Comparative Analysis of Italian Lettuce (Lactuca sativa L. var. ramose) Transcriptome Profiles Reveals the Molecular Mechanism on Exogenous Melatonin Preventing Cadmium Toxicity
Abstract
:1. Introduction
2. Materials and Methods
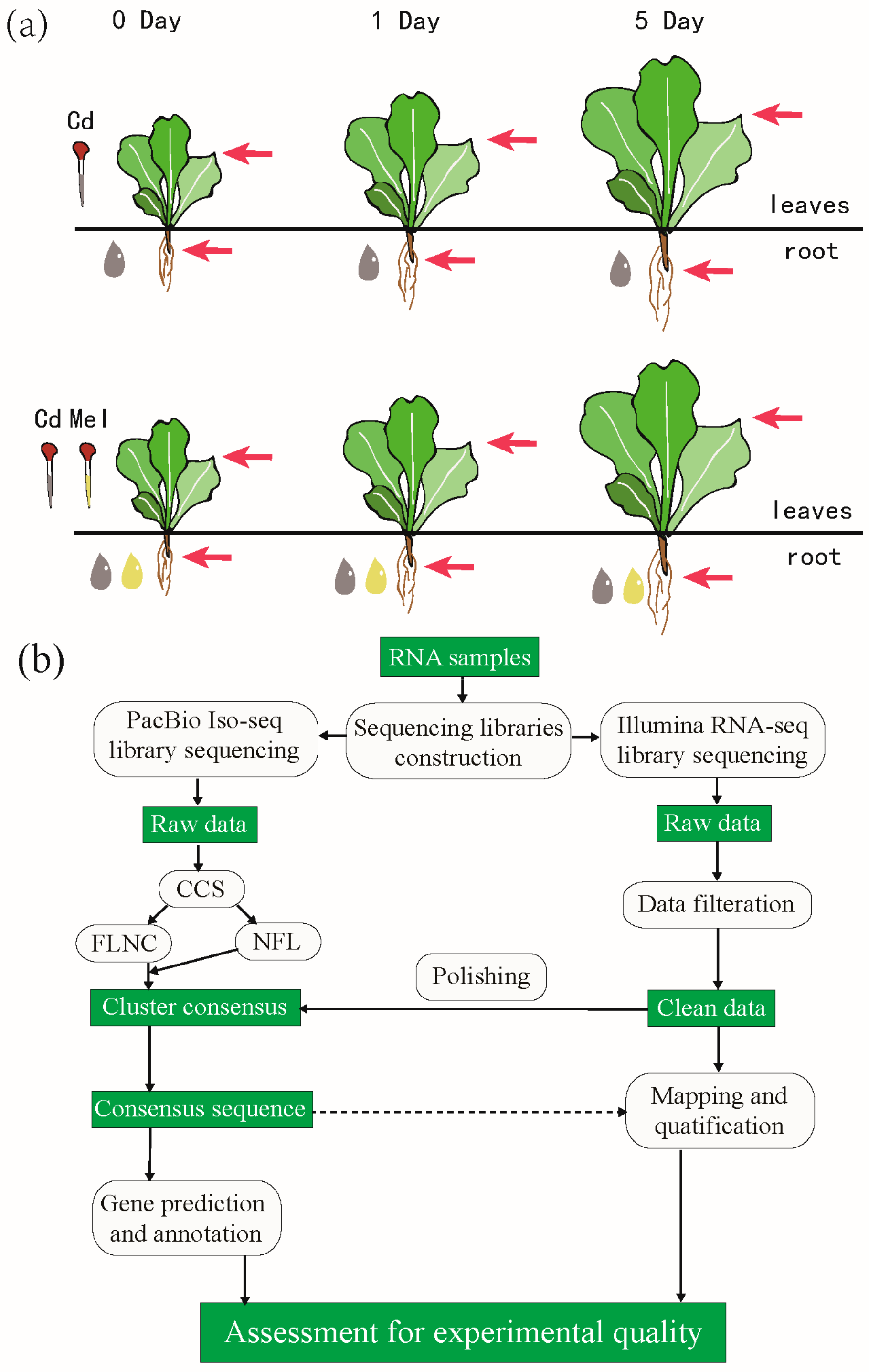
2.1. Sample Preparation and Treatment
2.2. Library Construction and Sequencing
2.3. Subread Processing and Error Correction
2.4. Transcriptome Characterizing and Annotation
2.5. Differential Expression Analysis and Function Enrichment
3. Results
3.1. Transcriptome Features and Description
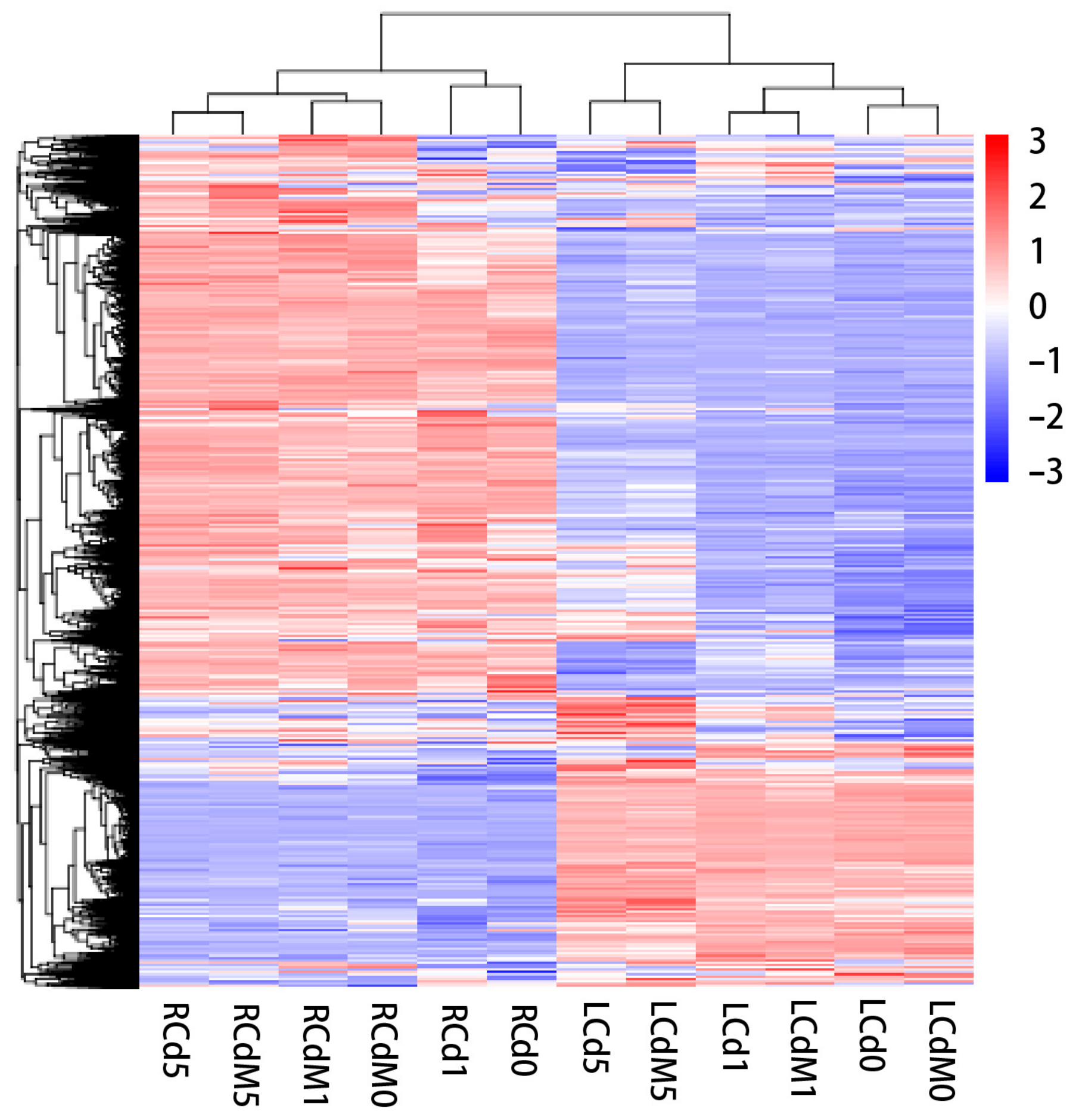
3.2. Expression Pattern during Cd Stress and Melatonin Treatment
3.3. Functional Enrichment during Cd Response and Melatonin Treatment
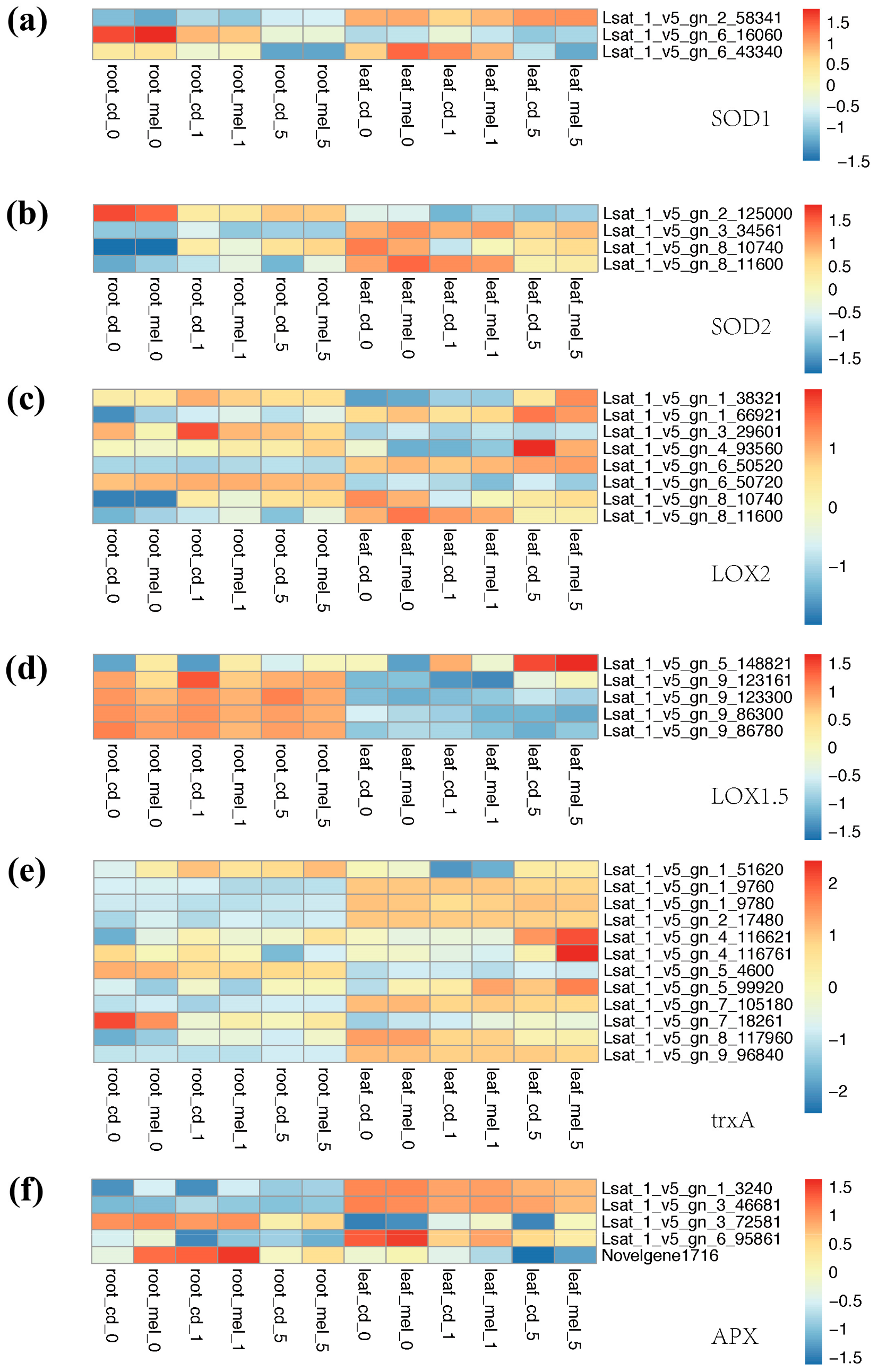
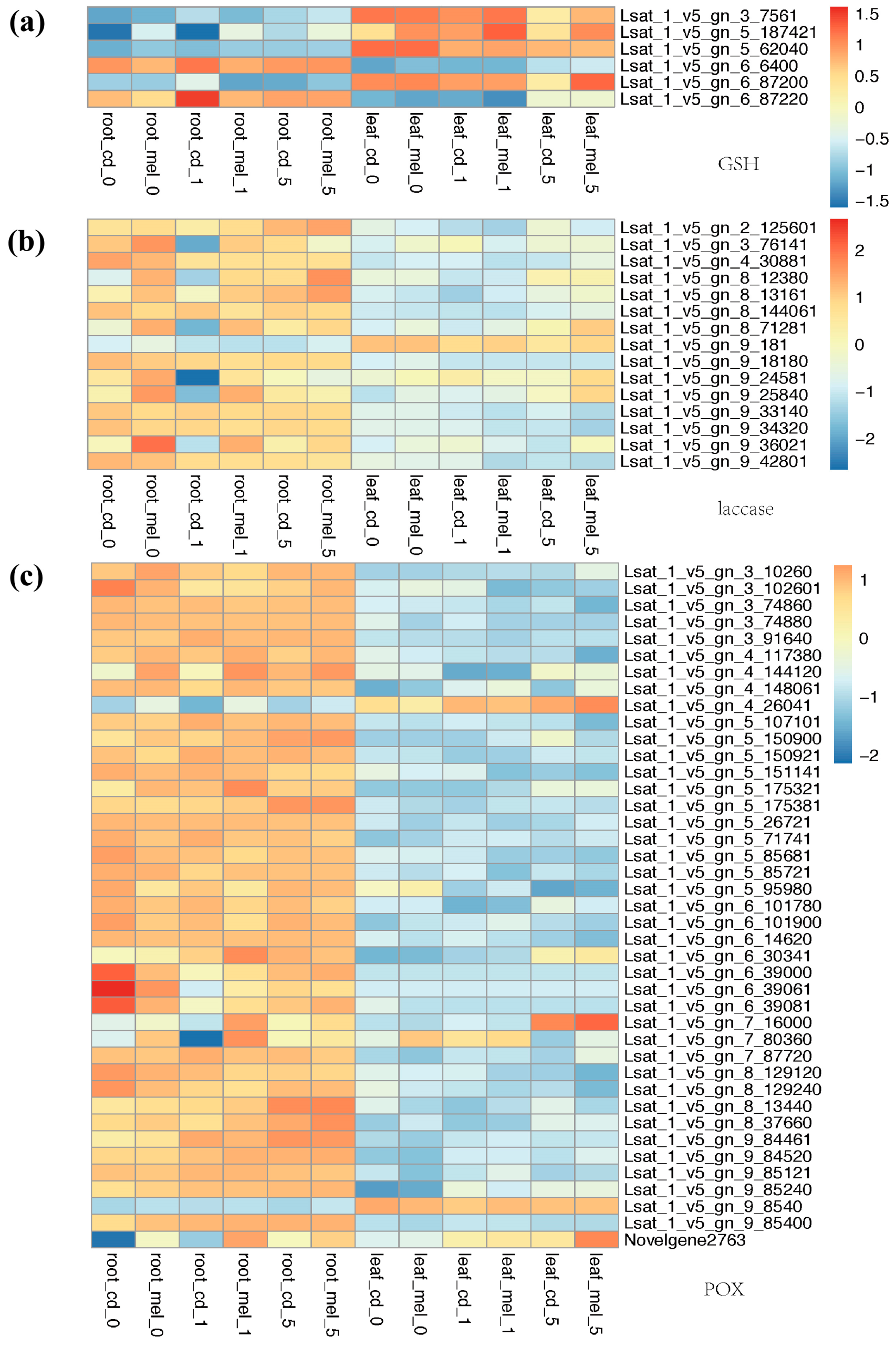
3.4. Functional Divergence of Genes Involved in Antioxidant System
3.5. Functional Divergence of Genes Involved in Hormone Signaling Transduction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chaney, R.L. How does contamination of rice soils with Cd and Zn cause high incidence of human Cd disease in subsistence rice farmers. Curr. Pollut. Rep. 2015, 1, 13–22. [Google Scholar] [CrossRef]
- Khan, A.; Khan, S.; Alam, M.; Khan, M.A.; Aamir, M.; Qamar, Z.; Rehman, Z.U.; Perveen, S. Toxic metal interactions affect the bioaccumulation and dietary intake of macro- and micro-nutrients. Chemosphere 2016, 146, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Adrees, M.; Ali, S.; Rizwan, M.; Ibrahim, M.; Abbas, F.; Farid, M.; Zia-Ur-Rehman, M.; Irshad, M.K.; Bharwana, S.A. The effect of excess copper on growth and physiology of important food crops: A review. Environ. Sci. Pollut. Res. 2015, 22, 8148–8162. [Google Scholar] [CrossRef] [PubMed]
- Yousaf, B.; Liu, G.; Wang, R.; Imtiaz, M.; Zia-Ur-Rehman, M.; Munir, M.A.; Niu, Z. Bioavailability evaluation, uptake of heavy metals and potential health risks via dietary exposure in urban-industrial areas. Environ. Sci. Pollut. Res. Int. 2016, 23, 22443–22453. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Ali, S.; Abbas, T.; Zia-Ur-Rehman, M.; Hannan, F.; Keller, C.; Al-Wabel, M.I.; Ok, Y.S. Cadmium minimization in wheat: A critical review. Ecotoxicol. Environ. Saf. 2016, 130, 43–53. [Google Scholar] [CrossRef]
