Antioxidant Activity of Phenolic Extraction from Different Sweetpotato (Ipomoea batatas (L.) Lam.) Blades and Comparative Transcriptome Analysis Reveals Differentially Expressed Genes of Phenolic Metabolism in Two Genotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Determination of Antioxidant Activity of Blade
2.3. Determination of Total Polyphenol Content and Total Flavonoid Content
2.4. cDNA Library Construction and RNA-Seq
2.5. Transcriptome Assembly and Functional Annotation
2.6. Differentially Expressed Genes (DEGs) Analysis
2.7. Quantitative Real-Time PCR (qRT-PCR) Analysis of DEGs
2.8. Statistical Analysis
3. Results
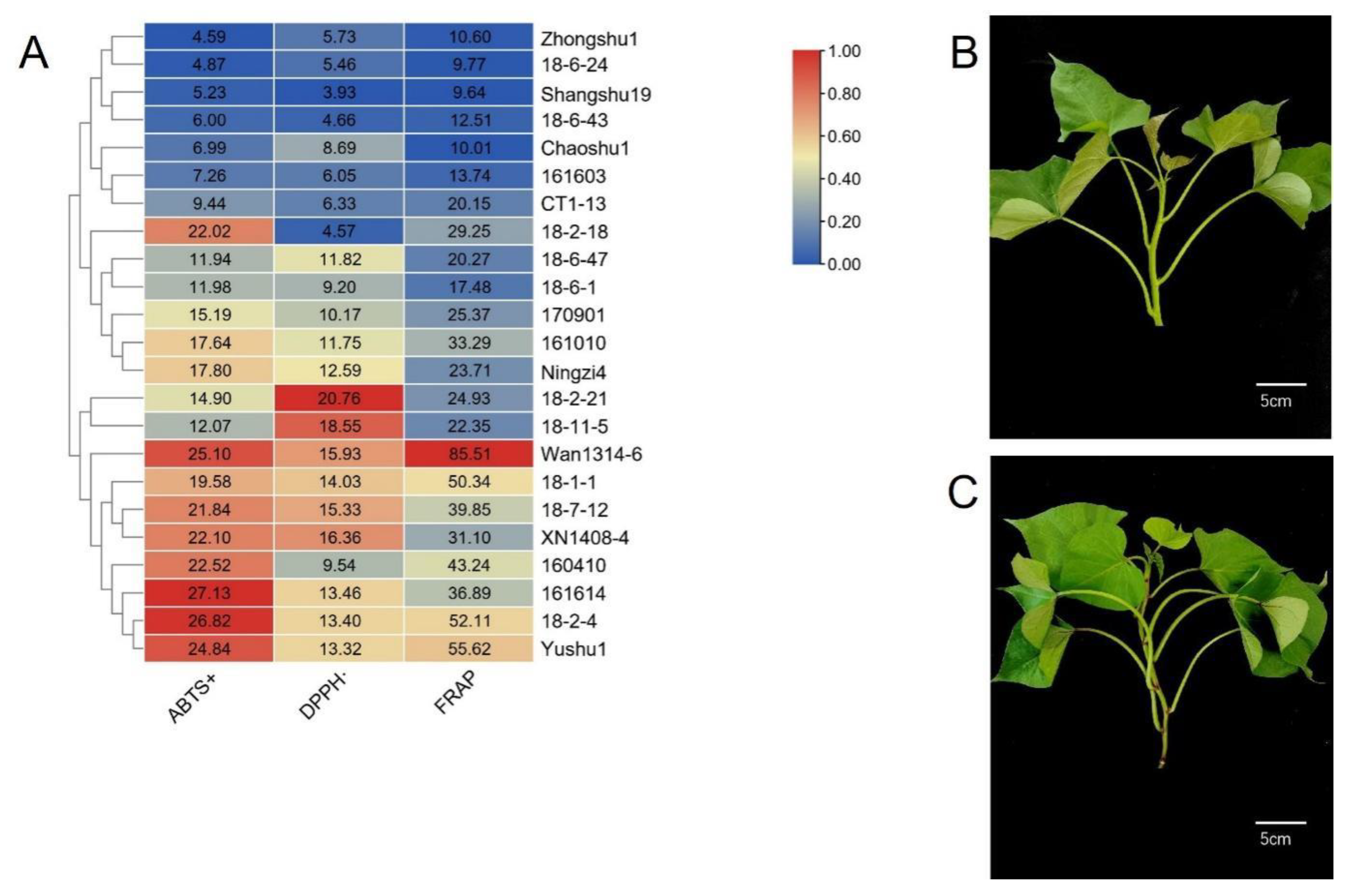
3.1. Antioxidant Activity of Sweetpotato Blades
3.2. TPC and TFC of Sweetpotato Blades
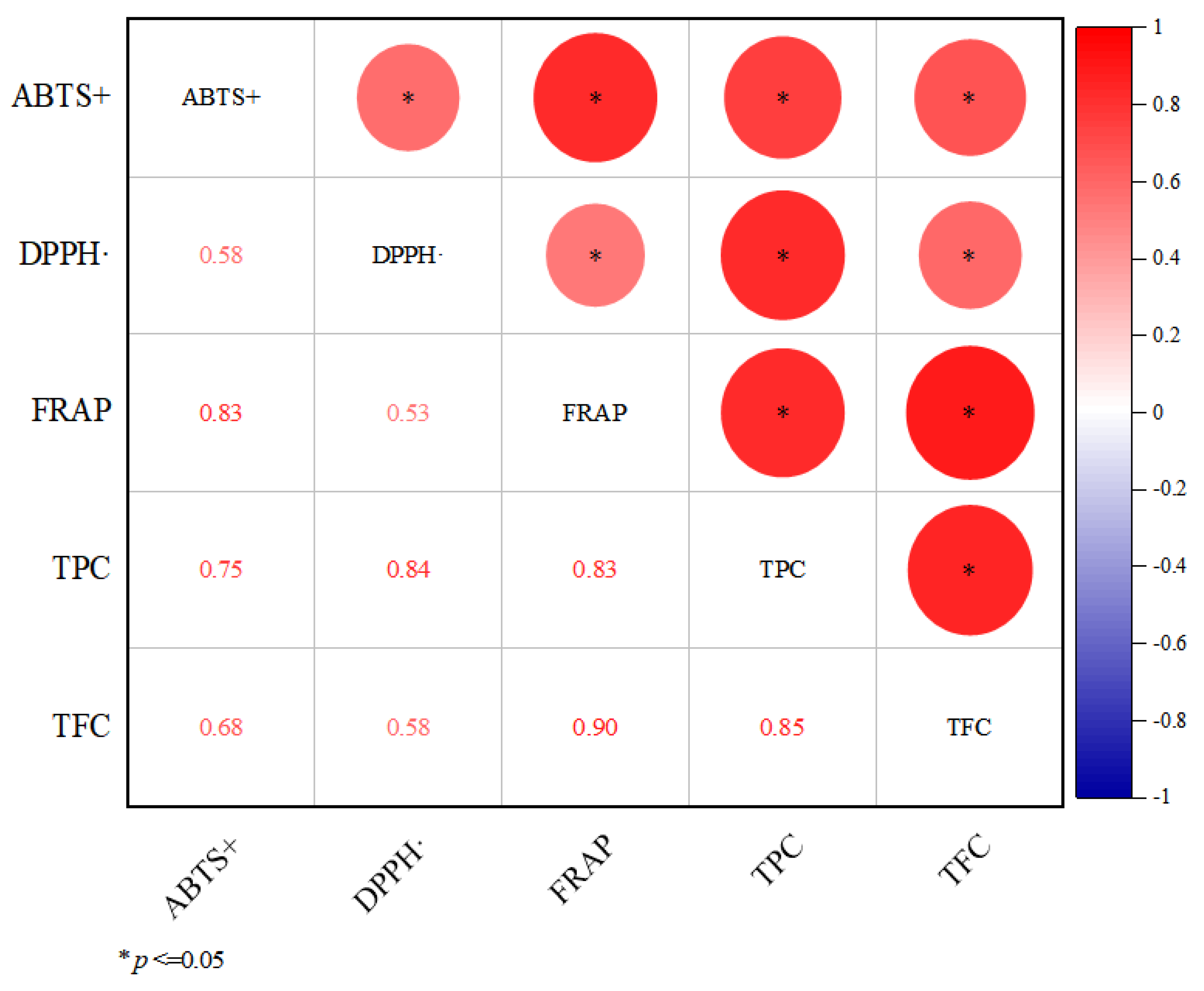
3.3. Correlation Analysis between Antioxidant Activity, TPC and TFC
