolar Disorder. Nat. Genet. 2019, 51, 793–803. [Google Scholar] [CrossRef]
- Murray, R.M.; Bhavsar, V.; Tripoli, G.; Howes, O. 30 Years on: How the Neurodevelopmental Hypothesis of Schizophrenia Morphed into the Developmental Risk Factor Model of Psychosis. Schizophr. Bull. 2017, 43, 1190–1196. [Google Scholar] [CrossRef]
- Schmitt, A.; Malchow, B.; Hasan, A.; Falkai, P. The Impact of Environmental Factors in Severe Psychiatric Disorders. Front. Neurosci. 2014, 8, 19. [Google Scholar] [CrossRef]
- Mullins, N.; Forstner, A.J.; O’Connell, K.S.; Coombes, B.; Coleman, J.R.I.; Qiao, Z.; Als, T.D.; Bigdeli, T.B.; Børte, S.; Bryois, J.; et al. Genome-Wide Association Study of More than 40,000 Bipolar Disorder Cases Provides New Insights into the Underlying Biology. Nat. Genet. 2021, 53, 817–829. [Google Scholar] [CrossRef]
- Uher, R. Gene-Environment Interactions in Severe Mental Illness. Front. Psychiatry 2014, 5, 48. [Google Scholar] [CrossRef] [Green Version]
- Craddock, N.; Sklar, P. Genetics of Bipolar Disorder. Lancet 2013, 381, 1654–1662. [Google Scholar] [CrossRef]
- Kieseppä, T.; Partonen, T.; Haukka, J.; Kaprio, J.; Lönnqvist, J. High Concordance of Bipolar I Disorder in a Nationwide Sample of Twins. Am. J. Psychiatry 2004, 161, 1814–1821. [Google Scholar] [CrossRef]
- Bienvenu, O.J.; Davydow, D.S.; Kendler, K.S. Psychiatric ‘Diseases’ versus Behavioral Disorders and Degree of Genetic Influence. Psychol. Med. 2011, 41, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, C.; Hernandez, M.; Faedda, G.L. The Role of Environmental Exposures as Risk Factors for Bipolar Disorder: A Systematic Review of Longitudinal Studies. J. Affect. Disord. 2016, 193, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Rowland, T.A.; Marwaha, S. Epidemiology and Risk Factors for Bipolar Disorder. Ther. Adv. Psychopharmacol. 2018, 8, 251–269. [Google Scholar] [CrossRef] [PubMed]
- Wendt, F.R.; Pathak, G.A.; Levey, D.F.; Nuñez, Y.Z.; Overstreet, C.; Tyrrell, C.; Adhikari, K.; De Angelis, F.; Tylee, D.S.; Goswami, A.; et al. Sex-Stratified Gene-by-Environment Genome-Wide Interaction Study of Trauma, Posttraumatic-Stress, and Suicidality. Neurobiol. Stress 2021, 14, 100309. [Google Scholar] [CrossRef]
- Dunn, E.C.; Wiste, A.; Radmanesh, F.; Almli, L.M.; Gogarten, S.M.; Sofer, T.; Faul, J.D.; Kardia, S.L.R.; Smith, J.A.; Weir, D.R.; et al. Genome-wide association study (gwas) and genome-wide by environment interaction study (GWEIS) of depressive symptoms in african american and hispanic/latina women. Depress. Anxiety 2016, 33, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Cheng, S.; Ye, J.; Chu, X.; Wen, Y.; Liu, L.; Qi, X.; Jia, Y.; Zhang, F. Evaluating the Genetic Effects of Sex Hormone Traits on the Development of Mental Traits: A Polygenic Score Analysis and Gene-Environment-Wide Interaction Study in UK Biobank Cohort. Mol. Brain 2021, 14, 3. [Google Scholar] [CrossRef]
- Liang, X.; Ye, J.; Wen, Y.; Li, P.; Cheng, B.; Cheng, S.; Liu, L.; Zhang, L.; Ma, M.; Qi, X.; et al. Long-Term Antibiotic Use during Early Life and Risks to Mental Traits: An Observational Study and Gene–Environment-Wide Interaction Study in UK Biobank Cohort. Neuropsychopharmacology 2021, 46, 1086–1092. [Google Scholar] [CrossRef]
- Werme, J.; van der Sluis, S.; Posthuma, D.; de Leeuw, C.A. Genome-Wide Gene-Environment Interactions in Neuroticism: An Exploratory Study across 25 Environments. Transl. Psychiatry 2021, 11, 180. [Google Scholar] [CrossRef]
- Nagel, M.; Speed, D.; Sluis, S.; Østergaard, S.D. Genome-wide Association Study of the Sensitivity to Environmental Stress and Adversity Neuroticism Cluster. Acta Psychiatr. Scand. 2020, 141, 476–478. [Google Scholar] [CrossRef] [PubMed]