- Rizwan, M.; Ali, S.; Adrees, M.; Rizvi, H.; Zia-Ur-Rehman, M.; Hannan, F.; Qayyum, M.F.; Hafeez, F.; Ok, Y.S. Cadmium stress in rice: Toxic effects, tolerance mechanisms, and management: A critical review. Environ. Sci. Pollut. Res. Int. 2016, 23, 17859–17879. [Google Scholar] [CrossRef]
- Rizwan, M.; Ali, S.; Adrees, M.; Ibrahim, M.; Tsang, D.C.W.; Zia-Ur-Rehman, M.; Zahir, Z.A.; Rinklebe, J.; Tack, F.M.G.; Ok, Y.S. A critical review on effects, tolerance mechanisms and management of cadmium in vegetables. Chemosphere 2017, 182, 90–105. [Google Scholar] [CrossRef]
- Huang, B.; Xin, J.; Dai, H.; Liu, A.; Zhou, W.; Yi, Y.; Liao, K. Root morphological responses of three hot pepper cultivars to Cd exposure and their correlations with Cd accumulation. Environ. Sci. Pollut. Res. Int. 2015, 22, 1151–1159. [Google Scholar] [CrossRef]
- Xin, J.; Huang, B. Subcellular distribution and chemical forms of cadmium in two hot pepper cultivars differing in cadmium accumulation. J. Agric. Food Chem. 2014, 62, 508–515. [Google Scholar] [CrossRef]
- Hassan, W.; Bano, R.; Bashir, S.; Aslam, Z. Cadmium toxicity and soil biological index under potato (Solanum tuberosum L.) cultivation. Soil Res. 2016, 54, 460–468. [Google Scholar] [CrossRef]
- Hédiji, H.; Djebali, W.; Belkadhi, A.; Cabasson, C.; Moing, A.; Rolin, D.; Brouquisse, R.; Gallusci, P.; Chaïbi, W. Impact of long-term cadmium exposure on mineral content of Solanum lycopersicum plants: Consequences on fruit production. S. Afr. J. Bot. 2015, 97, 176–181. [Google Scholar] [CrossRef]
- Zhou, Q.; Guo, J.J.; He, C.T.; Shen, C.; Huang, Y.Y.; Chen, J.X.; Guo, J.H.; Yuan, J.G.; Yang, Z.Y. Comparative Transcriptome Analysis between Low- and High-Cadmium-Accumulating Genotypes of Pakchoi (Brassica chinensis L.) in Response to Cadmium Stress. Environ. Sci. Technol. 2016, 50, 6485–6494. [Google Scholar] [CrossRef] [PubMed]
- Zorrig, W.; Khouni, A.E.; Ghnaya, T.; Davidian, J.C.; Berthomieu, P. Lettuce (lactuca sativa): A species with a high capacity for cadmium (Cd) accumulation and growth stimulation in the presence of low cd concentrations. J. Hortic. Sci. Biotechnol. 2013, 88, 783–789. [Google Scholar] [CrossRef]