3.4. RNA-Seq and Data Pre-Processing
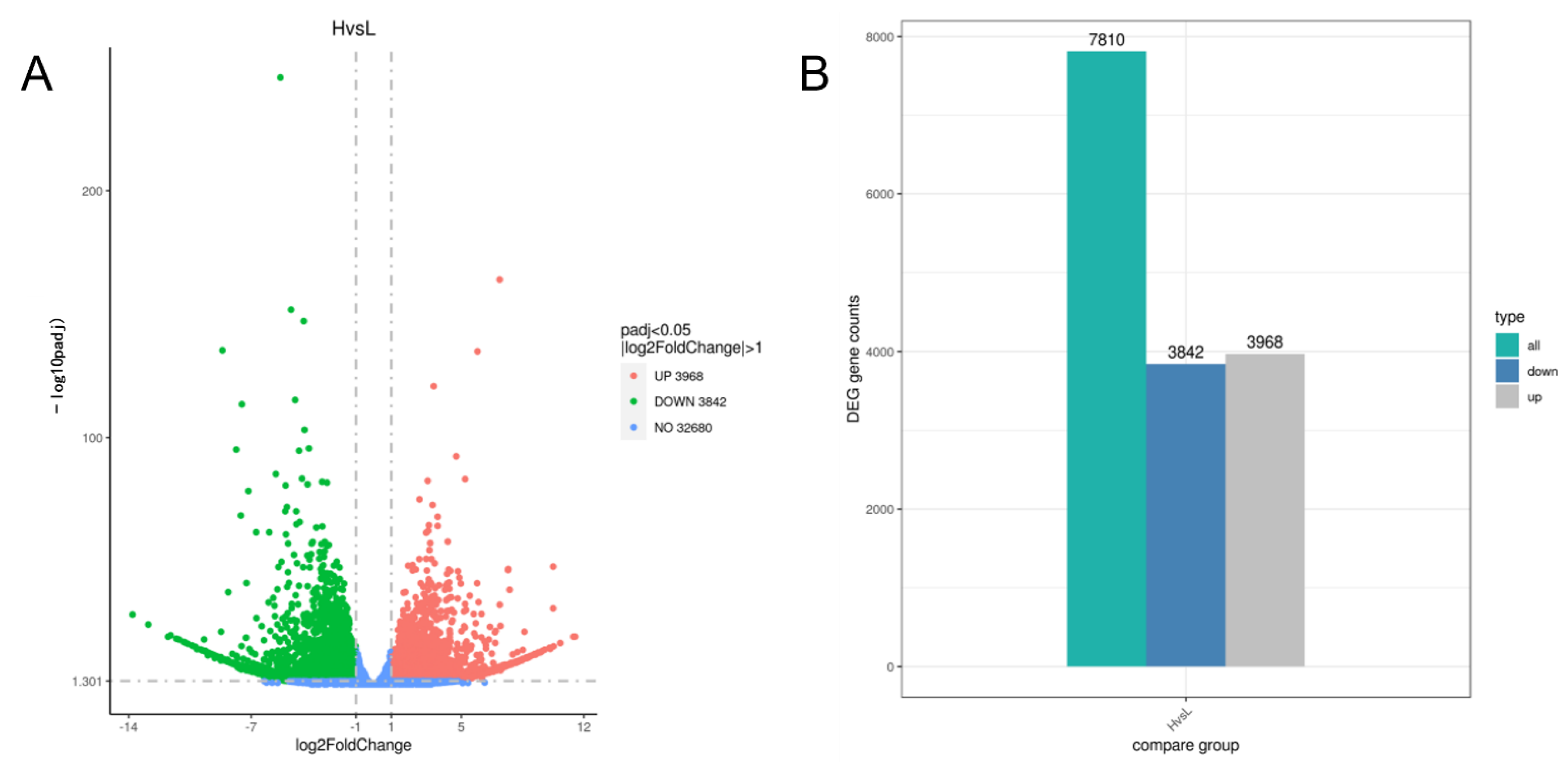
3.5. Analysis of DEGs
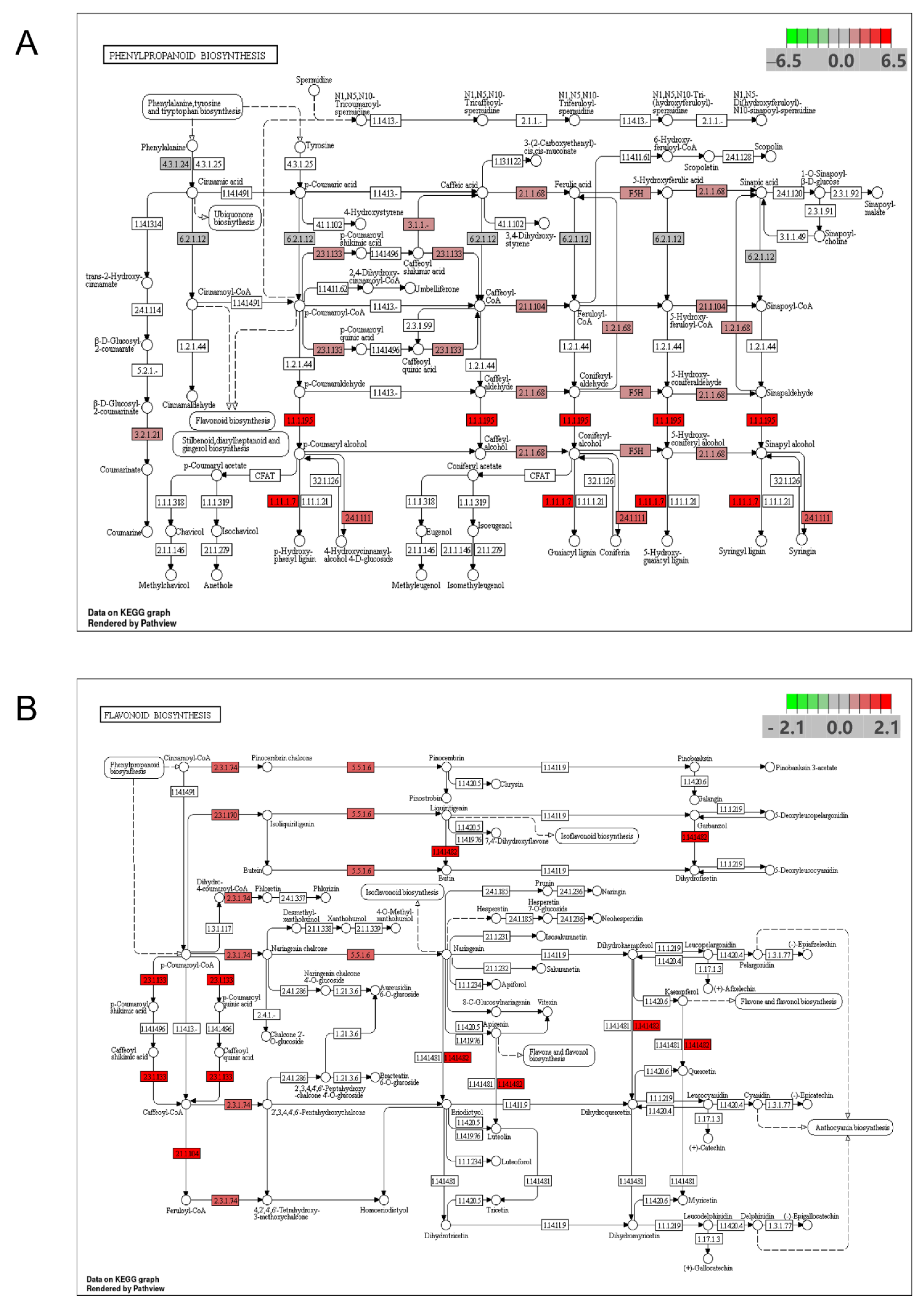
3.6. qRT-PCR Verification of the Phenylpropanoid Biosynthesis Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhen, S.; van Iersel, M.W. Photochemical Acclimation of Three Contrasting Species to Different Light Levels: Implications for Optimizing Supplemental Lighting. J. Am. Soc. Heretic. Sci. 2017, 142, 346–354. [Google Scholar] [CrossRef] [Green Version]
- Kivuva, B.M.; Githiri, S.M.; Yencho, G.C.; Sibiya, J. Screening Sweetpotato Genotypes for Tolerance to Drought Stress. F. Crop. Res. 2015, 171, 11–22. [Google Scholar] [CrossRef]
- FAO. FAO Statistical Databases; FAO: Rome, Italy, 2020; Available online: http://www.fao.org/faostat/zh/#dataQC (accessed on 1 November 2021).