- Hosang, G.M.; Uher, R.; Keers, R.; Cohen-Woods, S.; Craig, I.; Korszun, A.; Perry, J.; Tozzi, F.; Muglia, P.; McGuffin, P.; et al. Stressful Life Events and the Brain-Derived Neurotrophic Factor Gene in Bipolar Disorder. J. Affect. Disord. 2010, 125, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Hallmayer, J.; Wang, P.W.; Hill, S.J.; Johnson, S.L.; Ketter, T.A. Brain-Derived Neurotrophic Factor Val66met Genotype and Early Life Stress Effects upon Bipolar Course. J. Psychiatr. Res. 2013, 47, 252–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickerson, F.B.; Boronow, J.J.; Stallings, C.; Origoni, A.E.; Cole, S.; Leister, F.; Krivogorsky, B.; Yolken, R.H. The Catechol O-Methyltransferase Val158Met Polymorphism and Herpes Simplex Virus Type 1 Infection Are Risk Factors for Cognitive Impairment in Bipolar Disorder: Additive Gene-Environmental Effects in a Complex Human Psychiatric Disorder. Bipolar Disord. 2006, 8, 124–132. [Google Scholar] [CrossRef]
- Savitz, J.; van der Merwe, L.; Newman, T.K.; Stein, D.J.; Ramesar, R. Catechol-o-Methyltransferase Genotype and Childhood Trauma May Interact to Impact Schizotypal Personality Traits. Behav. Genet. 2010, 40, 415–423. [Google Scholar] [CrossRef]
- Bortolasci, C.C.; Vargas, H.O.; Souza-Nogueira, A.; Barbosa, D.S.; Moreira, E.G.; Nunes, S.O.V.; Berk, M.; Dodd, S.; Maes, M. Lowered Plasma Paraoxonase (PON)1 Activity Is a Trait Marker of Major Depression and PON1 Q192R Gene Polymorphism–Smoking Interactions Differentially Predict the Odds of Major Depression and Bipolar Disorder. J. Affect. Disord. 2014, 159, 23–30. [Google Scholar] [CrossRef]
- Debnath, M.; Busson, M.; Jamain, S.; Etain, B.; Hamdani, N.; Oliveira, J.; Boukouaci, W.; Amokrane, K.; Moins-Teisserenc, H.; Lajnef, M.; et al. The HLA-G Low Expressor Genotype Is Associated with Protection against Bipolar Disorder. Hum. Immunol. 2013, 74, 593–597. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, J.; Etain, B.; Lajnef, M.; Hamdani, N.; Bennabi, M.; Bengoufa, D.; Sundaresh, A.; Chaabane, A.B.; Bellivier, F.; Henry, C.; et al. Combined Effect of TLR2 Gene Polymorphism and Early Life Stress on the Age at Onset of Bipolar Disorders. PLoS ONE 2015, 10, e0119702. [Google Scholar] [CrossRef]
- De Pradier, M.; Gorwood, P.; Beaufils, B.; Adès, J.; Dubertret, C. Influence of the Serotonin Transporter Gene Polymorphism, Cannabis and Childhood Sexual Abuse on Phenotype of Bipolar Disorder: A Preliminary Study. Eur. Psychiatry 2010, 25, 323–327. [Google Scholar] [CrossRef]
- Misiak, B.; Stramecki, F.; Gawęda, Ł.; Prochwicz, K.; Sąsiadek, M.M.; Moustafa, A.A.; Frydecka, D. Interactions Between Variation in Candidate Genes and Environmental Factors in the Etiology of Schizophrenia and Bipolar Disorder: A Systematic Review. Mol. Neurobiol. 2018, 55, 5075–5100. [Google Scholar] [CrossRef]
- Musci, R.J.; Augustinavicius, J.L.; Volk, H. Gene-Environment Interactions in Psychiatry: Recent Evidence and Clinical Implications. Curr. Psychiatry Rep. 2019, 21, 81. [Google Scholar] [CrossRef] [PubMed]
- Cuéllar-Barboza, A.B.; McElroy, S.L.; Veldic, M.; Singh, B.; Kung, S.; Romo-Nava, F.; Nunez, N.A.; Cabello-Arreola, A.; Coombes, B.J.; Prieto, M.; et al. Potential Pharmacogenomic Targets in Bipolar Disorder: Considerations for Current Testing and the Development of Decision Support Tools to Individualize Treatment Selection. Int. J. Bipolar Disord. 2020, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.H.P.H.; Anttila, V.; Won, H.; Feng, Y.-C.A.Y.-C.A.; Rosenthal, J.; Zhu, Z.; Tucker-Drob, E.M.E.M.; Nivard, M.G.M.G.; Grotzinger, A.D.; Posthuma, D.; et al. Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 2019, 179, 1469–1482.