- Xue, Z.; Gao, H.; Zhao, S. Effects of cadmium on the photosynthetic activity in mature and young leaves of soybean plants. Environ. Sci. Pollut. Res. Int. 2014, 21, 4656–4664. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wang, Q.; Shah, F.A.; Liu, W.; Wang, D.; Huang, S.; Fu, S.; Wu, L. Exogenous Melatonin Confers Cadmium Tolerance by Counterbalancing the Hydrogen Peroxide Homeostasis in Wheat Seedlings. Molecules 2018, 23, 799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, D.X.; Manchester, L.C.; Esteban-Zubero, E.; Zhou, Z.; Reiter, R.J. Melatonin as a Potent and Inducible Endogenous Antioxidant: Synthesis and Metabolism. Molecules 2015, 20, 8886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawaz, M.A.; Huang, Y.; Bie, Z.; Ahmed, W.; Reiter, R.J.; Niu, M.; Hameed, S. Melatonin: Current Status and Future Perspectives in Plant Science. Front. Plant Sci. 2016, 6, 1230. [Google Scholar] [CrossRef] [Green Version]
- Arnao, M.B.; Hernández-Ruiz, J. Functions of melatonin in plants: A review. J. Pineal Res. 2015, 59, 133–150. [Google Scholar] [CrossRef] [Green Version]
- Cui, G.; Zhao, X.; Liu, S.; Sun, F.; Zhang, C.; Xi, Y. Beneficial effects of melatonin in overcoming drought stress in wheat seedlings. Plant Physiol. Biochem. 2017, 118, 138–149. [Google Scholar] [CrossRef]
- Qi, Z.Y.; Wang, K.X.; Yan, M.Y.; Kanwar, M.K.; Li, D.Y.; Wijaya, L.; Alyemeni, M.N.; Ahmad, P.; Zhou, J. Melatonin Alleviates High Temperature-Induced Pollen Abortion in Solanum lycopersicum. Molecules 2018, 23, 386. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wei, J.P.; Scott, E.R.; Liu, J.W.; Guo, S.; Li, Y.; Zhang, L.; Han, W.Y. Exogenous Melatonin Alleviates Cold Stress by Promoting Antioxidant Defense and Redox Homeostasis in Camellia sinensis L. Molecules 2018, 23, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, C.; Zheng, G.; Li, W.; Wang, Y.; Hu, B.; Wang, H.; Wu, H.; Qian, Y.; Zhu, X.G.; Tan, D.X.; et al. Melatonin delays leaf senescence and enhances salt stress tolerance in rice. J. Pineal Res. 2015, 59, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.K.; Ahammed, G.J.; Yin, L.; Shi, K.; Xia, X.; Zhou, Y.; Yu, J.; Zhou, J. Melatonin mitigates cadmium phytotoxicity through modulation of phytochelatins biosynthesis, vacuolar sequestration, and antioxidant potential in Solanum lycopersicum L. Front. Plant Sci. 2015, 6, 601. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Q.; Hasan, M.K.; Li, C.X.; Ahammed, G.J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Reiter, R.J.; Yu, J.Q.; Xu, M.X.; et al. Melatonin mediates selenium-induced tolerance to cadmium stress in tomato plants. J. Pineal Res. 2016, 61, 291–302. [Google Scholar] [CrossRef]
- Wei, W.; Li, Q.T.; Chu, Y.N.; Reiter, R.J.; Yu, X.M.; Zhu, D.H.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, J.S.; et al. Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 2015, 66, 695–707. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.J.; Zhang, N.; Yang, R.C.; Wang, L.; Sun, Q.Q.; Li, D.B.; Cao, Y.Y.; Weeda, S.; Zhao, B.; Ren, S.; et al. Melatonin promotes seed germination under high salinity by regulating antioxidant systems, ABA and GA4 interaction in cucumber (Cucumis sativus L.). J. Pineal Res. 2014, 57, 269–279. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, Y.; Zhang, X.Z.; Du, H.M.; Xu, B.; Huang, B.R. Melatonin suppression of heat-induced leaf senescence involves changes in abscisic acid and cytokinin biosynthesis and signaling pathways in perennial ryegrass (Lolium perenne L.). Environ. Exp. Bot. 2017, 138, 36–45. [Google Scholar] [CrossRef]
- Rodriguez, C.; Mayo, J.C.; Sainz, R.M.; Antolín, I.; Herrera, F.; Martín, V.; Reiter, R.J. Regulation of antioxidant enzymes: A significant role for melatonin. J. Pineal Res. 2004, 36, 1–9. [Google Scholar] [CrossRef]
- Zhang, N.; Sun, Q.; Zhang, H.; Cao, Y.; Weeda, S.; Ren, S.; Guo, Y.D. Roles of melatonin in abiotic stress resistance in plants. J. Exp. Bot. 2015, 66, 647–656. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Yin, L.; Liang, D.; Li, C.; Ma, F.; Yue, Z. Delayed senescence of apple leaves by exogenous melatonin treatment: Toward regulating the ascorbate-glutathione cycle. J. Pineal Res. 2012, 53, 11–20. [Google Scholar] [CrossRef]