- Ishida, H.; Suzuno, H.; Sugiyama, N.; Innami, S.; Tadokoro, T.; Maekawa, A. Nutritive Evaluation on Chemical Components of Leaves, Stalks and Stems of Sweet Potatoes (Ipomoea batatas Poir). Food Chem. 2000, 68, 359–367. [Google Scholar] [CrossRef]
- Islam, M.S.; Yoshimoto, M.; Yahara, S.; Okuno, S.; Ishiguro, K.; Yamakawa, O. Identification and Characterization of Foliar Polyphenolic Composition in Sweetpotato (Ipomoea Batatas L.) Genotypes. J. Agric. Food Chem. 2002, 50, 3718–3722. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Nie, S.; Zhu, F. Chemical Constituents and Health Effects of Sweet Potato. Food Res. Int. 2016, 89, 90–116. [Google Scholar] [CrossRef]
- Sun, H.; Mu, T.; Xi, L.; Zhang, M.; Chen, J. Sweet Potato (Ipomoea batatas L.) Leaves as Nutritional and Functional Foods. Food Chem. 2014, 156, 380–389. [Google Scholar] [CrossRef]
- Jiang, Y.; Sun, C. Advances in Studies on Nutritious Components of Sweet Potato and the Health-promoting Functions. Sci. Technol. A 2010, 12, 56–61. [Google Scholar]
- Gessner, D.K.; Ringseis, R.; Eder, K. Potential of Plant Polyphenols to Combat Oxidative Stress and Inflammatory Processes in Farm Animals. J. Anim. Physiol. Anim. Nutr. 2017, 101, 605–628. [Google Scholar] [CrossRef]
- Islam, S. Sweetpotato (Ipomoea batatas L.) Leaf: Its Potential Effect on Human Health and Nutrition. J. Food Sci. 2006, 71, R13–R121. [Google Scholar] [CrossRef]
- Ojong, P.B.; Njiti, V.; Guo, Z.; Gao, M.; Besong, S.; Barnes, S.L. Variation of Flavonoid Content Among Sweetpotato Accessions. J. Amer. Soc. Hort. Sci. 2008, 133, 819–824. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Mu, B.; Song, Z.; Ma, Z.; Mu, T. The in Vitro Antioxidant Activity and Inhibition of Intracellular Reactive Oxygen Species of Sweet Potato Leaf Polyphenols. Oxid. Med. Cell. Longev. 2018, 2018, 9017828. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Wang, W.; Wu, J.; Huang, S.; Zhang, X.; Zhang, Z.; Yu, S. Yield Composition Analysis of Vine Tip of Leaf-vegetable Sweetpotato. Agric. Sci. Technol. 2009, 10, 88–91. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, C.; Zhao, Y.; Zhao, W.; Liu, X.; Zeng, L.; Liao, Z.; Zhang, Q. Chlorogenic Acid Contents in Shoot-Tips of Different Vegetable-Use Sweetpotato Varieties and Their DPPH·Scavenging Capacities. Sci. Agric. Sin. 2010, 43, 4814–4822. [Google Scholar] [CrossRef]
- Tao, X.; Wu, Q.; Aalim, H.; Li, L.; Mao, L.; Luo, Z.; Ying, T. Effects of Exogenous Abscisic Acid on Bioactive Components and Antioxidant Capacity of Postharvest Tomato during Ripening. Molecules 2020, 25, 1346. [Google Scholar] [CrossRef] [Green Version]
- Løvdal, T.; Olsen, K.M.; Slimestad, R.; Verheul, M.; Lillo, C. Synergetic Effects of Nitrogen Depletion, Temperature, and Light on the Content of Phenolic Compounds and Gene Expression in Leaves of Tomato. Phytochemistry 2010, 71, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, L.; Liang, X.; Dai, P.; Zhang, Y.; Li, B.; Lin, X.; Sun, C. Enhancement of Polyphenolic Metabolism as an Adaptive Response of Lettuce (Lactuca sativa) Roots to Aluminum Stress. Environ. Pollut. 2020, 261, 114230. [Google Scholar] [CrossRef] [PubMed]
