Vasa Is a Potential Germ Cell Marker in Leopard Coral Grouper (Plectropomus leopardus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Ethics
2.2. RNA Extraction and cDNA Synthesis
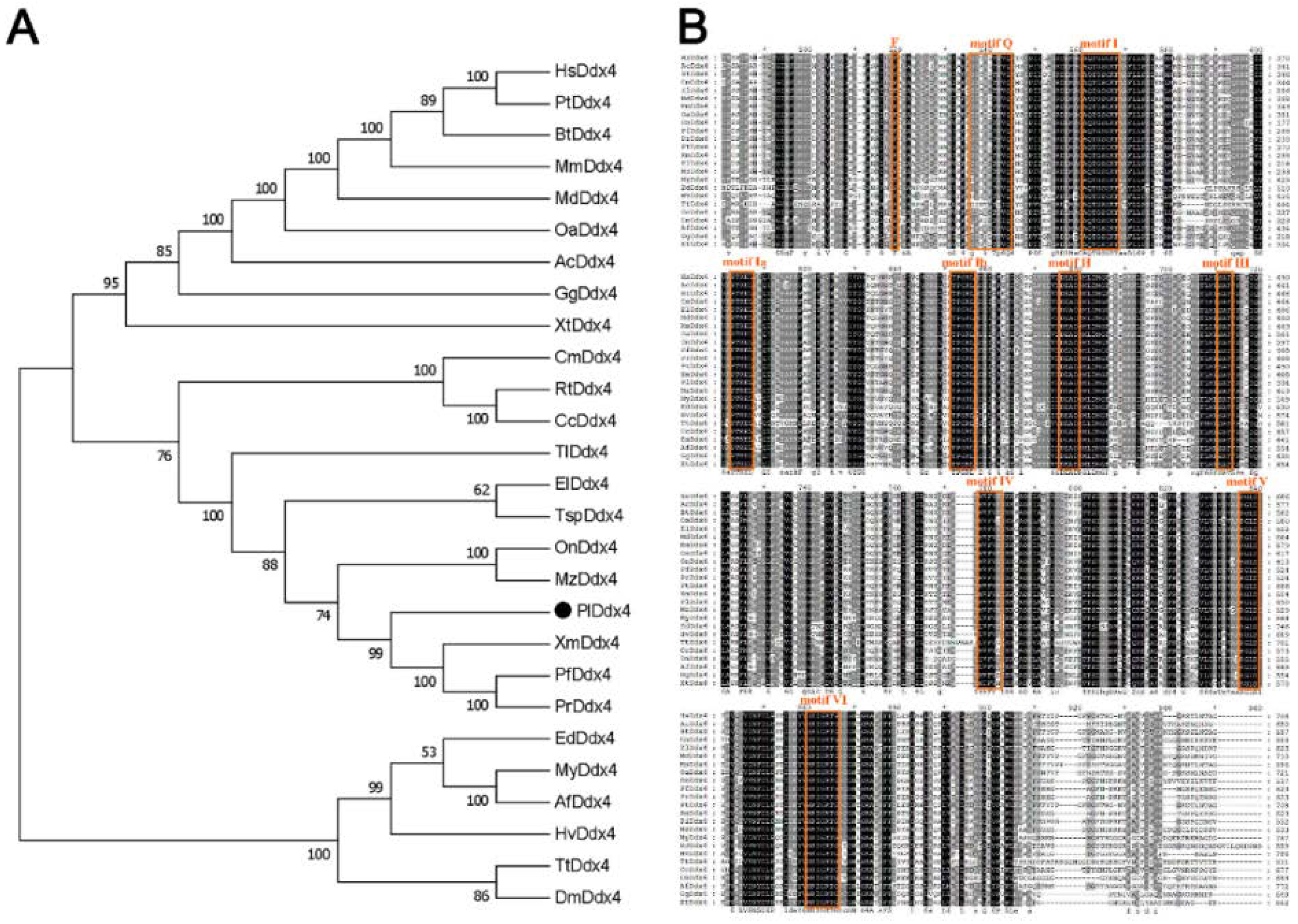
2.3. Phylogenetic Analysis
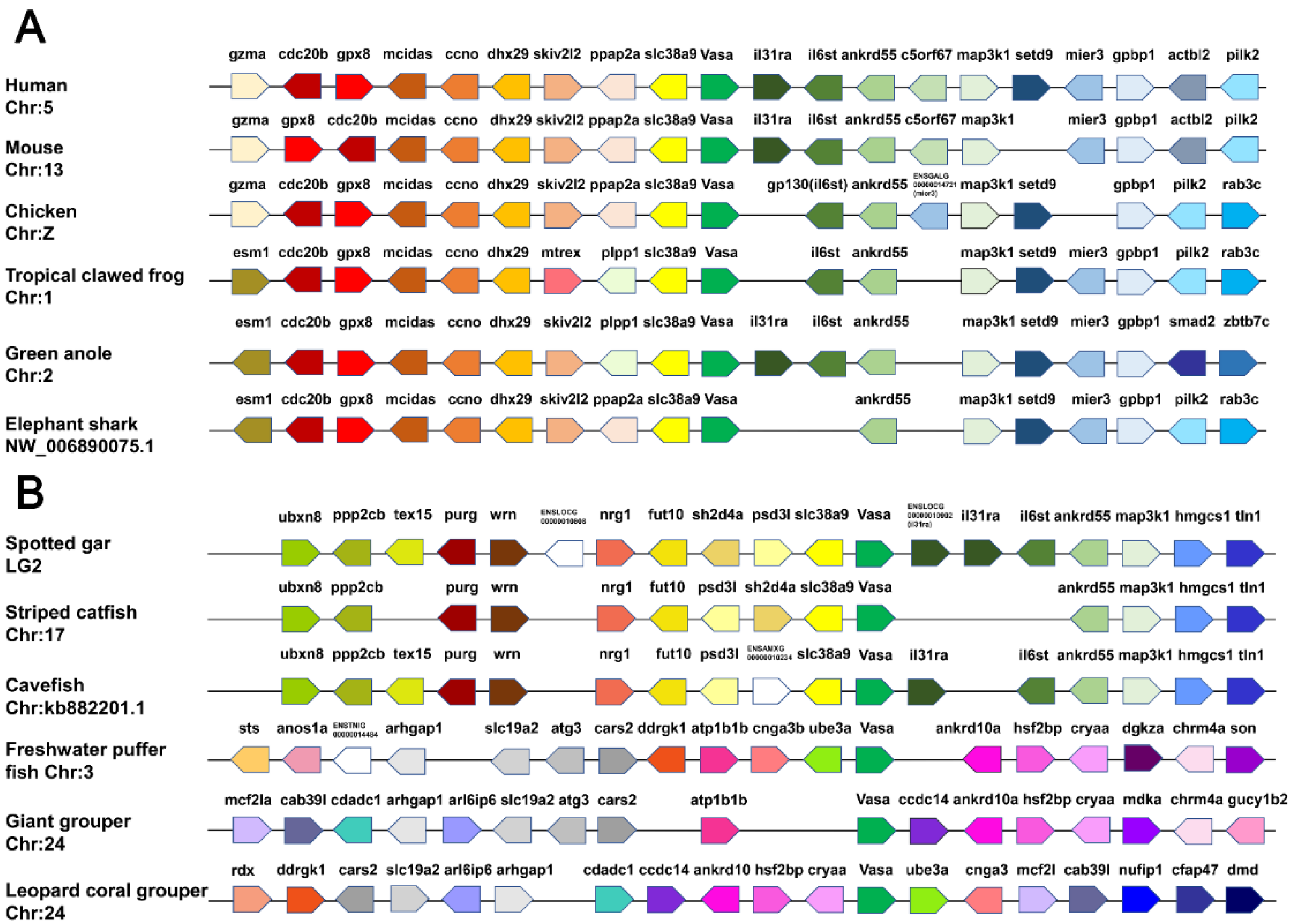
2.4. Synteny Analysis
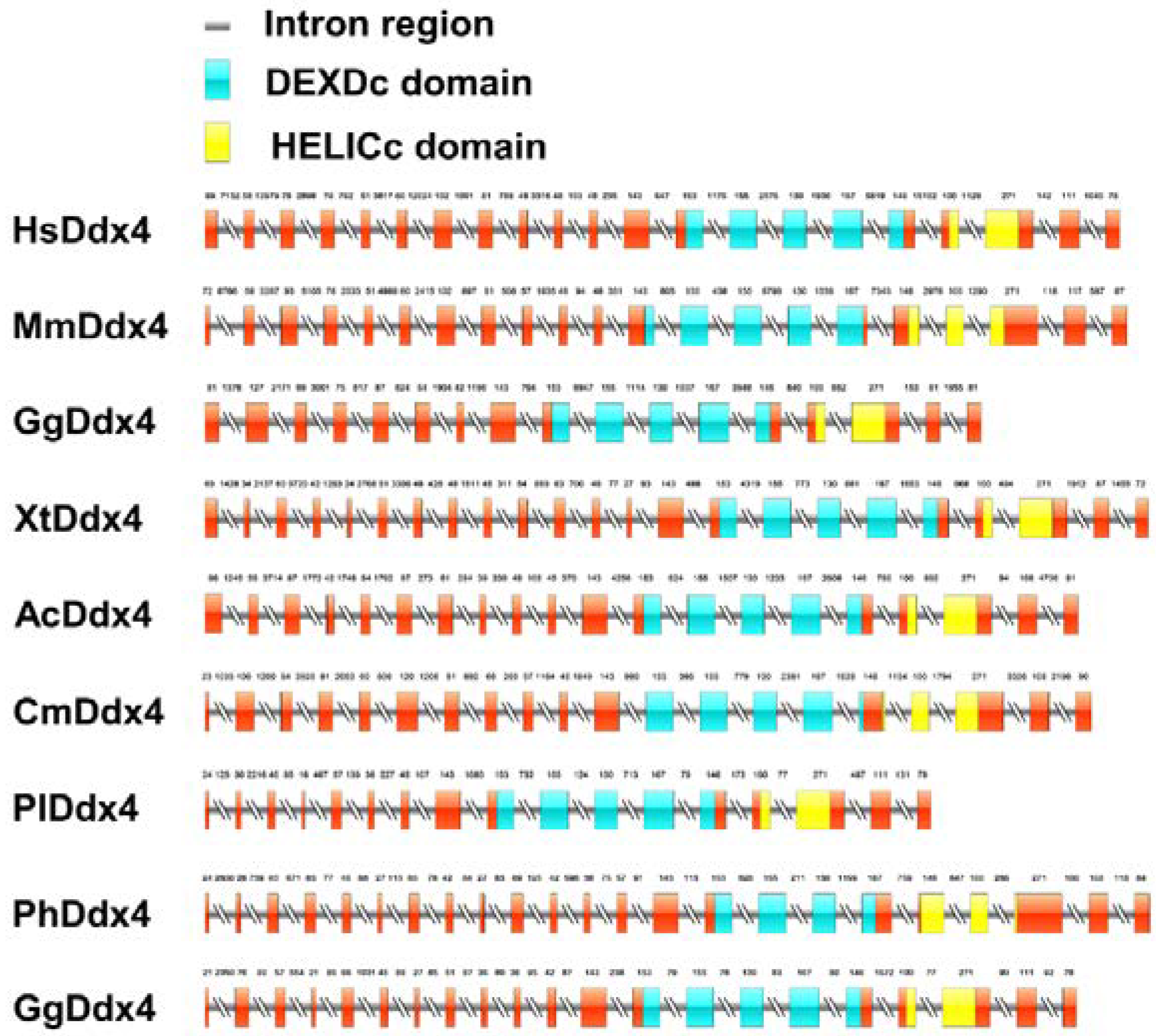
2.5. Genomic Structure Analysis
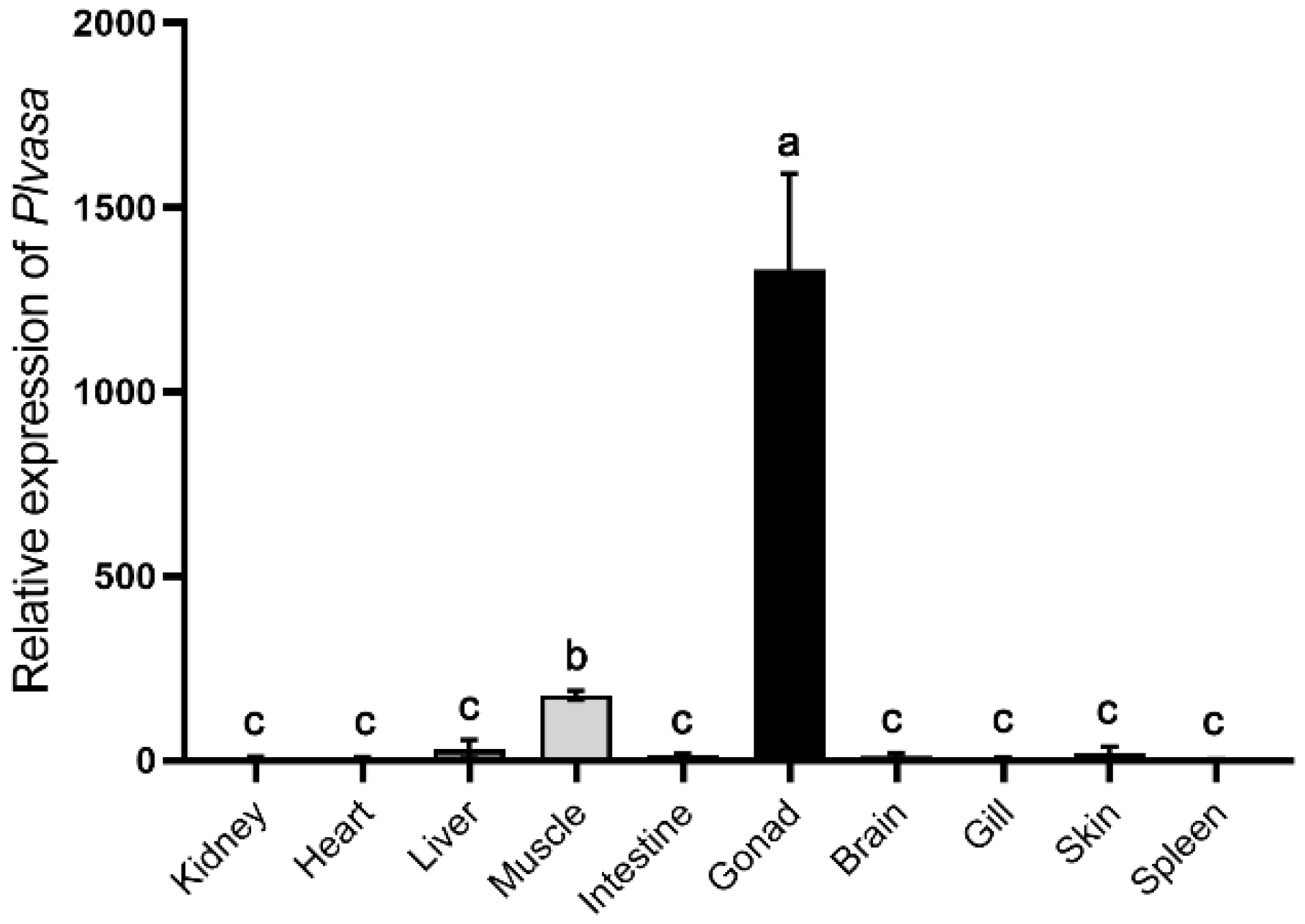
2.6. Quantitative Real-Time PCR (qPCR)
2.7. Fluorescence In Situ Hybridization (FISH)
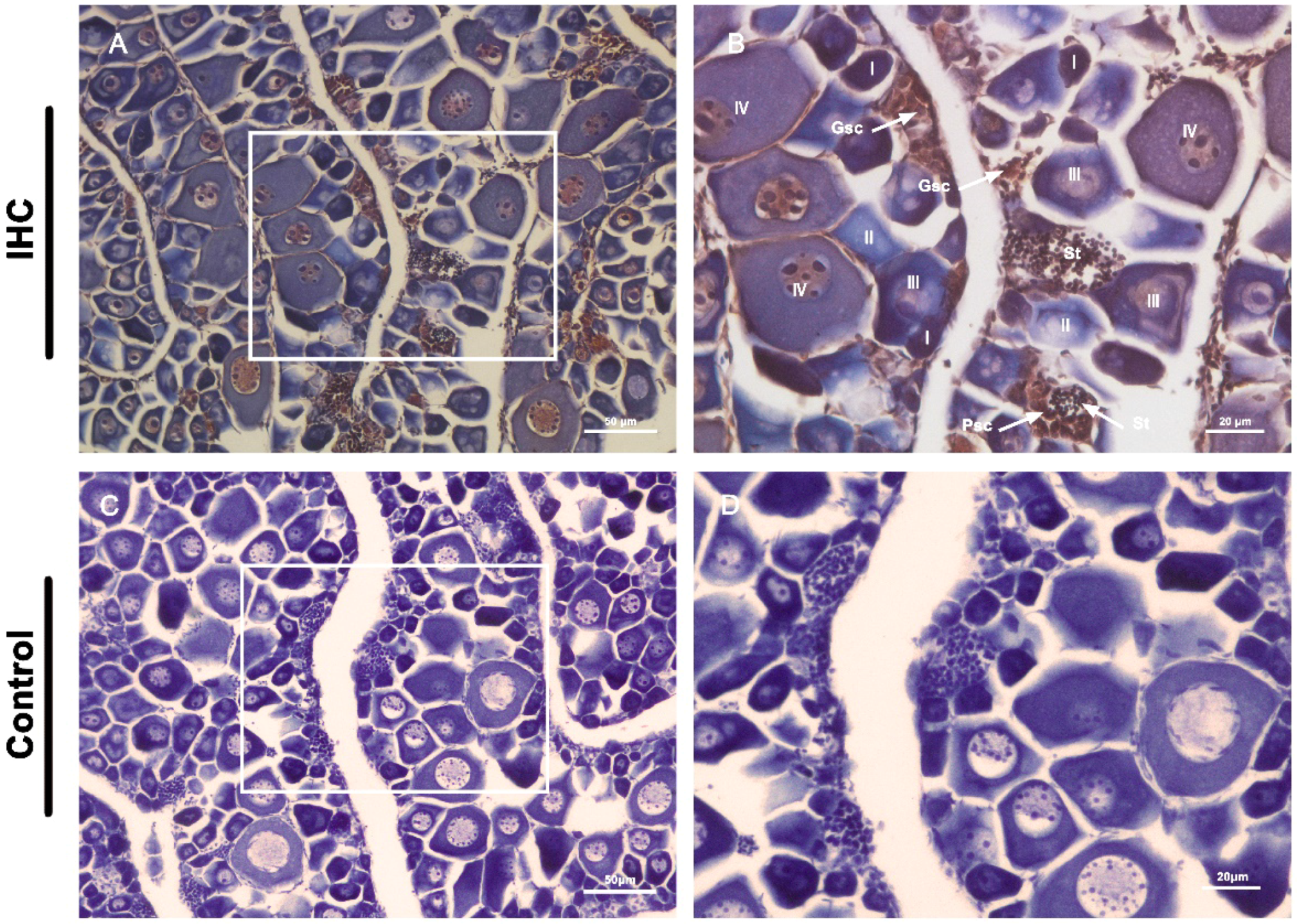
2.8. Immunohistochemistry (IHC)
2.9. Density Gradient Centrifugation and qPCR Analysis
3. Results
3.1. Phylogenetic Analysis of Vasa Genes
3.2. Gene Structure Analysis of Vasa Genes
3.3. Synteny Analyses of Vasa Genes
3.4. Gonad-Specific Expression of Vasa in Leopard Coral Grouper
3.5. Germline Cell-Specific Expression of Vasa in Leopard Coral Grouper
3.6. Density Gradient Centrifugation Enrichment and Relative Expression of Vasa in Different Types
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Campos-Ramos, R.; Garza-Torres, R.; Guerrero-Tortolero, D.A.; Maeda-Martinez, A.M.; Obregon-Barboza, H. Environmental sex determination, external sex differentiation and structure of the androgenic gland in the Pacific white shrimp Litopenaeus vannamei (Boone). Aquac. Res. 2006, 37, 1583–1593. [Google Scholar] [CrossRef]
- Okutsu, T.; Yano, A.; Nagasawa, K.; Shikina, S.; Kobayashi, T.; Takeuchi, Y.; Yoshizaki, G. Manipulation of fish germ cell: Visualization, cryopreservation and transplantation. J. Reprod. Dev. 2006, 52, 685–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Dong, J.; Yan, L.; Yong, J.; Liu, X.; Hu, Y.; Fan, X.; Wu, X.; Guo, H.; Wang, X.; et al. Single-Cell RNA-Seq Analysis Maps Development of Human Germline Cells and Gonadal Niche Interactions. Cell Stem Cell 2017, 20, 858–873.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Du, X.; Wang, H.; Jin, C.; Gao, C.; Liu, J.; Zhang, Q. Comparative studies on duplicated tdrd7 paralogs in teleosts: Molecular evolution caused neo-functionalization. Comp. Biochem. Physiol. D-Genom. Proteom. 2019, 30, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, M.; Gui, J.; Hong, Y. Fish germ cells. Sci. China-Life Sci. 2010, 53, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Ikenishi, K. Germ plasm in Caenorhabditis elegans, Drosophila and Xenopus. Dev. Growth Differ. 1998, 40, 1–10. [Google Scholar] [CrossRef]