- Weeda, S.; Zhang, N.; Zhao, X.; Ndip, G.; Guo, Y.; Buck, G.A.; Fu, C.; Ren, S. Arabidopsis transcriptome analysis reveals key roles of melatonin in plant defense systems. PLoS ONE 2014, 9, 93462. [Google Scholar] [CrossRef] [Green Version]
- Gu, Q.; Chen, Z.; Yu, X.; Cui, W.; Pan, J.; Zhao, G.; Xu, S.; Wang, R.; Shen, W. Melatonin confers plant tolerance against cadmium stress via the decrease of cadmium accumulation and reestablishment of microRNA-mediated redox homeostasis. Plant Sci. 2017, 261, 28–37. [Google Scholar] [CrossRef]
- Cai, S.Y.; Zhang, Y.; Xu, Y.P.; Qi, Z.Y.; Li, M.Q.; Ahammed, G.J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Reiter, R.J.; et al. HsfA1a upregulates melatonin biosynthesis to confer cadmium tolerance in tomato plants. J. Pineal Res. 2017, 62, e12387. [Google Scholar] [CrossRef] [PubMed]
- Kolukisaoglu, H.U.; Bovet, L.; Klein, M.; Eggmann, T.; Geisler, M.; Wanke, D.; Martinoia, E.; Schulz, B. Family business: The multidrug-resistance related protein (MRP) ABC transporter genes in Arabidopsis thaliana. Planta 2002, 216, 107–119. [Google Scholar] [CrossRef]
- Kim, D.Y.; Bovet, L.; Maeshima, M.; Martinoia, E.; Lee, Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007, 50, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Vijver, M.G.; Peijnenburg, W.J. Impacts of major cations (K(+), Na (+), Ca (2+), Mg (2+)) and protons on toxicity predictions of nickel and cadmium to lettuce (Lactuca sativa L.) using exposure models. Ecotoxicology 2014, 23, 385–395. [Google Scholar] [CrossRef]
- Kim, S.J.; Chang, A.C.; Page, A.L.; Warneke, J.E. Relative Concentrations of Cadmium and Zinc in Tissue of Selected Food Plants Grown on Sludge-Treated Soils. J. Environ. Qual. 1988, 17, 568–573. [Google Scholar] [CrossRef]
- Monteiro, C.; Santos, C.; Pinho, S.; Oliveira, H.; Pedrosa, T.; Dias, M.C. Cadmium-induced cyto- and genotoxicity are organ-dependent in lettuce. Chem. Res. Toxicol. 2012, 25, 1423–1434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Yuan, J.; Kong, W.; Yang, Z. Genotype variations in cadmium and lead accumulations of leafy lettuce (Lactuca sativa L.) and screening for pollution-safe cultivars for food safety. Environ. Sci. Process Impacts. 2013, 15, 1245–1255. [Google Scholar] [CrossRef]