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; Ripke, S.; Neale, B.M.; Faraone, S.V.; Purcell, S.M.; Perlis, R.H.; Mowry, B.J.; Thapar, A.; Goddard, M.E.; Witte, J.S.; et al. Genetic Relationship between Five Psychiatric Disorders Estimated from Genome-Wide SNPs. Nat. Genet. 2013, 45, 984–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smeland, O.B.; Bahrami, S.; Frei, O.; Shadrin, A.; O’Connell, K.; Savage, J.; Watanabe, K.; Krull, F.; Bettella, F.; Steen, N.E.; et al. Genome-Wide Analysis Reveals Extensive Genetic Overlap between Schizophrenia, Bipolar Disorder, and Intelligence. Mol. Psychiatry 2020, 25, 844–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wray, G.A.; Hahn, M.W.; Abouheif, E.; Balhoff, J.P.; Pizer, M.; Rockman, M.V.; Romano, L.A. The Evolution of Transcriptional Regulation in Eukaryotes. Mol. Biol. Evol. 2003, 20, 1377–1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef] [Green Version]
- Strazisar, M.; Cammaerts, S.; van der Ven, K.; Forero, D.A.; Lenaerts, A.-S.; Nordin, A.; Almeida-Souza, L.; Genovese, G.; Timmerman, V.; Liekens, A.; et al. MIR137 Variants Identified in Psychiatric Patients Affect Synaptogenesis and Neuronal Transmission Gene Sets. Mol. Psychiatry 2015, 20, 472–481. [Google Scholar] [CrossRef]
- Duan, J.; Shi, J.; Fiorentino, A.; Leites, C.; Chen, X.; Moy, W.; Chen, J.; Alexandrov, B.S.; Usheva, A.; He, D.; et al. A Rare Functional Noncoding Variant at the GWAS-Implicated MIR137/MIR2682 Locus Might Confer Risk to Schizophrenia and Bipolar Disorder. Am. J. Hum. Genet. 2014, 95, 744–753. [Google Scholar] [CrossRef] [Green Version]
- Davis, C.J.; Bohnet, S.G.; Meyerson, J.M.; Krueger, J.M. Sleep Loss Changes MicroRNA Levels in the Brain: A Possible Mechanism for State-Dependent Translational Regulation. Neurosci. Lett. 2007, 422, 68–73. [Google Scholar] [CrossRef] [Green Version]
- Maccani, M.A.; Knopik, V.S. Cigarette Smoke Exposure-Associated Alterations to Non-Coding RNA. Front. Genet. 2012, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Flemington, E.K. MiRNAs in the Pathogenesis of Oncogenic Human Viruses. Cancer Lett. 2011, 305, 186–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocerha, J.; Dwivedi, Y.; Brennand, K.J. Noncoding RNAs and Neurobehavioral Mechanisms in Psychiatric Disease. Mol. Psychiatry 2015, 20, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Dunbar, M.; Shelton, R.C.; Dwivedi, Y. Identification of MicroRNA-124-3p as a Putative Epigenetic Signature of Major Depressive Disorder. Neuropsychopharmacology 2017, 42, 864–875. [Google Scholar] [CrossRef] [Green Version]
- Roy, B.; Wang, Q.; Palkovits, M.; Faludi, G.; Dwivedi, Y. Altered MiRNA Expression Network in Locus Coeruleus of Depressed Suicide Subjects. Sci. Rep. 2017, 7, 4387. [Google Scholar] [CrossRef] [Green Version]
- Serafini, G.; Pompili, M.; Hansen, K.F.; Obrietan, K.; Dwivedi, Y.; Shomron, N.; Girardi, P. The Involvement of MicroRNAs in Major Depression, Suicidal Behavior, and Related Disorders: A Focus on MiR-185 and MiR-491-3p. Cell. Mol. Neurobiol. 2014, 34, 17–30. [Google Scholar] [CrossRef]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory Network of MiRNA on Its Target: Coordination between Transcriptional and Post-Transcriptional Regulation of Gene Expression. Cell. Mol. Life Sci. 2019, 76, 441–451. [Google Scholar] [CrossRef]
- Uhlmann, S.; Mannsperger, H.; Zhang, J.D.; Horvat, E.; Schmidt, C.; Küblbeck, M.; Henjes, F.; Ward, A.; Tschulena, U.; Zweig, K.; et al. Global MicroRNA Level Regulation of EGFR-driven Cell-cycle Protein Network in Breast Cancer. Mol. Syst. Biol. 2012, 8, 570. [Google Scholar] [CrossRef]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of MicroRNA–Target Recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef]