- Martinez-Arroyo, A.M.; Miguez-Forjan, J.M.; Remohi, J.; Pellicer, A.; Medrano, J.V. Understanding Mammalian Germ Line Development with In Vitro Models. Stem Cells Dev. 2015, 24, 2101–2113. [Google Scholar] [CrossRef]
- Ubeda-Manzanaro, M.; Rebordinos, L.; Sarasquete, C. Cloning and characterization of Vasa gene expression pattern in adults of the Lusitanian toadfish Halobatrachus didactylus. Aquat. Biol. 2014, 21, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Knaut, H.; Pelegri, F.; Bohmann, K.; Schwarz, H.; Nusslein-Volhard, C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J. Cell Biol. 2000, 149, 875–888. [Google Scholar] [CrossRef] [Green Version]
- Fairman-Williams, M.E.; Guenther, U.-P.; Jankowsky, E. SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 2010, 20, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Linder, P.; Fuller-Pace, F.V. Looking back on the birth of DEAD-box RNA helicases. Biochim. Biophys. Acta-Gene Regul. Mech. 2013, 1829, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Rocak, S.; Linder, P. Dead-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004, 5, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Schupbach, T.; Wieschaus, E. Maternal-effect mutations altering the anterior-posterior pattern of the Drosophila embryo. Rouxs Arch. Dev. Biol. 1986, 195, 302–317. [Google Scholar] [CrossRef] [PubMed]
- Castrillon, D.H.; Quade, B.J.; Wang, T.Y.; Quigley, C.; Crum, C.P. The human VASA gene is specifically expressed in the germ cell lineage. Proc. Natl. Acad. Sci. USA 2000, 97, 9585–9590. [Google Scholar] [CrossRef] [Green Version]
- Toyooka, Y.; Tsunekawa, N.; Takahashi, Y.; Matsui, Y.; Satoh, M.; Noce, T. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech. Dev. 2000, 93, 139–149. [Google Scholar] [CrossRef]
- Komiya, T.; Itoh, K.; Ikenishi, K.; Furusawa, M. Isolation and Characterization of a Novel Gene of the DEAD Box Protein Family Which Is Specifically Expressed in Germ Cells of Xenopus laevis. Dev. Biol. 1994, 162, 354–363. [Google Scholar] [CrossRef]