- Yan, Z.S.; Li, M.; Tang, Y. Effects of Exogenous Melatonin on Photosynthetic Characteristics of Lettuce Seedlings under NaCl Stress. In Proceedings of the 5th International Conference on Energy and Environmental Protection, Shenzhen, China, 17–18 September 2016. [Google Scholar]
- Zhou, X.T.; Yang, T.; Jiang, Z.L.; He, Z.Q.; Zou, Z.R. Regulation of melatonin on chlorophyll fluorescence and nitrate accumulation in lettuce seedlings under excess nitrate stress. IOP Conf. Ser. Earth Environ. Sci. 2019, 330, 042043. [Google Scholar] [CrossRef]
- Biosciences, P. SMRT Link User Guide. 2017. Available online: https://www.pacb.com/wp-content/uploads/SMRT-Link-User-Guide-v8.0.pdf (accessed on 4 May 2020).
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhang, J.; Zhou, Z. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform. 2014, 15, 311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.J.V.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes-Chin-Wo, S.; Wang, Z.; Yang, X.; Kozik, A.; Arikit, S.; Song, C.; Xia, L.; Froenicke, L.; Lavelle, D.O.; Truco, M.J.; et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 2017, 8, 14953. [Google Scholar] [CrossRef]
- Gao, J.; Sun, L.; Yang, X.E.; Liu, J.X. Transcriptomic analysis of cadmium stress response in the heavy metal hyperaccumulator Sedum alfredii Hance. PLoS ONE 2013, 8, e64643. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Zeng, X.N.; Song, Q.L.; Sun, Y.; Feng, Y.J.; Lai, Y.C. Identification of key genes and modules in response to Cadmium stress in different rice varieties and stem nodes by weighted gene co-expression network analysis. Sci. Rep. 2020, 10, 9525. [Google Scholar] [CrossRef]
- Wei, T.; Treuren, R.V.; Liu, X.; Zhang, Z.; Chen, J.; Liu, Y.; Dong, S.; Sun, P.; Yang, T.; Lan, T.; et al. Whole-genome resequencing of 445 Lactuca accessions reveals the domestication history of cultivated lettuce. Nat. Genet. 2021, 53, 752–760. [Google Scholar] [CrossRef]