- Nestler, E.J.; Pena, C.J.; Kundakovic, M.; Mitchell, A.; Akbarian, S. Epigenetic Basis of Mental Illness. Neuroscience 2016, 22, 447–463. [Google Scholar] [CrossRef]
- Fries, G.R.; Carvalho, A.F.; Quevedo, J. The MiRNome of Bipolar Disorder. J. Affect. Disord. 2018, 233, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Adlakha, Y.K.; Saini, N. Brain MicroRNAs and Insights into Biological Functions and Therapeutic Potential of Brain Enriched MiRNA-128. Mol. Cancer 2014, 13, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, J.S.; Michlewski, G. MiRNAs in Development and Pathogenesis of the Nervous System. Biochem. Soc. Trans. 2013, 41, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Krichevsky, A.M.; King, K.S.; Donahue, C.P.; Khrapko, K.; Kosik, K.S. A MicroRNA Array Reveals Extensive Regulation of MicroRNAs during Brain Development. RNA 2003, 9, 1274–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes Provide a Protective and Enriched Source of MiRNA for Biomarker Profiling Compared to Intracellular and Cell-Free Blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef] [PubMed]
- Issler, O.; Chen, A. Determining the Role of MicroRNAs in Psychiatric Disorders. Nat. Rev. Neurosci. 2015, 16, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-Y.; Lu, R.-B.; Wang, L.-J.; Chang, C.-H.; Lu, T.; Wang, T.-Y.; Tsai, K.-W. Serum MiRNA as a Possible Biomarker in the Diagnosis of Bipolar II Disorder. Sci. Rep. 2020, 10, 1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gotzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Healthcare Interventions: Explanation and Elaboration. BMJ 2009, 339, b2700. [Google Scholar] [CrossRef] [Green Version]
- Cuschieri, S. The STROBE Guidelines. Saudi J. Anaesth. 2019, 13, 31. [Google Scholar] [CrossRef]
- Watanabe, K.; Taskesen, E.; Van Bochoven, A.; Posthuma, D. Functional Mapping and Annotation of Genetic Associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef] [Green Version]
- Smedley, D.; Haider, S.; Durinck, S.; Pandini, L.; Provero, P.; Allen, J.; Arnaiz, O.; Awedh, M.H.; Baldock, R.; Barbiera, G.; et al. The BioMart Community Portal: An Innovative Alternative to Large, Centralized Data Repositories. Nucleic Acids Res. 2015, 43, W589–W598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; van den Beek, M.; Blankenberg, D.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2016 Update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boughton, A.P.; Welch, R.P.; Flickinger, M.; VandeHaar, P.; Taliun, D.; Abecasis, G.R.; Boehnke, M. LocusZoom. Js: Interactive and Embeddable Visualization of Genetic Association Study Results. Bioinformatics 2021, 37, 3017–3018. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Miller, J.A.; Ding, S.-L.; Sunkin, S.M.; Smith, K.A.; Ng, L.; Szafer, A.; Ebbert, A.; Riley, Z.L.; Royall, J.J.; Aiona, K.; et al. Transcriptional Landscape of the Prenatal Human Brain. Nature 2014, 508, 199–206. [Google Scholar] [CrossRef]
- Hoss, A.G.; Labadorf, A.; Latourelle, J.C.; Kartha, V.K.; Hadzi, T.C.; Gusella, J.F.; MacDonald, M.E.; Chen, J.-F.; Akbarian, S.; Weng, Z.; et al. MiR-10b-5p Expression in Huntington’s Disease Brain Relates to Age of Onset and the Extent of Striatal Involvement. BMC Med. Genom. 2015, 8, 10. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Hodge, R.D.; Bakken, T.E.; Miller, J.A.; Smith, K.A.; Barkan, E.R.; Graybuck, L.T.; Close, J.L.; Long, B.; Johansen, N.; Penn, O.; et al. Conserved Cell Types with Divergent Features in Human versus Mouse Cortex. Nature 2019, 573, 61–68. [Google Scholar] [CrossRef]
- Transcriptomics Explorer. Available online: https://celltypes.brain-map.org/rnaseq/human_ctx_smart-seq (accessed on 5 December 2021).