- Olsen, L.C.; Aasland, R.; Fjose, A. A vasa-like gene in zebrafish identifies putative primordial germ cells. Mech. Dev. 1997, 66, 95–105. [Google Scholar] [CrossRef]
- Özhan-Kizil, G.; Havemann, J.; Gerberding, M. Germ cells in the crustacean Parhyale hawaiensis depend on Vasa protein for their maintenance but not for their formation. Dev. Biol. 2009, 327, 230–239. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Kajiura-Kobayashi, H.; Nagahama, Y. Differential expression of vasa homologue gene in the germ cells during oogenesis and spermatogenesis in a teleost fish, tilapia, Oreochromis niloticus. Mech. Dev. 2000, 99, 139–142. [Google Scholar] [CrossRef]
- Li, W.; Zhang, P.; Wu, X.; Zhu, X.; Xu, H. A Novel Dynamic Expression of vasa in Male Germ Cells during Spermatogenesis in the Chinese Soft-Shell Turtle (Pelidiscus sinensis). J. Exp. Zool. Part B-Mol. Dev. Evol. 2017, 328, 230–239. [Google Scholar] [CrossRef]
- Lasko, P.F.; Ashburner, M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature 1988, 335, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Hay, B.; Jan, L.Y.; Jan, Y.N. Localization of Vasa, a Component of Drosophila Polar Granules, in Maternal-Effect Mutants that alter Embryonic Anteroposterior Polarity. Development 1990, 109, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Noce, T.; Okamoto-Ito, S.; Tsunekawa, N. Vasa homolog genes in mammalian germ cell development. Cell Struct. Funct. 2001, 26, 131–136. [Google Scholar] [CrossRef] [Green Version]
- Braat, A.K.; van de Water, S.; Korving, J.; Zivkovic, D. A zebrafish vasa morphant abolishes vasa protein but does not affect the establishment of the germline. Genesis 2001, 30, 183–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartung, O.; Forbes, M.M.; Marlow, F.L. Zebrafish Vasa is Required for Germ-Cell Differentiation and Maintenance. Mol. Reprod. Dev. 2014, 81, 946–961. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Hong, N.; Xu, H.; Yi, M.; Li, C.; Gui, J.; Hong, Y. Medaka vasa is required for migration but not survival of primordial germ cells. Mech. Dev. 2009, 126, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Yajima, M.; Wessel, G.M. The multiple hats of Vasa: Its functions in the germline and in cell cycle progression. Mol. Reprod. Dev. 2011, 78, 861–867. [Google Scholar] [CrossRef] [Green Version]