- Chen Han, C.; Jin, P.; Li, M.; Wang, L.; Zheng, Y. Physiological and Transcriptomic Analysis Validates Previous Findings of Changes in Primary Metabolism for the Production of Phenolic Antioxidants in Wounded Carrots. J. Agric. Food Chem. 2017, 65, 7159–7167. [Google Scholar] [CrossRef]
- Grimalt, M.; Legua, P.; Hernández, F.; Amorós, A.; Almansa, M.S. Antioxidant Activity and Bioactive Compounds Contents in Different Stages of Flower Bud Development from Three Spanish Caper (Capparis Spinosa) Cultivars. Hortic. J. 2019, 88, 410–419. [Google Scholar] [CrossRef] [Green Version]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C.A. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing Ability of Plasma (FRAP) as a Measure of “Antioxidant Power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.-H.; Huang, T.-C. Volatile and Nonvolatile Constituents and Antioxidant Capacity of Oleoresins in Three Taiwan Citrus Varieties as Determined by Supercritical Fluid Extraction. Molecules 2016, 21, 1735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventós, R.M. 1999. Analysis of total phenols and Other Oxidation Substrates and Antioxidants by means of Folin-ciocalteu reagent. Meth. Enzymol. 1999, 299, 152–178. [Google Scholar] [CrossRef]
- Nie, J.; Lu, D.; Li, J.; Liu, F.; Li, H.; Wang, K. A Preliminary Study on the Flavonoids in Fruits of 22 Apple Germplasm Resources. Sci. Agric. Sin. 2010, 43, 4455–4462. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real–time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TB tools: An Integrative Toolkit Developed for in-Teractive Analyses of Big Biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, X.; Li, Q.; Cao, Q.; Ma, D. Current Status and Future Prospective of Sweetpotato Production and Seed Industry in China. Sci. Agric. Sin. 2021, 54, 483–492. [Google Scholar]
- Cevallos-Casals, B.A.; Cisneros-Zevallos, L. Stoichiometric and Kinetic Studies of Phenolic Antioxidants from Andean Purple Corn and Red-Fleshed Sweetpotato. J. Agric. Food Chem. 2003, 51, 3313–3319. [Google Scholar] [CrossRef]
- Truong, V.D.; McFeeters, R.F.; Thompson, R.T.; Dean, L.L.; Shofran, B. Phenolic Acid Content and Composition in Leaves and Roots of Common Commercial Sweetpotato (Ipomoea Batatas L.) Cultivars in the United States. J. Food Sci. 2007, 72, 343–349. [Google Scholar] [CrossRef]
- Xie, X.; Zhao, Y.; Luo, W.; Sen, X.; Zeng, L.; Yang, C.; Fu, Y. Behavioral Differences of Ethanol Extracts from Sprouting Shoot Tips of Different Sweetpotato Varieties in the Process of DPPH Scavenging Reaction. Sci. Agric. Sin. 2013, 46, 270–281. [Google Scholar] [CrossRef]
- Nakagawa, S.; Ohmura, R.; Toshima, S.; Park, H.; Narasako, Y.; Hirano, T.; Otani, M.; Kunitake, H. Changes in Polyphenols, Anthocyanins, and DPPH Radical-Scavenging Activities in Sweetpotato (Ipomoea batatas L.) during Tuber Growth. Sci. Hortic. 2021, 284, 110100. [Google Scholar] [CrossRef]