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-MicroT Web Server v5.0: Service Integration into MiRNA Functional Analysis Workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [Green Version]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLOS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Camkurt, M.A.; Karababa, İ.F.; Erdal, M.E.; Kandemir, S.B.; Fries, G.R.; Bayazıt, H.; Ay, M.E.; Kandemir, H.; Ay, Ö.I.; Coşkun, S.; et al. MicroRNA Dysregulation in Manic and Euthymic Patients with Bipolar Disorder. J. Affect. Disord. 2020, 261, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Camkurt, M.A.; Karababa, F.; Erdal, M.E.; Bayazıt, H.; Kandemir, S.B.; Ay, M.E.; Kandemir, H.; Ay, Ö.İ.; Çiçek, E.; Selek, S.; et al. Investigation of Dysregulation of Several MicroRNAs in Peripheral Blood of Schizophrenia Patients. Clin. Psychopharmacol. Neurosci. 2016, 14, 256–260. [Google Scholar] [CrossRef] [Green Version]
- Edbauer, D.; Neilson, J.R.; Foster, K.A.; Wang, C.-F.; Seeburg, D.P.; Batterton, M.N.; Tada, T.; Dolan, B.M.; Sharp, P.A.; Sheng, M. Regulation of Synaptic Structure and Function by FMRP-Associated MicroRNAs MiR-125b and MiR-132. Neuron 2010, 65, 373–384. [Google Scholar] [CrossRef] [Green Version]
- Kandemir, H.; Erdal, M.E.; Selek, S.; Ay, Ö.İ.; Karababa, İ.F.; Kandemir, S.B.; Ay, M.E.; Yılmaz, Ş.G.; Bayazıt, H.; Taşdelen, B. Evaluation of Several Micro RNA (MiRNA) Levels in Children and Adolescents with Attention Deficit Hyperactivity Disorder. Neurosci. Lett. 2014, 580, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, L.; Cheng, K.; Wang, X.; Ren, G.; Xie, P. Identification of Suitable Plasma-Based Reference Genes for MiRNAome Analysis of Major Depressive Disorder. J. Affect. Disord. 2014, 163, 133–139. [Google Scholar] [CrossRef]
- Perkins, D.O.; Jeffries, C.D.; Jarskog, L.F.; Thomson, J.M.; Woods, K.; Newman, M.A.; Parker, J.S.; Jin, J.; Hammond, S.M. MicroRNA Expression in the Prefrontal Cortex of Individuals with Schizophrenia and Schizoaffective Disorder. Genome Biol. 2007, 8, R27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Smalheiser, N.R.; Lugli, G.; Zhang, H.; Rizavi, H.; Cook, E.H.; Dwivedi, Y. Expression of MicroRNAs and Other Small RNAs in Prefrontal Cortex in Schizophrenia, Bipolar Disorder and Depressed Subjects. PLoS ONE 2014, 9, e86469. [Google Scholar] [CrossRef]
- Fries, G.R.; Lima, C.N.C.; Valvassori, S.S.; Zunta-Soares, G.; Soares, J.C.; Quevedo, J. Preliminary Investigation of Peripheral Extracellular Vesicles’ MicroRNAs in Bipolar Disorder. J. Affect. Disord. 2019, 255, 10–14. [Google Scholar] [CrossRef]
- Ceylan, D.; Tufekci, K.U.; Keskinoglu, P.; Genc, S.; Özerdem, A. Circulating Exosomal MicroRNAs in Bipolar Disorder. J. Affect. Disord. 2020, 262, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, N.; Burmeister, M.; McInnis, M.G. MicroRNA Expression Changes in Lymphoblastoid Cell Lines in Response to Lithium Treatment. Int. J. Neuropsychopharmacol. 2009, 12, 975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, C.H.; Zainal, N.Z.; Kanagasundram, S.; Zain, S.M.; Mohamed, Z. Preliminary Examination of MicroRNA Expression Profiling in Bipolar Disorder I Patients during Antipsychotic Treatment. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2016, 171, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Pisanu, C.; Merkouri Papadima, E.; Melis, C.; Congiu, D.; Loizedda, A.; Orrù, N.; Calza, S.; Orrù, S.; Carcassi, C.; Severino, G.; et al. Whole Genome Expression Analyses of MiRNAs and MRNAs Suggest the Involvement of MiR-320a and MiR-155-3p and Their Targeted Genes in Lithium Response in Bipolar Disorder. Int. J. Mol. Sci. 2019, 20, 6040. [Google Scholar] [CrossRef] [Green Version]