- Poon, J.; Wessel, G.M.; Yajima, M. An unregulated regulator: Vasa expression in the development of somatic cells and in tumorigenesis. Dev. Biol. 2016, 415, 24–32. [Google Scholar] [CrossRef]
- Yajima, M.; Wessel, G.M. Essential elements for translation: The germline factor Vasa functions broadly in somatic cells. Development 2015, 142, 1960–1970. [Google Scholar] [CrossRef] [Green Version]
- Kuiter, R.H.; Tonozuka, T. Pictorial guide to Indonesian reef fishes. Part 3. Jawfishes—Sunfishes, Opistognathidae—Molidae. Zoonetics. Aust. Ref. 2001, 8637, 623–893. [Google Scholar]
- Ferreira, B.P. Reproduction of the Common Coral Trout Plectropomus Leopardus (Serranidae: Epinephelinae) from the Central and Northern Great Barrier Reef, Australia. Bull. Mar. Sci. 1995, 56, 653–669. [Google Scholar]
- Sadovy de Mitcheson, Y.; Liu, M. Functional hermaphroditism in teleosts. Fish Fish. 2008, 9, 1–43. [Google Scholar] [CrossRef]
- Allsop, D.J.; West, S.A. Constant relative age and size at sex change for sequentially hermaphroditic fish. J. Evol. Biol. 2003, 16, 921–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zupa, R.; Martino, N.A.; Marzano, G.; Dell’Aquila, M.E.; Corriero, A. Meagre Argyrosomus regius (Asso, 1801) Stem Spermatogonia: Histological Characterization, Immunostaining, In Vitro Proliferation, and Cryopreservation. Animals 2020, 10, 851. [Google Scholar] [CrossRef]
- He, Y.-Q.; Chen, J.; Lu, X.-J.; Shi, Y.-H. Characterization of P2 × 7R and Its Function in the Macrophages of ayu, Plecoglossus altivelis. PLoS ONE 2013, 8, e57505. [Google Scholar] [CrossRef] [Green Version]
- Lanave, C.; Licciulli, F.; De Robertis, M.; Marolla, A.; Attimonelli, M. Update of AMmtDB: A database of multi-aligned Metazoa mitochondrial DNA sequences. Nucleic Acids Res. 2002, 30, 174–175. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Muffato, M.; Louis, A.; Poisnel, C.E.; Roest Crollius, H. Genomicus: A database and a browser to study gene synteny in modern and ancestral genomes. Bioinformatics 2010, 26, 1119–1121. [Google Scholar] [CrossRef]