- Padda, M.S.; Picha, D.H. Quantification of Phenolic Acids and Antioxidant Activity in Sweetpotato Genotypes. Sci. Hortic. 2008, 119, 17–20. [Google Scholar] [CrossRef]
- Luo, C.; Wang, X.; Gao, G.; Wang, L.; Li, Y.; Sun, C. Identification and Quantification of Free, Conjugate and Total Phenolic Compounds in Leaves of 20 Sweetpotato Cultivars by HPLC-DAD and HPLC-ESI-MS/MS. Food Chem. 2013, 141, 2697–2706. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Lau, K.H.; Cao, Q.; Hamilton, J.P.; Sun, H.; Zhou, C.; Eserman, L.; Gemenet, D.C.; Olukolu, B.A.; Wang, H.; et al. Genome Sequences of Two Diploid Wild Relatives of Cultivated Sweetpotato Reveal Targets for Genetic Improvement. Nat. Commun. 2018, 9, 4580. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of Phenylpropanoid Pathway and the Role of Polyphenols in Plants under Abiotic Stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for genetics, biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995, 7, 1085–1097. [Google Scholar] [CrossRef]
- Cheng, G.W.; Breen, P.J. Activity of Phenylalanine Ammonia-Lyase (PAL) and Concentrations of Anthocyanins and Phenolics in Developing Strawberry Fruit. J. Am. Soc. Hortic. Sci. 2022, 116, 865–869. [Google Scholar] [CrossRef]
- Walker, K.D.; Klettke, K.; Akiyama, T.; Croteau, R. Cloning, heterologous expression and characterization of a phenylalanine aminomutase involved in taxol biosynthesis. J. Biol. Chem. 2004, 279, 56854–63947. [Google Scholar] [CrossRef] [Green Version]
- Clifford, M.N.; Jaganath, I.B.; Ludwig, I.A.; Crozier, A. Chlorogenic Acids and the Acyl-quinic Acids: Discovery, Biosynthesis, bioavailability and bioactivity. Nat. Prod. Rep. 2017, 34, 1391–1421. [Google Scholar] [CrossRef] [Green Version]
- Niggeweg, R.; Michael, A.J.; Martin, C. Engineering Plants with Increased Levels of the Antioxidant Chlorogenic Acid. Nat. Biotechnol. 2004, 22, 746–754. [Google Scholar] [CrossRef]
- Chen, J.H.; Ho, C.T. Antioxidant Activities of Caffeic Acid and Its Related Hydroxycinnamic Acid Compounds. J. Agric. Food Chem. 1997, 45, 2374–2378. [Google Scholar] [CrossRef]
- Ozgen, M.; Reese, R.N.; Tulio, A.Z.; Scheerens, J.C.; Miller, A.R. Modified 2,2-Azino-Bis-3-Ethylbenzothiazoline-6-Sulfonic Acid (ABTS) Method to Measure Antioxidant Capacity of Selected Small Fruits and Comparison to Ferric Reducing Antioxidant Power (FRAP) and 2,2′-Diphenyl-1- Picrylhydrazyl (DPPH) Methods. J. Agric. Food Chem. 2006, 54, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Mohanraj, R.; Sivasankar, S. Sweet Potato (Ipomoea batatas [L.] Lam)—A Valuable Medicinal Food: A Review. J. Med. Food. 2014, 17, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Routaboul, J.M.; Kerhoas, L.; Debeaujon, I.; Pourcel, L.; Caboche, M.; Einhorn, J.; Lepiniec, L. Flavonoid Diversity and Biosynthesis in Seed of Arabidopsis Thaliana. Planta 2006, 224, 96–107. [Google Scholar] [CrossRef]