- Tang, X.; Pang, Y.; Ji, P.; Gao, P.; Nguyen, T.H.; Tong, Y. Cadmium uptake in above-ground parts of lettuce (Lactuca sativa L.). Ecotoxicol. Environ. Saf. 2016, 125, 102–106. [Google Scholar] [CrossRef]
- Sharma, S.S.; Dietz, K.J. The relationship between metal toxicity and cellular redox imbalance. Trends. Plant Sci. 2009, 14, 43–50. [Google Scholar] [CrossRef]
- Yan, Y.Y.; Jing, X.; Tang, H.M.; Li, X.T.; Gong, B.; Shi, Q.H. Using transcriptome to discover a novel melatonin-induced sodic alkaline stress resistant pathway in Solanum lycopersicum L. Plant Cell Physiol. 2019, 60, 2051–2064. [Google Scholar] [CrossRef]
- Hu, Z.C.; Fu, Q.S.; Zheng, J.; Zhang, A.A.; Wang, H.S. Transcriptomic and metabolomic analyses reveal that melatonin promotes melon root development under copper stress by inhibiting jasmonic acid biosynthesis. Hortic. Res. 2020, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Younesi-Melerdi, E.; Nematzadeh, G.A.; Pakdin-Parizi, A.; Bakhtiarizadeh, M.R.; Motahari, S.A. De novo RNA sequencing analysis of Aeluropus littoralis halophyte plant under salinity stress. Sci. Rep. 2020, 10, 9148. [Google Scholar] [CrossRef]
- Zhou, M.; Zheng, S.; Liu, R.; Lu, J.; Lu, L.; Zhang, C.; Liu, Z.; Luo, C.; Zhang, L.; Wu, Y. Comparative analysis of root transcriptome profiles between low- and high-cadmium-accumulating genotypes of wheat in response to cadmium stress. Funct. Integr. Genomics. 2019, 19, 281–294. [Google Scholar] [CrossRef]
- DalCorso, G.; Farinati, S.; Furini, A. Regulatory networks of cadmium stress in plants. Plant Signal Behav. 2010, 5, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Zhang, Y.; Han, L.; Guan, Z.; Chai, T. A novel WRKY transcriptional factor from Thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Rep. 2008, 27, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends. Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Tang, W.; Charles, T.M.; Newton, R.J. Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus Virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol. Biol. 2005, 59, 603–617. [Google Scholar] [CrossRef]
- Mortel, J.E.V.D.; Schat, H.; Moerland, P.D.; Themaat, E.V.L.V.; Ent, S.V.D.; Blankestijn, H.; Ghandilyan, A.; Tsiatsiani, S.; Aarts, M.G. Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 2008, 31, 301–324. [Google Scholar] [CrossRef]