- Zhou, Q.; Guo, X.; Huang, Y.; Gao, H.; Xu, H.; Liu, S.; Zheng, W.; Zhang, T.; Tian, C.; Zhu, C.; et al. De novo sequencing and chromosomal-scale genome assembly of leopard coral grouper, Plectropomus leopardus. Mol. Ecol. Resour. 2020, 20, 1403–1413. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Chen, X.; Sun, Y.; Zhang, P.; Li, J.; Li, H.; Wei, C.; Cao, Z.; Zhou, Y. Screening of stable internal reference genes by quantitative real-time PCR in humpback grouper Cromileptes altivelis. J. Oceanol. Limnol. 2021, 39, 1985–1999. [Google Scholar] [CrossRef]
- Chi, L.; Li, X.; Liu, Q.; Liu, Y. Photoperiod regulate gonad development via kisspeptin/kissr in hypothalamus and saccus vasculosus of Atlantic salmon (Salmo solar). PLoS ONE 2017, 12, e0169569. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Wang, X.; Zhang, Q. Evolutionary Conservation of pou5f3 Genomic Organization and Its Dynamic Distribution during Embryogenesis and in Adult Gonads in Japanese Flounder Paralichthys olivaceus. Int. J. Mol. Sci. 2017, 18, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.-C.; Hsu, H.-J.; Lin, G.-W.; Wang, T.-F.; Chang, C.-C.; Lin, M.-D. Germ plasm localisation of the HELICc of Vasa in Drosophila: Analysis of domain sufficiency and amino acids critical for localisation. Sci. Rep. 2015, 5, 14703. [Google Scholar] [CrossRef] [Green Version]
- Hickford, D.E.; Frankenberg, S.; Pask, A.J.; Shaw, G.; Renfree, M.B. DDX4 (VASA) Is Conserved in Germ Cell Development in Marsupials and Monotremes. Biol. Reprod. 2011, 85, 733–743. [Google Scholar] [CrossRef] [Green Version]
- Boonanuntanasarn, S.; Bunlipatanon, P.; Ichida, K.; Yoohat, K.; Mengyu, O.; Detsathit, S.; Yazawa, R.; Yoshizaki, G. Characterization of a vasa homolog in the brown-marbled grouper (Epinephelus fuscoguttatus) and its expression in gonad and germ cells during larval development. Fish Physiol. Biochem. 2016, 42, 1621–1636. [Google Scholar] [CrossRef]
- Sengoku, T.; Nureki, O.; Dohmae, N.; Nakamura, A.; Yokoyama, S. Crystallization and preliminary X-ray analysis of the helicase domains of Vasa complexed with RNA and an ATP analogue. Acta Crystallogr. Sect. D-Struct. Biol. 2004, 60, 320–322. [Google Scholar] [CrossRef] [Green Version]