- Xu, J.; Zhu, J.; Wang, X.; Han, Y.; Tang, L.; Zhu, H. Cloning and Expression Analysis of Chalcone Synthase Gene IbCHS1 in Ipomoea Batatas. Mol. Plant Breed. 2018, 16, 1752–1757. [Google Scholar] [CrossRef]
- Guo, J.; Zhou, W.; Lu, Z.; Li, H.; Li, H.; Gao, F. Isolation and Functional Analysis of Chalcone Isomerase Gene from Purple-Fleshed Sweet Potato. Plant Mol. Biol. Rep. 2015, 33, 1451–1463. [Google Scholar] [CrossRef]
- Zhou, W.; Gong, Y.; Lu, X.; Huang, C.; Gao, F. Molecular Cloning and Characterization of a Flavonoid 3′-Hydroxylase Gene from Purple-Fleshed Sweet Potato (Ipomoea Batatas). Mol. Biol. Rep. 2012, 39, 295–302. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin Biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Xiang, Z.; Ye, F.; Zhao, G. Composition and Structure of an Antioxidant Acetic Acid Lignin Isolated from Shoot Shell of Bamboo (Dendrocalamus Latiforus). Ind. Crops Prod. 2016, 91, 340–349. [Google Scholar] [CrossRef]




| Raw Reads | Clean Reads | Clean Bases | Error Rate | Q20 | Q30 | GC Content | |
|---|---|---|---|---|---|---|---|
| W1 | 47094680 | 44240870 | 6.64G | 0.03 | 97.57 | 93.45 | 46.59 |
| W2 | 45400034 | 44564352 | 6.68G | 0.03 | 96.99 | 92.00 | 46.13 |
| W3 | 43525644 | 41821630 | 6.27G | 0.03 | 97.66 | 93.62 | 46.27 |
| S1 | 42353168 | 40573440 | 6.09G | 0.03 | 97.65 | 93.60 | 46.48 |
| S2 | 42027724 | 40358894 | 6.05G | 0.03 | 97.67 | 93.64 | 46.33 |
| S3 | 44622468 | 42746376 | 6.41G | 0.03 | 97.61 | 93.46 | 46.37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, P.; Ran, H.; Li, J.; Zong, J.; Luo, Q.; Zhao, T.; Liao, Z.; Tang, Y.; Fu, Y. Antioxidant Activity of Phenolic Extraction from Different Sweetpotato (Ipomoea batatas (L.) Lam.) Blades and Comparative Transcriptome Analysis Reveals Differentially Expressed Genes of Phenolic Metabolism in Two Genotypes. Genes 2022, 13, 1078. https://doi.org/10.3390/genes13061078
Chen P, Ran H, Li J, Zong J, Luo Q, Zhao T, Liao Z, Tang Y, Fu Y. Antioxidant Activity of Phenolic Extraction from Different Sweetpotato (Ipomoea batatas (L.) Lam.) Blades and Comparative Transcriptome Analysis Reveals Differentially Expressed Genes of Phenolic Metabolism in Two Genotypes. Genes. 2022; 13(6):1078. https://doi.org/10.3390/genes13061078
Chicago/Turabian StyleChen, Peitao, Hairong Ran, Jiaxin Li, Jikai Zong, Qingqing Luo, Tengfei Zhao, Zhihua Liao, Yueli Tang, and Yufan Fu. 2022. "Antioxidant Activity of Phenolic Extraction from Different Sweetpotato (Ipomoea batatas (L.) Lam.) Blades and Comparative Transcriptome Analysis Reveals Differentially Expressed Genes of Phenolic Metabolism in Two Genotypes" Genes 13, no. 6: 1078. https://doi.org/10.3390/genes13061078
APA StyleChen, P., Ran, H., Li, J., Zong, J., Luo, Q., Zhao, T., Liao, Z., Tang, Y., & Fu, Y. (2022). Antioxidant Activity of Phenolic Extraction from Different Sweetpotato (Ipomoea batatas (L.) Lam.) Blades and Comparative Transcriptome Analysis Reveals Differentially Expressed Genes of Phenolic Metabolism in Two Genotypes. Genes, 13(6), 1078. https://doi.org/10.3390/genes13061078