- Manchia, M.; Adli, M.; Akula, N.; Ardau, R.; Aubry, J.-M.; Backlund, L.; Banzato, C.E.; Baune, B.T.; Bellivier, F.; Bengesser, S.; et al. Assessment of Response to Lithium Maintenance Treatment in Bipolar Disorder: A Consortium on Lithium Genetics (ConLiGen) Report. PLoS ONE 2013, 8, e65636. [Google Scholar] [CrossRef] [Green Version]
- Hudder, A.; Novak, R.F. MiRNAs: Effectors of Environmental Influences on Gene Expression and Disease. Toxicol. Sci. 2008, 103, 228–240. [Google Scholar] [CrossRef]
- Forstner, A.J.; Hofmann, A.; Maaser, A.; Sumer, S.; Khudayberdiev, S.; Mühleisen, T.W.; Leber, M.; Schulze, T.G.; Strohmaier, J.; Degenhardt, F.; et al. Genome-Wide Analysis Implicates MicroRNAs and Their Target Genes in the Development of Bipolar Disorder. Transl. Psychiatry 2015, 5, e678. [Google Scholar] [CrossRef] [Green Version]
- Sanei, M.; Chen, X. Mechanisms of MicroRNA Turnover. Curr. Opin. Plant Biol. 2015, 27, 199–206. [Google Scholar] [CrossRef] [Green Version]
- Makeyev, E.V.; Zhang, J.; Carrasco, M.A.; Maniatis, T. The MicroRNA MiR-124 Promotes Neuronal Differentiation by Triggering Brain-Specific Alternative Pre-MRNA Splicing. Mol. Cell 2007, 27, 435–448. [Google Scholar] [CrossRef] [Green Version]
- Martins, H.C.; Schratt, G. MicroRNA-Dependent Control of Neuroplasticity in Affective Disorders. Transl. Psychiatry 2021, 11, 263. [Google Scholar] [CrossRef]
- Choo, K.B.; Soon, Y.L.; Nguyen, P.N.N.; Hiew, M.S.Y.; Huang, C.-J. MicroRNA-5p and -3p Co-Expression and Cross-Targeting in Colon Cancer Cells. J. Biomed. Sci. 2014, 21, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of Post-Transcriptional Regulation by MicroRNAs: Are the Answers in Sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, M.M.J.; Krauskopf, J.; Ramaekers, J.G.; Kleinjans, J.C.S.; Prickaerts, J.; Briedé, J.J. Circulating MicroRNAs as Potential Biomarkers for Psychiatric and Neurodegenerative Disorders. Prog. Neurobiol. 2020, 185, 101732. [Google Scholar] [CrossRef]
- Redis, R.S.; Calin, S.; Yang, Y.; You, M.J.; Calin, G.A. Cell-to-Cell MiRNA Transfer: From Body Homeostasis to Therapy. Pharmacol. Ther. 2012, 136, 169–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; How Huang, K.; Jen Lee, M.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Yu, Z.; Yuan, S.; Xie, W.; Li, C.; Hu, Z.; Xiang, Y.; Wu, N.; Wu, L.; Bai, L.; et al. Circulating Exosomal MicroRNAs as Prognostic Biomarkers for Non-Small-Cell Lung Cancer. Oncotarget 2017, 8, 13048–13058. [Google Scholar] [CrossRef] [Green Version]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. MiRNA-Based Biomarkers, Therapies, and Resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef]
- Momi, N.; Kaur, S.; Rachagani, S.; Ganti, A.K.; Batra, S.K. Smoking and MicroRNA Dysregulation: A Cancerous Combination. Trends Mol. Med. 2014, 20, 36–47. [Google Scholar] [CrossRef] [Green Version]
- Dwivedi, Y. Emerging Role of MicroRNAs in Major Depressive Disorder: Diagnosis and Therapeutic Implications. Dialogues Clin. Neurosci. 2014, 16, 43–61. [Google Scholar] [CrossRef]