- Sengoku, T.; Nureki, O.; Nakamura, A.; Kobayashi, S.; Yokoyama, S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 2006, 125, 287–300. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Liu, W.; Zhou, N.; Wang, H.; Li, P.; Wang, M.; Zhang, Q. Molecular characterization, origin, and evolution of teleost p68 gene family: Insights from Japanese flounder, Paralichthys olivaceus. Mar. Genom. 2015, 24, 363–370. [Google Scholar] [CrossRef]
- Glasauer, S.M.K.; Neuhauss, S.C.F. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef] [Green Version]
- Pasquier, J.; Cabau, C.; Thaovi, N.; Jouanno, E.; Severac, D.; Braasch, I.; Journot, L.; Pontarotti, P.; Klopp, C.; Postlethwait, J.H.; et al. Gene evolution and gene expression after whole genome duplication in fish: The PhyloFish database. BMC Genom. 2016, 17, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davesne, D.; Friedman, M.; Schmitt, A.D.; Fernandez, V.; Carnevale, G.; Ahlberg, P.E.; Sanchez, S.; Benson, R.B.J. Fossilized cell structures identify an ancient origin for the teleost whole-genome duplication. Proc. Natl. Acad. Sci. USA 2021, 118, e2101780118. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-J.; Liu, L.; Chen, X.-H.; Zhang, T.; Gan, F.; Cheng, B.-L. Identification of a vasa homologue gene in grass carp and its expression pattern in tissues and during embryogenesis. Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 2010, 157, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Jung, H.J.; Yoon, M.J. VASA (DDX4) is a Putative Marker for Spermatogonia, Spermatocytes and Round Spermatids in Stallions. Reprod. Domest. Anim. 2015, 50, 1032–1038. [Google Scholar] [CrossRef]
- Du, S.; Zhou, L.; Wang, X.; Xu, S.; Li, J.; Song, Z.; Liu, Q. Characterization of vasa and dnd homologs in summer flounder, Paralichthys dentatus: Expression analysis and colocalization of PGCs during embryogenesis. Theriogenology 2022, 181, 180–189. [Google Scholar] [CrossRef]
- Ikenishi, K.; Tanaka, T.S. Spatio-temporal expression of Xenopus vasa homolog, XVLG1, in oocytes and embryos: The presence of XVLG1 RNA in somatic cells as well as germline cells. Dev. Growth Differ. 2000, 42, 95–103. [Google Scholar] [CrossRef]