| Comparing Group | Control | Treatment | Up | Down | Total DEG Number |
|---|---|---|---|---|---|
| G1 | LCd0 | LCd1 | 360 | 418 | 778 |
| G1 | LCd1 | LCd5 | 706 | 777 | 1483 |
| G2 | LCdM0 | LCdM1 | 92 | 81 | 173 |
| G2 | LCdM1 | LCdM5 | 405 | 536 | 941 |
| G3 | LCd0 | LCdM0 | 7 | 24 | 31 |
| G3 | LCd1 | LCdM1 | 0 | 0 | 0 |
| G3 | LCd5 | LCdM5 | 7 | 10 | 17 |
| G4 | LCd0 | LCd1 | 752 | 994 | 1746 |
| G4 | LCd1 | LCd5 | 282 | 192 | 474 |
| G5 | LCdM0 | LCdM1 | 177 | 327 | 504 |
| G5 | LCdM1 | LCdM5 | 245 | 84 | 329 |
| G6 | RCd0 | RCdM0 | 704 | 436 | 1140 |
| G6 | RCd1 | LCdM1 | 2178 | 1349 | 3527 |
| G6 | LCd5 | LCdM5 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Liang, L.; Xie, Y.; Tang, Y.; Tan, H.; Zhang, J.; Lin, L.; Sun, B.; Huang, Z.; Liu, J.; et al. Comparative Analysis of Italian Lettuce (Lactuca sativa L. var. ramose) Transcriptome Profiles Reveals the Molecular Mechanism on Exogenous Melatonin Preventing Cadmium Toxicity. Genes 2022, 13, 955. https://doi.org/10.3390/genes13060955
Yu X, Liang L, Xie Y, Tang Y, Tan H, Zhang J, Lin L, Sun B, Huang Z, Liu J, et al. Comparative Analysis of Italian Lettuce (Lactuca sativa L. var. ramose) Transcriptome Profiles Reveals the Molecular Mechanism on Exogenous Melatonin Preventing Cadmium Toxicity. Genes. 2022; 13(6):955. https://doi.org/10.3390/genes13060955
Chicago/Turabian StyleYu, Xuena, Le Liang, Yongdong Xie, Yi Tang, Huaqiang Tan, Jianwei Zhang, Lijin Lin, Bo Sun, Zhi Huang, Ji Liu, and et al. 2022. "Comparative Analysis of Italian Lettuce (Lactuca sativa L. var. ramose) Transcriptome Profiles Reveals the Molecular Mechanism on Exogenous Melatonin Preventing Cadmium Toxicity" Genes 13, no. 6: 955. https://doi.org/10.3390/genes13060955
APA StyleYu, X., Liang, L., Xie, Y., Tang, Y., Tan, H., Zhang, J., Lin, L., Sun, B., Huang, Z., Liu, J., Li, X., Tu, L., & Li, H. (2022). Comparative Analysis of Italian Lettuce (Lactuca sativa L. var. ramose) Transcriptome Profiles Reveals the Molecular Mechanism on Exogenous Melatonin Preventing Cadmium Toxicity. Genes, 13(6), 955. https://doi.org/10.3390/genes13060955