- Bocchio-Chiavetto, L.; Maffioletti, E.; Bettinsoli, P.; Giovannini, C.; Bignotti, S.; Tardito, D.; Corrada, D.; Milanesi, L.; Gennarelli, M. Blood MicroRNA Changes in Depressed Patients during Antidepressant Treatment. Eur. Neuropsychopharmacol. 2013, 23, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Vieta, E.; Montes, J.M. A Review of Asenapine in the Treatment of Bipolar Disorder. Clin. Drug Investig. 2018, 38, 87–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alda, M. Lithium in the Treatment of Bipolar Disorder: Pharmacology and Pharmacogenetics. Mol. Psychiatry 2015, 20, 661–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinbold, C.S.; Forstner, A.J.; Hecker, J.; Fullerton, J.M.; Hoffmann, P.; Hou, L.; Heilbronner, U.; Degenhardt, F.; Adli, M.; Akiyama, K.; et al. Analysis of the Influence of MicroRNAs in Lithium Response in Bipolar Disorder. Front. Psychiatry 2018, 9. [Google Scholar] [CrossRef]
- Almeida, P.G.C.; Nani, J.V.; Oses, J.P.; Brietzke, E.; Hayashi, M.A.F. Neuroinflammation and Glial Cell Activation in Mental Disorders. Brain Behav. Immun. Health 2020, 2, 100034. [Google Scholar] [CrossRef]
- Brisch, R.; Wojtylak, S.; Saniotis, A.; Steiner, J.; Gos, T.; Kumaratilake, J.; Henneberg, M.; Wolf, R. The Role of Microglia in Neuropsychiatric Disorders and Suicide. Eur. Arch. Psychiatry Clin. Neurosci. 2021. [Google Scholar] [CrossRef]
- Xu, N.; Shinohara, K.; Saunders, K.E.A.; Geddes, J.R.; Cipriani, A. Effect of Lithium on Circadian Rhythm in Bipolar Disorder: A Systematic Review and Meta-analysis. Bipolar Disord. 2021, 23, 445–453. [Google Scholar] [CrossRef]
- McCarthy, M.J.; Wei, H.; Nievergelt, C.M.; Stautland, A.; Maihofer, A.X.; Welsh, D.K.; Shilling, P.; Alda, M.; Alliey-Rodriguez, N.; Anand, A.; et al. Chronotype and Cellular Circadian Rhythms Predict the Clinical Response to Lithium Maintenance Treatment in Patients with Bipolar Disorder. Neuropsychopharmacology 2019, 44, 620–628. [Google Scholar] [CrossRef]
- Papadima, E.M.; Niola, P.; Melis, C.; Pisanu, C.; Congiu, D.; Cruceanu, C.; Lopez, J.P.; Turecki, G.; Ardau, R.; Severino, G.; et al. Evidence towards RNA Binding Motif (RNP1, RRM) Protein 3 (RBM3) as a Potential Biomarker of Lithium Response in Bipolar Disorder Patients. J. Mol. Neurosci. 2017, 62, 304–308. [Google Scholar] [CrossRef]
- Geoffroy, P.A.; Curis, E.; Courtin, C.; Moreira, J.; Morvillers, T.; Etain, B.; Laplanche, J.-L.; Bellivier, F.; Marie-Claire, C. Lithium Response in Bipolar Disorders and Core Clock Genes Expression. World J. Biol. Psychiatry 2018, 19, 619–632. [Google Scholar] [CrossRef]
- Hunsberger, J.G.; Chibane, F.L.; Elkahloun, A.G.; Henderson, R.; Singh, R.; Lawson, J.; Cruceanu, C.; Nagarajan, V.; Turecki, G.; Squassina, A.; et al. Novel Integrative Genomic Tool for Interrogating Lithium Response in Bipolar Disorder. Transl. Psychiatry 2015, 5, e504. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| miRNA Expression in BD Group Compared to Control Group | Camkurt et al. [75] | Fries et al. [83] | Ceylan et al. [84] | Lee et al. [59] |
|---|---|---|---|---|
| Setting | Patients recruited from the outpatient psychiatry clinic of Harran University Faculty of Medicine, Turkey. | Subjects were recruited at the Center of Excellence in Mood Disorders at UTHealth, USA. | Manic patients from Shenzhen Kang Ning Hospital, China | Subjects were recruited at the Department of psychiatry, Kaohsiung Veterans General Hospital and National Cheng Kung University Hospital, Taiwan. |
| Participants | 58 medicated BD type I patients and 51 healthy controls | 20 medicated BD type I patients and 21 healthy controls. | 69 medicated BD type I patients and 41 healthy controls | 79 non-medicated BD type II patients and 95 healthy controls. |
| Diagnostic instrument | SCID-1 | SCID-1 | SCID-1 YMRS HAM-D | SADS-L |
| Study material | Extracellular miRNA from peripheral, whole blood. | miRNA from exosomes in peripheral blood | miRNA from exosomes in peripheral blood. | Intra- and extracellular miRNA from peripheral blood, serum. |