- Wang, Z.; Gao, J.; Song, H.; Wu, X.; Sun, Y.; Qi, J.; Yu, H.; Wang, Z.; Zhang, Q. Sexually dimorphic expression of vasa isoforms in the tongue sole (Cynoglossus semilaevis). PLoS ONE 2014, 9, e93380. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.H.; Davie, A.; Migaud, H. Expression pattern of nanos, piwil, dnd, vasa and pum genes during ontogenic development in Nile tilapia Oreochromis niloticus. Gene 2019, 688, 62–70. [Google Scholar] [CrossRef] [Green Version]
- Qu, L.; Wu, X.; Liu, M.; Zhong, C.; Xu, H.; Li, S.; Lin, H.; Liu, X. Identification and characterization of germ cell genes vasa and dazl in a protogynous hermaphrodite fish, orange-spotted grouper (Epinephelus coioides). Gene Expr. Patterns 2020, 35, 119095. [Google Scholar] [CrossRef]
- Bhat, N.; Hong, Y. Cloning and expression of boule and dazl in the Nile tilapia (Oreochromis niloticus). Gene 2014, 540, 140–145. [Google Scholar] [CrossRef]
- Xu, H.; Lim, M.; Dwarakanath, M.; Hong, Y. Vasa Identifies Germ Cells and Critical Stages of Oogenesis in the Asian Seabass. Int. J. Biol. Sci. 2014, 10, 225–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, T.; Yu, M.; Pan, Q.; Wang, Y.; Fang, J.; Li, L.; Deng, Y.; Chen, K.; Wang, Q.; Chen, T. Black carp vasa identifies embryonic and gonadal germ cells. Dev. Genes Evol. 2017, 227, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Rzepkowska, M.; Ostaszewska, T. Proliferating cell nuclear antigen and Vasa protein expression during gonadal development and sexual differentiation in cultured Siberian (Acipenser baerii Brandt, 1869) and Russian (Acipenser gueldenstaedtii Brandt & Ratzeburg, 1833) sturgeon. Rev. Aquac. 2014, 6, 75–88. [Google Scholar] [CrossRef]
- Ye, H.; Yue, H.-M.; Yang, X.-G.; Li, C.-J.; Wei, Q. Identification and sexually dimorphic expression of vasa isoforms in Dabry’s sturgeon (Acipenser dabryanus), and functional analysis of vasa 3’-untranslated region. Cell Tissue Res. 2016, 366, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Yang, Y.; Xu, H.; Duan, J.; Cheng, N.; Wang, J.; Hu, W.; Zhao, H. Germ cell specific expression of Vasa in rare minnow, Gobiocypris rarus. Comp. Biochem. Physiol. A-Mol. Integr. Physiol. 2012, 162, 163–170. [Google Scholar] [CrossRef]
- Ye, H.; Li, C.-J.; Yue, H.-M.; Yang, X.-G.; Wei, Q.-W. Differential expression of fertility genes boule and dazl in Chinese sturgeon (Acipenser sinensis), a basal fish. Cell Tissue Res. 2015, 360, 413–425. [Google Scholar] [CrossRef]
- Strome, S.; Updike, D. Specifying and protecting germ cell fate. Nat. Rev. Mol. Cell Biol. 2015, 16, 406–416. [Google Scholar] [CrossRef]
- Vasconcelos, A.C.N.; Streit, D.P., Jr.; Octavera, A.; Miwa, M.; Kabeya, N.; Freitas Garcia, R.R.; Rotili, D.A.; Yoshizaki, G. Isolation and characterization of a germ cell marker in teleost fish Colossoma macropomum. Gene 2019, 683, 54–60. [Google Scholar] [CrossRef]
- Yoshizaki, G.; Tago, Y.; Takeuchi, Y.; Sawatari, E.; Kobayashi, T.; Takeuchi, T. Green fluorescent protein labeling of primordial germ cells using a nontransgenic method and its application for germ cell transplantation in Salmonidae. Biol. Reprod. 2005, 73, 88–93. [Google Scholar] [CrossRef]
- Huang, J.; Chen, S.; Liu, Y.; Shao, C.; Lin, F.; Wang, N.; Hu, Q. Molecular characterization, sexually dimorphic expression, and functional analysis of 3’-untranslated region of vasa gene in half-smooth tongue sole (Cynoglossus semilaevis). Theriogenology 2014, 82, 213–224. [Google Scholar] [CrossRef]


| Purpose | 5′ to 3′ Sequence |
|---|---|
| Vasa-PCR-Fw | ATGGACGAGTGGGATGAA |
| Vasa-PCR-Rv | CACCGTTGTCCTGGAAAG |
| Vasa-qRT-PCR-Fw | AAGACAGACTACTTGTTCTTGGCTG |
| Vasa-qRT-PCR-Rv | CAAGGAGCTGATCTCTCTTGCA |
| Vasa-ISH-Fw | ATTTAGGTGACACTATAGCTGATTTCCTCGCCGCTT |
| Vasa-ISH-Rv | TAATACGACTCACTATAGGGTGGCTCTTCACACCGTTGTC |
| Rpl13-qRT-PCR-Fw | CAGCGTCTGAAGGAGTACCG |
| Rpl13-qRT-PCR-Rv | ACCAGCGAGCTGAGTGGC |
| B2m-qRT-PCR-Fw | GGCAATTCCACCTGACCAAG |
| B2m-qRT-PCR-Rv | CAACCCAGGCATATTCCTTAACT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Ding, H.; Wu, S.; Wang, M.; Wei, C.; Wang, B.; Bao, Z.; Hu, J. Vasa Is a Potential Germ Cell Marker in Leopard Coral Grouper (Plectropomus leopardus). Genes 2022, 13, 1077. https://doi.org/10.3390/genes13061077
Wang M, Ding H, Wu S, Wang M, Wei C, Wang B, Bao Z, Hu J. Vasa Is a Potential Germ Cell Marker in Leopard Coral Grouper (Plectropomus leopardus). Genes. 2022; 13(6):1077. https://doi.org/10.3390/genes13061077
Chicago/Turabian StyleWang, Mingyi, Hui Ding, Shaoxuan Wu, Mengya Wang, Cun Wei, Bo Wang, Zhenmin Bao, and Jingjie Hu. 2022. "Vasa Is a Potential Germ Cell Marker in Leopard Coral Grouper (Plectropomus leopardus)" Genes 13, no. 6: 1077. https://doi.org/10.3390/genes13061077