| Method | qRT-PCR | Microarray | qRT-PCR | qRT-PCR |
| Upregulated | miR-29a-3p | miR-185–5p | miR-7-5p | |
| miR-106b-5p | miR-23b-3p | |||
| miR-107 | miR-142-3p | |||
| miR-125a-3p | miR-221-5p | |||
| miR-370-3p | ||||
| Downregulated | miR-142-3p | |||
| miR-484 | ||||
| miR-652-3p |
| Chen et al. [85] | Lim et al. [86] | Lim et al. [86] | Pisanu et al. [87] | |
|---|---|---|---|---|
| Setting | Recruited from the University of Michigan Depression Center Prechter Bipolar repository, USA. | Recruited from the University Malaya Medical Centre (UMMC) inpatient psychiatric Ward, Malaysia. | Recruited from the University Malaya Medical Centre (UMMC) inpatient psychiatric Ward, Malaysia. | Recruited at the Lithium Clinic of the Clinical Psychopharmacology Centre of the University Hospital of Cagliari, Italy. |
| Participants | 10 BD I patients | 5 BD I (manic phase) patients | 5 BD I (manic phase) patients | BD patients. 12 Excellent responders and 12 Non-Responders |
| Diagnostic instrument | Not reported | YMRS | YMRS | SADS-L |
| Exposure | Lithium (treated vs. non-treated) | Asenapine (drug free vs. medicated) | Risperidone (drug free vs. medicated) | Lithium (responders vs. non-responders) |
| Study material | Lymphoblastoid cell lines | Whole blood | Whole blood | Lymphoblastoid cell lines |
| Method | qRT-PCR | Microarray | Microarray | Next Generation Sequencing (NGS) |
| Upregulated | miR-221 | miR-18a-5p | miR-148a-3p | |
| miR-152 | miR-19b-3p | miR-22-3p | ||
| miR-15a | miR-145-5p | miR-26b-5p | ||
| miR-27a-3p | miR-223-3p | |||
| miR-148b-3p | miR-155-3p | |||
| miR-210-3p | miR-744-5p | |||
| miR-17-3p | miR-181a-3p | |||
| miR-30b-5p | miR-15a-5p | |||
| miR-106b-5p | miR-148b-3p | |||
| miR-339-5p | miR-15b-3p | |||
| miR-106a-5p | miR-454-5p | |||
| miR-20a-5p | miR-4677-3p | |||
| miR-17-5p | miR-374a-3p | |||
| miR-15a-5p | miR-19b-3p | |||
| miR-101-3p | ||||
| miR-148a-5p | ||||
| miR-142-3p | ||||
| miR-454-3p | ||||
| miR-142-5p | ||||
| let-7f-5p | ||||
| miR-27a-5p | ||||
| let-7a-5p | ||||
| miR-146a-5p | ||||
| miR-15b-5p | ||||
| miR-335-3p | ||||
| miR-421 | ||||
| miR-26a-5p | ||||
| Downregulated | miR-494 | miR-92b-5p | miR-664b-5p | miR-320a |
| miR-1343-5p | miR-6778-5p | miR-125a-5p | ||
| miR-146b-5p | miR-574-3p | |||
| miR-1273h-3p | ||||
| miR-9-5p | ||||
| miR-378a-5p | ||||
| miR-505-3p | ||||
| let-7e-5p | ||||
| miR-138-5p | ||||
| miR-941 | ||||
| miR-652-3p | ||||
| miR-130b-3p | ||||
| miR-345-5p | ||||
| let-7d-3p | ||||
| miR-181d-5p | ||||
| miR-629-5p | ||||
| miR-574-5p | ||||
| miR-378a-3p | ||||
| miR-598-3p | ||||
| miR-23a-3p | ||||
| miR-425-5p | ||||
| miR-197-3p | ||||
| miR-194-5p |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clausen, A.R.; Durand, S.; Petersen, R.L.; Staunstrup, N.H.; Qvist, P. Circulating miRNAs as Potential Biomarkers for Patient Stratification in Bipolar Disorder: A Combined Review and Data Mining Approach. Genes 2022, 13, 1038. https://doi.org/10.3390/genes13061038
Clausen AR, Durand S, Petersen RL, Staunstrup NH, Qvist P. Circulating miRNAs as Potential Biomarkers for Patient Stratification in Bipolar Disorder: A Combined Review and Data Mining Approach. Genes. 2022; 13(6):1038. https://doi.org/10.3390/genes13061038
Chicago/Turabian StyleClausen, Alexandra R., Simon Durand, Rasmus L. Petersen, Nicklas H. Staunstrup, and Per Qvist. 2022. "Circulating miRNAs as Potential Biomarkers for Patient Stratification in Bipolar Disorder: A Combined Review and Data Mining Approach" Genes 13, no. 6: 1038. https://doi.org/10.3390/genes13061038






